Abstract
Alfalfa sprouts and other seed sprouts have been implicated in numerous outbreaks of salmonellosis. The source of these epidemics appears to have been low-level contamination of seeds by Salmonella bacteria that developed into clinically significant populations during the seed germination process. To test the possibility that Salmonella enterica strains carry host range determinants that allow them to grow on alfalfa, strains isolated from alfalfa or other sources were surveyed for their ability to grow on germinating alfalfa seeds. An S. enterica serovar Cubana strain originally isolated from contaminated alfalfa sprouts multiplied most rapidly during the initial 24 h of the seed germination process. Germinating alfalfa seeds supported the multiplication of S. enterica cells prior to the emergence of the root radicle at 72 h. Thereafter, much lower rates of multiplication were apparent. The ability of S. enterica to grow on germinating alfalfa seeds was independent of the serovar, isolation source, or virulence of the strain. Isolates obtained from alfalfa attained population levels similar to those observed for strains isolated from contaminated meat products or stools. Each of the strains could be detected in the waste irrigation water, with populations being strongly correlated with those detected on the germinating alfalfa seeds. The S. enterica strains were capable of utilizing the waste irrigation water as a sole carbon and nitrogen source. S. enterica strains thus appear to grow saprophytically on soluble organics released from seeds during early phases of germination. The ability to detect S. enterica in the waste irrigation water early in the germination process indicates that this method may be used as a simple way to monitor the contamination of sprouts during commercial operations.
In recent years, numerous outbreaks of salmonellosis have been associated with the consumption of contaminated alfalfa sprouts (30). A California study implicated the consumption of contaminated alfalfa sprouts in nearly 50% of the documented cases of salmonellosis occurring prior to the institution of U.S. Food and Drug Administration guidelines for the production of alfalfa sprouts in 1998 (22). These outbreaks have involved a variety of Salmonella enterica serotypes, including strains of Bovismorbificans, Stanley, Newport, Montevideo, Meleagridis, Infantalis, Anatum, Senftenberg, Havana, Cubana, Tennessee, Saint-Paul (United Kingdom), Gold-Coast (United Kingdom), Mbandaka, and Enteritidis (5, 20, 27). Related outbreaks involving Escherichia coli O157:H7 (10, 22) and Bacillus cereus (24) have also been reported.
In each of the aforementioned sprout-associated outbreaks, the seeds appear to have been the source of the contamination (20). Contaminating populations were usually undetectable in seed lots prior to germination. In the cases where clinically significant populations developed, S. enterica was found growing epiphytically on sprout surfaces without producing obvious signs of contamination (6, 26). Alfalfa sprouts and other seed sprouts are capable of supporting significant microbial populations. Naturally occurring microbial populations on sprouts, which are primarily root tissue, form biofilms, and numbers of CFU can reach 108/g fresh weight (gfw) (11). Biofilms can be observed on alfalfa sprout hypocotyls as early as 2 days postgermination, and by day 4 they can be found on all parts of the plant (11). Several S. enterica strains have been shown to reach clinically significant levels (106 to 107 CFU/gfw) 2 days after germination (31). Little is known about how any of these organisms grow in or on developing alfalfa sprouts. Charkowski et al. (8) recently reported that S. enterica strains appear to preferentially colonize the emerging root radicle.
The colonization of roots by bacteria is genetically complex (19), but type III protein secretion systems (TTSS) have recently been implicated in the growth and survival of Pseudomonas fluorescens, a rhizosphere-associated organism (25). It is thought that P. fluorescens utilizes the TTSS to achieve an ecological advantage over other rhizosphere-associated bacteria by translocating effector molecules into the cytoplasm of host cells, where they promote nutrient efflux into the rhizosphere. TTSS are required for the growth of a number of plant-associated bacteria, such as Pseudomonas syringae (9, 15). Effector proteins translocated by the TTSS appear to interfere with host cell signaling pathways in order to promote nutrient efflux and suppress the host's cellular-defense responses. Specific strains of P. syringae have limited host ranges that appear to be linked to the effectors that they produce (9, 16). The asymptomatic epiphytic growth of a P. syringae strain under field conditions has also been linked to the activities of a TTSS (14).
