Abstract
Phylogenetic relationships, diversity, and in situ identification of spirochetes in the gut of the termite Neotermes koshunensis were examined without cultivation, with an emphasis on ectosymbionts attached to flagellated protists. Spirochetes in the gut microbial community investigated so far are related to the genus Treponema and divided into two phylogenetic clusters. In situ hybridizations with a 16S rRNA-targeting consensus oligonucleotide probe for one cluster (known as termite Treponema cluster I) detected both the ectosymbiotic spirochetes on gut protists and the free-swimming spirochetes in the gut fluid of N. koshunensis. The probe for the other cluster (cluster II), which has been identified as ectosymbionts on gut protists of two other termite species, Reticulitermes speratus and Hodotermopsis sjoestedti, failed to detect any spirochete population. The absence of cluster II spirochetes in N. koshunensis was confirmed by intensive 16S ribosomal DNA (rDNA) clone analysis, in which remarkably diverse spirochetes of 45 phylotypes were identified, almost all belonging to cluster I. Ectosymbiotic spirochetes of the three gut protist species Devescovina sp., Stephanonympha sp., and Oxymonas sp. in N. koshunensis were identified by their 16S rDNA and by in situ hybridizations using specific probes. The probes specific for these ectosymbionts did not receive a signal from the free-swimming spirochetes. The ectosymbionts were dispersed in cluster I of the phylogeny, and they formed distinct phylogenetic lineages, suggesting multiple origins of the spirochete attachment. Each single protist cell harbored multiple spirochete species, and some of the spirochetes were common among protist species. The results indicate complex relationships of the ectosymbiotic spirochetes with the gut protists.
The relationship between termites and the microorganisms inhabiting their guts is one of the most remarkable examples of symbiosis. The relationship enables termites to feed on lignocelluloses. The gut microbial community consists of both protists (single-cell eukaryotes) and prokaryotes and is known to have several beneficial roles (6). However, culture-independent molecular sequence studies reveal that the majority of microbes in the gut are yet to be characterized (8, 18-23, 27, 31), which limits our understanding of the symbiosis.
Spirochetes are one of the most abundant and morphologically distinct groups of bacteria that are common in the termite gut (5, 16). Although these spirochetes had long been uncultivated, two strains (Treponema sp. strains ZAS-1 and ZAS-2) have recently been isolated from the gut of the termite Zootermopsis angusticollis and shown to be CO2-reducing acetogens, whose metabolism is beneficial for termites, since acetate is their major carbon and energy source (12). It has been demonstrated that some of the spirochetes, including the gut isolates, have potential nitrogen fixation activity (13), an activity that contributes substantially to the nitrogen economy of termites that thrive on nitrogen-poor food. The findings imply important roles for symbiotic spirochetes in the nutrition of host termites.
The presence of diverse spirochetes has been reported from several termite species through analyses of the 16S rRNA gene (16S ribosomal DNA [rDNA]) amplified directly from DNA of a mixed microbial population in the termite gut (2, 3, 9, 14, 17-19, 24). These spirochetes were found to be affiliated with the genus Treponema, but none were closely related to any identified species of the genus. It has been reported that a single termite species alone harbors >20 phylotypes of Treponema species (14). Closely related phylotypes of gut spirochetes rarely occur among termite genera (14, 17). Given the existence of ∼280 termite genera, these observations suggest a great diversity of gut spirochetes of termites. They are divided into two phylogenetic clusters, which have been designated termite Treponema clusters I and II (9, 17). In this study, we refer to them simply as clusters I and II. Cluster I contains diverse phylotypes of the gut spirochetes and includes the strains isolated from the termite gut, whereas cluster II is smaller and belongs to the Treponema bryantii subgroup.
Spirochetes either exist freely in the gut fluid or are attached as ectosymbionts to the cell surfaces of gut protists. An example of the ectosymbiotic association is the fact that the ectosymbiotic spirochetes are linked with the motility of a protist cell (7), although most ectosymbionts observed to date do not appear to be involved in motility symbioses (4, 5, 10, 11). Nevertheless, we use the term ectosymbiont to mean simply that they are physically associated. Ultrastructural observations of the ectosymbiotic spirochetes reveal specialized attachment sites on the protists (4, 28-30). Despite extensive analyses of the 16S rDNA sequences of spirochetes in the termite gut, their distributions and locations have not yet been investigated fully. In situ hybridization with rRNA-targeted oligonucleotide probes has been used for the phylogenetic identification of gut spirochetes at the cellular level. Two specific 16S rDNA sequences belonging to spirochetes in cluster I were identified as those of large species existing freely in the gut fluid (2, 24). Members of cluster II are identified as ectosymbiotic spirochetes of oxymonad protists in the termites Reticulitermes speratus and Hodotermopsis sjoestedti (9). However, not all ectosymbiotic spirochetes are in cluster II, since some populations of ectosymbiotic bacteria that exhibit spirochete-like morphology give no positive signal with a cluster II consensus probe. Furthermore, various species of devescovinid, calonymphid, and hypermastigote protists, in addition to the oxymonad protists, also harbor dense populations of ectosymbiotic spirochetes. Thus, little is known about the nature of this impressive ectosymbiosis.