S. enterica strains have been shown to express two distinct TTSS that are encoded by pathogenicity islands known as SPI1 and SPI2 (13). The SPI1-associated TTSS functions in the invasion of host cells, whereas the SPI2-linked TTSS is expressed after the invasion of host cells (18). Interestingly, at least one of the secreted effector proteins translocated by the SPI1 TTSS shares homology with a secreted effector protein found in many plant pathogens that is required for plant-associated growth (29). There is also evidence that some virulence factors of mammalian pathogens are able to affect both animal and plant hosts. For example, a clinical isolate of Pseudomonas aeruginosa, which can cause opportunistic infections in humans, has been shown to produce symptoms in Arabidopsis thaliana (23). At least 17 P. aeruginosa genes have been associated with symptom production in both plants and mice.
In order to determine the basis for the growth of S. enterica on developing alfalfa sprouts, the growth of S. enterica strains isolated from contaminated alfalfa was compared to that of strains from nonplant sources and that of distinct serotypes. To evaluate the role of known pathogenicity determinants in the colonization of alfalfa by S. enterica, strains with inactivated genes or lacking genes for such determinants were obtained and analyzed. The analysis of various S. enterica isolates and mutants demonstrates the role of prior association with alfalfa and known pathogenicity factors in the survival of this pathogen on alfalfa sprouts and elucidates whether such growth is epiphytic or saprophytic.
MATERIALS AND METHODS
Strains.
Strains used in the experiments are shown in Table 1. Cultures were routinely grown in Trypticase soy agar medium, Luria broth (LB), or M63 minimal salts medium supplemented with 0.2% glucose (4), with shaking at 37°C as indicated in Results.
TABLE 1.
Properties of tested S. enterica strains
| Strain | Serovar | Source or relevant property | Source or reference |
|---|---|---|---|
| 31922 | Agona | Stool sample | J. Kapera |
| 2094 | Anatum | Stool sample | J. Kaper |
| 98E01362SH2 | Cubana | Alfalfa | S. Josephb |
| 32196 | Infantalis | Gelfelte fish | J. Kaper |
| Newport | Newport | Sardine | S. Joseph |
| 31625 | Senftenberg | Stool sample | J. Kaper |
| HO558 | Stanley | Stool sample | S. Joseph |
| 98E013E2SH1 | Tennessee | Alfalfa | S. Joseph |
| S4344 | Typhimurium | Wild-type genotype | C. Leec |
| 339 | Typhimurium | hilA339::Kan | C. Lee |
| SD11 | Typhimurium | SPI1 deletion | 28 |
| RM69 | Typhimurium | SPI1 deletion | 28 |
| EE656 | Typhimurium | prgH020::Tn5lacZY | 28 |
Department of Microbiology and Immunology, School of Medicine, University of Maryland, Baltimore.
Department of Cell Biology and Molecular Genetics, University of Maryland, College Park.
Department of Microbiology and Molecular Genetics, Harvard Medical School, Boston, Mass.
Surface sterilization of alfalfa seeds.
Alfalfa seeds were surface sterilized by using a modification of the procedures recommended by the U.S. Food and Drug Administration (2). Seeds were soaked in a 10% sodium hypochlorite solution for 15 min with shaking, rinsed in distilled H2O, and allowed to air dry at room temperature. Seeds were stored at 4°C until use. Treated seed batches were free of contaminating S. enterica populations when they were tested as described below.
Infestation of alfalfa seeds with S. enterica.
Seeds were infested with S. enterica by vacuum infiltration. Surface-sterilized seeds and 1 drop of Tween 20 were added to an overnight broth culture of an S. enterica strain, and the culture was placed under a vacuum for 5 min. The seeds were then collected, rinsed with distilled water three times to remove surface-associated S. enterica, and allowed to air dry.
Seed germination.
Surface-sterilized alfalfa seeds were soaked in tap water for 30 min with shaking, rinsed with tap water, and spread evenly in a 5- by 32-cm sprouting tray. Infested and surface-sterilized seeds were treated separately. Fifteen grams of seeds was placed in each of the seed trays, which were incubated in an Easy Green sprouting unit (Seed and Grain Technologies, Albuquerque, N.Mex.) and maintained at 25°C. This sprouting apparatus provided intermittent spray irrigation of developing sprouts and single-pass, flowthrough irrigation similar to that used by large-scale commercial operations. Sprouts were misted for 30 min every 3 h at an average irrigation rate of 500 μl/cm2 of tray/day. Infected seed batches contained six inoculated seeds and were prepared by randomly adding inoculated seeds by hand to seed trays containing surface-sterilized seeds of the same age and stage of germination. This method led to an appropriate ratio of 1 infested seed per 1,000 sterile seeds.
Quantification of S. enterica on germinating alfalfa seeds.
Approximately 0.5-g samples of germinating seeds were removed in triplicate from random positions within the seed batch at the times indicated in Results and placed into 2 ml of sterile saline. Adherent S. enterica cells were released by vortex treatment for 2 min. Washes were serially diluted and plated onto XLT4 agar (21). After incubation at 25°C for 2 days, black colonies exhibiting characteristic phenotypes of S. enterica were enumerated.
Quantification of reducing sugars in waste irrigation water.
Reducing sugars were quantified by the method of Garcia et al. (12). Fresh bicinchoninic acid (BCA) reagent was prepared by mixing equal parts of BCA solution A (per liter, 54.28 g of Na2CO3, 24.2 g of NaHCO3, 1.942 g of disodium 2,2′-bicinchoninate) and BCA solution B (per liter, 1.248 g of CuSO4 · 5H2O, 1.262 g of l-serine). Five hundred-microliter samples of waste irrigation water were mixed with an equal volume of BCA reagent and incubated at 80°C for 30 min. After the samples cooled to room temperature, the optical densities at 560 nm were recorded and compared to a standard curve prepared by using diluted glucose solutions.
Sterilization of waste irrigation water.
Waste irrigation water was collected in batches for 24 h from surface-sterilized seeds. The waste irrigation water was centrifuged for 10 min at 8,000 × g and then filter sterilized with 0.22-μm-pore-size syringe filters. The doubling times of broth cultures were calculated during logarithmic growth by using 4- and 6-h time points and the standard equation.
RESULTS
Growth of S. enterica serovar Cubana on germinating alfalfa sprouts.
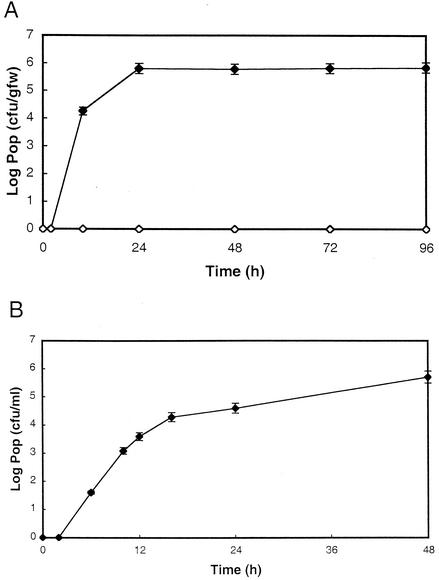
In order to define the basic growth characteristics of S. enterica on germinating alfalfa seeds under the experimental conditions, populations of an S. enterica strain previously isolated from contaminated alfalfa sprouts, S. enterica serovar Cubana 98E01362SH2, were monitored on germinating alfalfa seeds during the 5-day period typically used for commercial production of alfalfa sprouts. Initial populations of S. enterica in the infested seeds used to inoculate the seed batches were below the sensitivity of the plate assays used for determining resident populations on homogenized seeds. Populations of 98E01362SH2 could be detected on germinating seeds beginning approximately 6 h after the addition of the infested seeds and reached an apparent maximum by 48 h (Fig. 1A). The average maximum number of CFU detected on sprouts after 48 h was (5.0 ± 2.5) × 105/gfw. The doubling time for 98E01362SH2 during the initial phases of the interaction was determined to be approximately 47 min. S. enterica was not detected in germinating uninoculated surface-sterilized alfalfa seeds during the 5-day monitoring period. None of the samples obtained from infected batches had statistically larger populations of S. enterica cells than those of other samples collected at the same time from the same tray. This indicates that the initially inoculated seeds did not create limited zones of contamination.
FIG. 1.
Sprout-associated growth of S. enterica serovar Cubana. Batches of alfalfa seeds (15 g) were germinated and inoculated as described in Materials and Methods, and populations (Pop) that were adherent to germinating alfalfa seeds (A) or that were present in waste irrigation water (B) were monitored. The values reported are the means of results from three samples. The standard deviations for the reported values were less than 10% of the means. The experiments were repeated in triplicate, with similar results. Filled diamonds, S. enterica Cubana 98E01362SH2-inoculated seed batch; open diamonds, uninoculated seed batch.