In this study, we investigated the in situ localization of gut spirochetes in several termite species, using oligonucleotide probes that were specific for each cluster. The 16S rDNA-based community structure of the spirochetes was analyzed in the gut of Neotermes koshunensis, with an emphasis on the phylogenetic and in situ identification of the ectosymbiotic spirochetes of the gut protists.
MATERIALS AND METHODS
Termites.
Two subterranean termites, R. speratus (family Rhinotermitidae) and Coptotermes formosanus (Rhinotermitidae), the dry-wood termite N. koshunensis (Kalotermitidae), and the damp-wood termite H. sjoestedti (Termopsidae) were used in this study. They were collected in Japan in Saitama, Okinawa, Okinawa, and Kagoshima prefectures, respectively. The workers were used for the analyses, and they were cultured for a week on an artificial diet prior to use, as described previously (19). Only the termite N. koshunensis was used for DNA extraction of the gut microbial community and for micromanipulation of the protist cells.
DNA extraction and PCR amplification.
Approximately 30 workers of N. koshunensis were collected. After their exterior surfaces had been washed with distilled water, the entire guts were removed with forceps and gently squeezed. The cells of gut microorganisms were disrupted by freezing them in liquid nitrogen and thawing them at 60°C. The DNA was extracted using a DNA purification system (Qiagen) according to the supplier's instructions. The 16S rDNA was amplified from the extracted DNA by PCR using ExTaq DNA polymerase (Takara) according to the manufacturer's instructions. The PCR conditions were 20 cycles at 94°C for 30 s, 63°C for 45 s, and 72°C for 2 min. In order to reduce the occurrence of chimeric artifacts, we selected the minimum number of cycles that enabled us to obtain sufficient amplification products. The PCR primers used were S58F (5′-CGGCGCGTYTTAAGCATGC-3′; Y = C or T) and S1400R (14), which are specific for spirochete 16S rDNA, corresponding to the nucleotide positions 58 to 1400 of the Escherichia coli 16S rRNA sequence. PCR products of the expected size (∼1.4 kb) were isolated by electrophoresis, using a low-melting-point agarose gel (SeaPlaque GTG; FMC Bioproducts), and purified using the Wizard PCR Preps DNA purification system (Promega). The purified PCR products were cloned into the pGEM-T vector (Promega) according to the manufacturer's instructions. We repeated the PCR and cloning of the PCR products to construct two clone libraries, starting with independent PCRs.
Micromanipulation of protist cells.
Protist cells in the gut of N. koshunensis were fixed in solution U (32) containing 4% paraformaldehyde for 30 min. After being washed with solution U, the protists were resuspended in fresh solution U. Each protist cell was viewed under a microscope (DMIRB; Leica) and then picked up with a micromanipulator (Transfer-Man; Eppendorf) equipped with a handmade microcapillary that fitted the target cells. The separated cells were suspended in fresh solution U, and the micromanipulation was repeated three times to remove contaminating cells. Approximately 15 of the isolated protist cells were used as templates for the PCR described above, except that 30, instead of 20, cycles were used. The PCR products were isolated, purified, and cloned as described above.
Screening, sequencing, and sorting of the clones.
Spirochete clones containing inserts of the expected size were identified by PCR amplification of the inserts using the S58F-S1400R primer set. The PCR products were purified with a multiscreen PCR purification filter (Millipore), and their partial nucleotide sequences were determined with the primer EUB750R (18). Based on a comparison of the partial sequences, clones with >97% nucleotide identity were sorted into phylotypes. A representative clone of each phylotype that overlapped the two clone libraries was selected, and its complete nucleotide sequence was determined. Inserts of the clones derived from the protist cells collected by micromanipulation were PCR amplified and digested with either HhaI or HaeIII. Representative clones showing identical restriction patterns were selected, and in each instance, the complete nucleotide sequence was determined. Plasmid DNA was prepared using a Minipreps DNA purification kit (Promega) and then used as the template for the sequencing reaction. Nucleotide sequences were determined using an ABI PRISM Big Dye Terminator Cycle-Sequencing Ready Reaction kit (PE-Applied Biosystems) and an automatic sequence analyzer (ABI 3700). Complete sequences were determined using the sequencing primers described previously (18).