Detection of S. enterica in waste irrigation water.
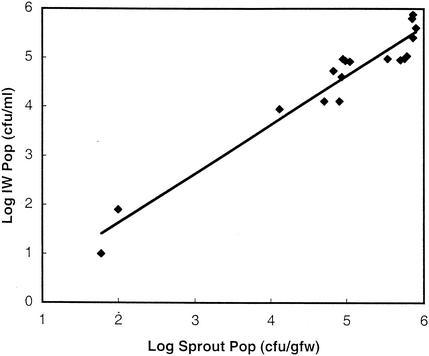
S. enterica could be detected in the waste irrigation water when S. enterica populations on developing sprouts were larger than 102 CFU/gfw. A strong linear correlation (R2 = 0.9136) was noted between S. enterica populations growing on sprouts and the populations detected in the waste irrigation water (Fig. 2). Average-size populations detected in the irrigation water were 33% ± 14% of the population sizes detected on sprouts, but by day 5, the population levels detected in the waste irrigation water decreased to 17% ± 2% of the population levels detected on germinating seeds.
FIG. 2.
Correlation between resident S. enterica serovar Cubana populations (Pop) on germinating alfalfa seeds (Sprout) and populations detected in waste irrigation water (IW). Seed batches were inoculated and populations were monitored as described in the legend to Fig. 1. The R2 value for the indicated correlation was 0.9136 as calculated by using the least-squares method.
To determine how early in the germination process S. enterica appears in waste irrigation water, populations of 98E01362SH2 in irrigation water collected from contaminated seed batches were monitored during the initial 48 h after inoculation. 98E01362SH2 could be detected in the waste irrigation water as early as 6 h after the initiation of germination (Fig. 1B), coincident with the appearance of detectable populations on alfalfa sprouts.
Evaluation of the ability of germinating alfalfa seeds to support S. enterica growth.
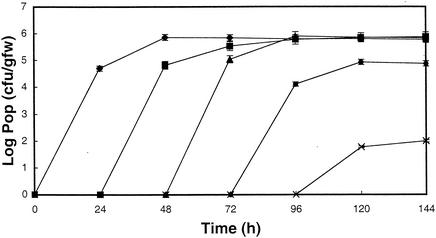
Since most of the increase in population levels occurred during the initial 24 h (>104-fold increase), the ability of S. enterica to colonize germinating seeds later in development was evaluated. A seed batch was inoculated as described above, and six germinating seeds from the inoculated seed batch were transferred at 24, 48, 72, and 96 h to batches of previously uncontaminated germinating seeds (approximately 6,000 seeds). Populations developing on germinating seeds and in the waste irrigation water were monitored as before. Seed batches inoculated at 0, 24, and 48 h after the initiation of germination all established roughly equivalent populations of 98E01362SH2 within 48 h after inoculation of the seed batch (Fig. 3). A 10-fold reduction in the size of maximum population was detected in seeds inoculated at 72 h, and little growth was detected in seed lots inoculated at 96 h. Similar results were obtained when waste irrigation water was sampled (data not shown).
FIG. 3.
Capability of germinating alfalfa seeds to support growth of S. enterica serovar Cubana. A batch of alfalfa seeds was inoculated and S. enterica populations were monitored as described in the legend to Fig. 1. At 24, 48, 72, and 96 h, six seeds or seedlings from the initially inoculated seed batch were transferred to concurrently grown batches of uninoculated surface-sterilized seeds. S. enterica populations (Pop) in the seed batches inoculated initially (filled diamonds), at 24 h (filled boxes), at 48 h (filled triangles), at 72 h (filled circles), and at 96 h (Xs) are shown. The experiment was repeated at least three times, with similar results.
Growth of S. enterica in waste irrigation water.
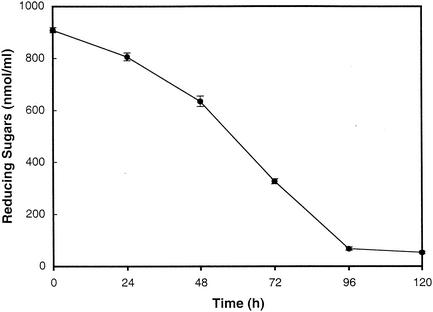
The rapid multiplication of 98E01362SH2 during the initial phase of seed germination coincided with a predicted period of nutrient efflux from germinating seeds. To estimate the rate of nutrient release from germinating seeds, levels of reducing sugars in collected waste irrigation water were monitored. The initial concentration of reducing sugars in the effluent was 0.9 mM and declined to less than 0.1 mM by 96 h after the initiation of germination (Fig. 4). Since these values represent sugar levels after dilution by the irrigation water, levels at the seed surface are likely to be substantially higher.
FIG. 4.
Residual reducing sugars in waste irrigation water obtained from germinating alfalfa seeds. Values reported are the means of results from three samples. The error bars represent the standard deviations. The experiment was repeated at least three times, with similar results.
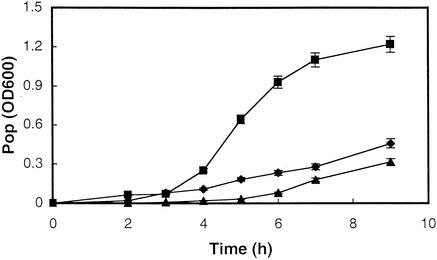
To evaluate whether S. enterica could grow on the nutrients released from germinating seeds, we monitored the growth of 98E01362SH2 populations in filter-sterilized waste irrigation water collected in batches from germinating surface-sterilized seeds during the initial 24 h of the germination process. After a lag of 2 h, the growth of 98E01362SH2 could be detected, with a doubling time of 83 min (Fig. 5). This rate was similar to the growth rate of 98E01362SH2 in minimal salts medium observed after a delay of 5 h. For comparison, the doubling time determined for the growth of 98E01362SH2 in a rich medium (LB) was approximately 54 min at this temperature. These experiments were performed in triplicate, with similar results.
FIG. 5.
Comparative levels of growth of S. enterica serovar Cubana in waste irrigation water, LB, and M63 glucose minimal salts medium. LB (filled boxes), M63 glucose minimal salts medium (filled triangles), and filter-sterilized irrigation water (filled diamonds) were inoculated with S. enterica serovar Cubana 98E01362SH2, and S. enterica populations (Pop) were monitored spectrophotometrically. The experiment was repeated three times, with similar results. Standard deviations for the reported means were less than 5% of the values. OD600, optical densities at 600 nm.
Evaluation of the ability of other S. enterica strains to grow on germinating alfalfa sprouts.
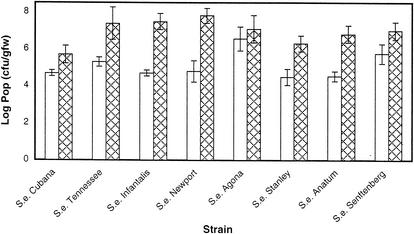
To determine if the ability of S. enterica strains to grow on germinating alfalfa seeds depended on the strain, the growth of isolates obtained from contaminated alfalfa sprouts (serovars Cubana and Tennessee) was compared to the growth of strains unlikely to have been in recent close association with plant tissue (Table 1). The latter strains were isolated from contaminated meat by-products or from stools of patients infected after consuming contaminated foodstuffs other than alfalfa sprouts. All nine strains tested were capable of growing on germinating alfalfa sprouts (Fig. 6). After 24 h of growth, the average population size for alfalfa-isolated strains was 1.2 × 105 CFU/gfw whereas the average population size for the other strains was 7.2 × 105 CFU/gfw. Based on a two-sided Student t test, however, there was no significant difference in the means at the 95% confidence level. Strains of both groups exhibited up to an additional 100-fold increase in population sizes after 48 h of growth on germinating alfalfa seeds. As before, the populations detected in the irrigation water mirrored the populations detected on the developing sprouts and the number of CFU was not less than 104/ml for any of the strains tested (data not shown). These results indicate that the S. enterica strains tested share the ability to grow on alfalfa sprouts, irrespective of their source, serotype, or previous history.
FIG. 6.
Comparison of the abilities of S. enterica (S.e.) strains to grow on germinating alfalfa seeds. Alfalfa seeds were inoculated with the indicated strain of S. enterica, and populations (Pop) were monitored as described in the legend to Fig. 1. Mean populations at 24 h (open bars, populations after exponential growth) and 48 h (diamond-filled bars, maximum populations) are shown. Error bars represent the standard deviations. The experiment was repeated at least three times, with similar results.
Role of S. enterica pathogenicity determinants in colonization of alfalfa sprouts.
To determine if there was any requirement for pathogenicity determinants during growth on alfalfa, S. enterica serovar Typhimurium SPI1 deletion mutants RM11 and SD69, prgH::Tn mutant EE656, which lacks a structural element of the SPI1-associated TTSS, and mutant 336 carrying a transposon mutation in a key transcriptional regulator of virulence gene expression (hilA) (17) were tested for their ability to grow on germinating alfalfa seeds. All strains were capable of growth on the germinating seeds during the initial 48 h (data not shown). The population sizes of the mutants detected on the sprouts were similar to those of the wild-type strain at all sampling times, and equivalent levels could be detected in the waste irrigation water (data not shown). Again, no statistically significant differences in the means were detected at the 95% confidence level. These results are consistent with the conclusion that early growth of S. enterica on sprouts appears to be saprophytic and utilizes nutrients released from seeds early in the germination process.
DISCUSSION
The contamination of alfalfa sprouts by S. enterica strains and other pathogenic bacteria has become a significant public health concern and has resulted in multiple warnings by regulatory agencies (e.g., references 2 and 3). Comparatively little was known about how S. enterica strains grow on germinating alfalfa sprouts and what aspects of that growth can be exploited to reduce the human health risk. We have shown here that the ability of S. enterica strains to grow on germinating alfalfa sprouts is unrelated to their pathogenicity. All the S. enterica strains tested were capable of growth up to potentially clinically significant populations, irrespective of each strain's isolation source, serovar, or virulence. The highest rates of multiplication were correlated with peak nutrient efflux from germinating seeds, suggesting saprophytic growth of the pathogen. The presence of S. enterica in the waste irrigation water was shown to be a key indicator of S. enterica contamination of germinating alfalfa sprouts.
Prior association with alfalfa did not appear to be a prerequisite for the growth of S. enterica strains on germinating alfalfa seeds. All the strains of S. enterica tested were capable of growth. These strains included representatives of nine serovars and had been collected from diverse sources. The two strains that had been isolated from contaminated alfalfa sprouts grew on germinating alfalfa seeds to levels similar to those of clinical isolates isolated from contaminated fish products and to those of other strains obtained from stools of patients not known to have consumed alfalfa sprouts. In view of the large number of distinct serovars that have been linked to alfalfa sprout-associated salmonellosis outbreaks, it appears that most, if not all, S. enterica strains are able to grow on germinating alfalfa seeds.
With other plant-associated bacteria, effectors secreted by the TTSS control the host range and individual strains are capable of colonizing only specific genotypes of the host. In these cases, population levels of the favored strain can be 1,000-fold higher than those of a nonfavored strain. No such specificity was observed with the S. enterica strains. In addition, SPI1 deletion mutants (28) that lack the genes to form the SPI1-encoded TTSS, a hilA::Tn mutant (with a mutation in a transcriptional regulator of the TTSS [17]), and mutants lacking a critical gene (prgH) required for the assembly of the TTSS (28) grew on germinating alfalfa seeds. These results indicate that the SPI1-encoded TTSS of S. enterica is not required for growth on alfalfa. Since the SPI2-encoded TTSS is expressed after the invasion of mammalian host cells (18), the growth of S. enterica strains appears to be independent of the known pathogenicity determinants.
The S. enterica strains tested appear to grow saprophytically on nutrients released by the germinating seeds during the germination process. The observation that S. enterica can utilize waste irrigation water as a growth medium indicates that these strains can metabolize the organic compounds released from germinating seeds. Seeds are known to release reducing sugars and other organic molecules into the medium during endosperm breakdown (7). The highest rates of multiplication of S. enterica strains on germinating alfalfa seeds correlated with the release of reducing sugars into waste irrigation water. Although the measured levels of reducing sugars in the waste irrigation water were relatively low, the irrigation process caused substantial dilution of the released nutrients. Concentrations at the seed surface were thus likely to be much higher. Consistent with this conclusion, the doubling time for an S. enterica strain growing on germinating alfalfa seeds was similar to the rate observed in the rich LB medium at this temperature.
Populations in waste irrigation water were strongly correlated with populations present on developing alfalfa sprouts. S. enterica could be detected in waste irrigation water from contaminated seed batches within 12 h after the initiation of the germination process. Each S. enterica strain tested could be detected in the waste irrigation water collected from germinating seeds inoculated with that strain. S. enterica populations in waste irrigation water from inoculated seed batches reached a maximum by 48 h, irrespective of the strain, but germinating alfalfa seeds could support the growth of S. enterica strains when they were inoculated at any time during the initial 72 h of the process. Thereafter, the rate of growth appeared to be significantly lower. Since waste irrigation water still contained comparatively high numbers of S. enterica CFU, the reduced ability of S. enterica to grow may have been related to the diminished release of sugars from the germinating seed, active defenses by the emerging root radicle, or a reduction in the ability of S. enterica strains to adhere to and colonize plant tissue. Interestingly, the relative numbers of CFU of S. enterica strains in waste irrigation water were reduced late in the seed germination process. The reduced levels may have been due to the formation of biofilms on the emerging root radicles, as reported previously by other groups (8, 11).
Our data lend support to the U.S. Food and Drug Administration's recommended testing methods to ensure that commercial sprout batches are free of S. enterica contamination (2). The testing of waste irrigation water 48 h after the initiation of seed germination was recommended (1), but supporting evidence was not provided. In our experiments, we attempted to mimic conditions that would be present in a commercial sprouting operation. Separately infested seeds were used to inoculate germinating seed batches to recreate the field conditions where only a few contaminated seeds were detected within a contaminated seed lot (2, 32). The irrigation conditions for germinating seeds were similar to those used by commercial operations. The finding that S. enterica was present in the waste irrigation water under these conditions indicates that our observations should be applicable to commercial operations utilizing similar growth conditions. The detection of S. enterica strains in the waste irrigation water at significant levels after 12 h suggests that the testing of wastewater can be performed very early in the sprouting process. Since S. enterica populations in waste irrigation water would be detectable by some commercial diagnostic systems relatively early in the seed germination process, monitoring S. enterica in the waste irrigation water provides a simple and comparatively inexpensive method for detecting the contamination of alfalfa sprouts during commercial production operations. This testing should be done early in the production cycle, as delayed sampling of waste irrigation water could diminish the chances of detecting contaminated seed batches. The early identification of S. enterica contamination of alfalfa sprouts should prevent contaminated products from reaching the consumer.
Acknowledgments
We thank S. Joseph, C. Lee, and J. Kaper for providing S. enterica strains, N. Schaad and S. Joseph for their technical advice on the experimental design, J. Charity for his assistance with the statistical analyses, and S. Joseph for his valuable comments on the manuscript.
This work was supported by funds provided by the Maryland Agricultural Experiment Station.
REFERENCES
- 1.Anonymous. 1999. Guidance for industry. I. Reducing microbial food safety hazards for sprouted seeds. And II. Sampling and microbial testing of spent irrigation water during sprout production. Fed. Regist. 64:57893-57902. [Google Scholar]
- 2.Anonymous. 1999. Microbiological safety evaluation and recommendations on sprouted seeds. Center for Food Safety and Nutrition, U.S. Food and Drug Administration, Washington, D.C.
- 3.Anonymous. 1997. Sanitary guidelines for the growing and packing for sale of fresh sprouts. International Sprout Growers Association, Seattle, Wash.
- 4.Atlas, R. 1993. Handbook of microbiological media. CRC Press, Boca Raton, Fla.
- 5.Backer, H., J. Mohle-Boetani, S. Werner, S. Abbott, J. Farrar, and D. Vugia. 2000. High incidence of extra-intestinal infections in a Salmonella Havana outbreak associated with alfalfa sprouts. Public Health Rep. 115:339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuchat, L., and J. Ryu. 1997. Produce handling and processing practices. Emerg. Infect. Dis. 3:459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan, B., W. Gruissem, and R. Jones. 2000. Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, Md.
- 8.Charkowski, A. O., J. D. Barak, C. Z. Sarreal, and R. E. Mandrell. 2002. Differences in growth of Salmonella enterica and Escherichia coli O157:H7 on alfalfa sprouts. Appl. Environ. Microbiol. 68:3114-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collmer, A., J. Badel, A. Charkowski, W. Deng, D. Fouts, A. Ramos, A. Rehm, D. Anderson, O. Schneewind, K. van Dijk, and J. Alfano. 2000. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. USA 97:8770-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Como-Sabetti, K. 1997. Outbreaks of E. coli O157:H7 infection associated with eating alfalfa sprouts—Michigan and Virginia, June-July 1997. Morb. Mortal. Wkly. Rep. 46:741-745. [Google Scholar]
- 11.Fett, W. 2000. Naturally occurring biofilms on alfalfa and other types of sprouts. J. Food Prot. 63:625-632. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, E., et al. 1993. Assessment of endo-1,4-beta-d-glucanase activity by a rapid colorimetric assay using disodium 2,2′-bicinchoninate. J. Food Biochem. 17:135-145. [Google Scholar]
- 13.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 14.Hirano, S., A. Charkowski, A. Collmer, D. Willis, and C. Upper. 1999. Role of the Hrp type III protein secretion system in growth of Pseudomonas syringae pv. syringae B728a on host plants in the field. Proc. Natl. Acad. Sci. USA 96:9851-9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano, S., and C. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a plant pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutcheson, S. 2001. The molecular biology of hypersensitivity to plant pathogenic bacteria. J. Plant Pathol. 83:151-172. [Google Scholar]
- 17.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas, R. L., and C. A. Lee. 2000. Unraveling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 19.Lustenberg, B., L. Dekkers, and G. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonads. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 20.Mahon, B. 1997. An international outbreak of Salmonella infections caused by alfalfa sprouts grown from contaminated seed. J. Infect. Dis. 175:876-882. [DOI] [PubMed] [Google Scholar]
- 21.Miller, R., C. Tate, E. Mallinson, and J. Scherrer. 1991. Xylose-lysine-tergitol 4: an improved selective agar medium for the isolation of Salmonella. Poult. Sci. 70:2429-2432. [DOI] [PubMed] [Google Scholar]
- 22.Mohle-Boetani, J., et al. 2001. E. coli O157:H7 and Salmonella infections associated with sprouts in California, 1996-1998. Ann. Intern. Med. 135:239-247. [DOI] [PubMed] [Google Scholar]
- 23.Plotnikova, J., L. Rahme, and F. Ausubel. 2000. Pathogenesis of the human opportunistic pathogen P. aeruginosa PA14 in Arabidopsis. Plant Physiol. 124:1766-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portnoy, B., J. Goepfert, and S. Harmon. 1976. An outbreak of B. cereus food poisoning resulting from contaminated vegetable sprouts. Am. J. Epidemiol. 103:589-594. [DOI] [PubMed] [Google Scholar]
- 25.Preston, G., N. Bertrand, and P. Rainey. 2001. Type III secretion in plant growth-promoting P. fluorescens SBW25. Mol. Microbiol. 41:999-1014. [DOI] [PubMed] [Google Scholar]
- 26.Proctor, M. E., M. Hamacher, M. L. Tortorello, J. R. Archer, and J. P. Davis. 2001. Multistate outbreak of Salmonella serovar Muenchen infections associated with alfalfa sprouts grown from seeds pretreated with calcium hypochlorite. J. Clin. Microbiol. 39:3461-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puohiniemi, R., T. Heiskanen, and A. Siitonen. 1997. Molecular epidemiology of two international sprout-borne Salmonella outbreaks. J. Clin. Microbiol. 35:2487-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schechter, L., S. Damrauer, and C. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 29.Staskawicz, B., M. Mudgett, J. Dangl, and J. Galan. 2001. Common and contrasting themes of plant and animal diseases. Science 292:2285-2289. [DOI] [PubMed] [Google Scholar]
- 30.Stewart, D., K. Reineke, J. Ulaszek, and M. Tortorello. 2001. Growth of Salmonella during sprouting of alfalfa seeds associated with salmonellosis outbreaks. J. Food Prot. 64:618-622. [DOI] [PubMed] [Google Scholar]
- 31.Taormina, P., C. Beuchat, and L. Slutsker. 1999. Infections associated with eating seed sprouts: an international concern. Emerg. Infect. Dis. 5:626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Beneden, C., W. Keene, R. Strang, D. Werke, A. King, B. Mahon, K. Hedberg, A. Bell, M. Kelly, V. Balan, W. MacKenzie, and B. Fleming. 1999. Multinational outbreak of Salmonella enterica serotype Newport infections due to contaminated alfalfa sprouts. JAMA 281:158-162. [DOI] [PubMed] [Google Scholar]