Chimera check.
Presumptive chimeric artifacts from the PCR were intensively searched for as follows. First, we analyzed the sequences with the CHIMERA CHECK program offered through the Ribosomal Database Project (15). We then analyzed putative secondary structures of the sequences by overlaying them on the secondary structure of Treponema pallidum 16S rRNA and checking for base pair compatibility of the regions. Signature nucleotides proposed by Paster et al. (25) and Lilburn et al. (14) were also checked to screen for chimeras. We independently examined the phylogenetic affiliations of the 5′- and 3′-end fragments of approximately one-third of the entire length of 16S rDNA and compared the affiliations of each with the other's length and with those based on the entire sequences. Finally, for each clone belonging to an identical phylotype but deriving from a different clone library, partial nucleotide sequences corresponding to the 5′- and 3′-end regions were determined using the primers EUB750R and EUB900F (18), respectively. This was done to confirm the significant sequence similarity of the phylotype with the complete sequence of the clone representing the phylotype in both DNA regions.
Phylogenetic analysis.
Sequence data used to infer a phylogenetic tree were retrieved from the databases (see Fig. 3 for accession numbers). The sequence data were aligned using the CLUSTAL W package and checked manually. Phylogenetic relationships were inferred using the PHYLIP package, as described previously (18).
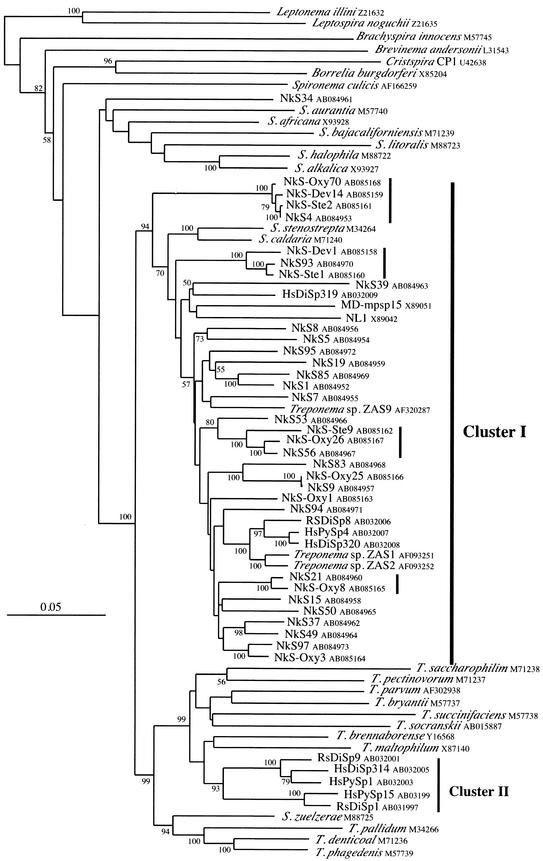
FIG.3.
Phylogenetic tree of 16S rDNA sequences of the gut spirochetes in N. koshunensis. The tree was inferred by the neighbor-joining method. Bootstrap values above 50 from 100 resamplings are shown for each node. The scale bar represents 0.05 nucleotide substitutions per position. The termite Treponema clusters I and II are indicated at the right of the tree. The database accession numbers are shown after the names of the clones and organisms. Genus abbreviations: T., Treponema; S., Spirochaeta. The sequences of Leptonema illini and Leptospira noguchii were used as outgroups. The four vertical bars indicate the sequences for which the cells of origin were identified by in situ hybridization.
In situ hybridization.
In situ hybridization was performed according to the method of Ohkuma et al. (23) with slight modifications. The gut contents were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 4 h at 4°C and then washed in PBS. The fixed cells were spotted onto a silane-coated glass slide (Matsunami Glass), air dried, and treated with 0.25 N HCl at room temperature for 30 min. The specimen was then washed twice with PBS, air dried, and sequentially dehydrated in 50, 80, and 100% ethanol. The hybridization solution (0.9 M NaCl, 0.1 M Tris-HCl) with fluorescently labeled probes was then applied, sealed in an incubation chamber (CoverWell; Grace Bio-Labs), and incubated for 3 h at 48°C. The specimen was washed for 15 min in washing buffer (0.2 M NaCl, 0.1 M Tris-HCl) at 48°C. It was then mounted in 90% glycerol with 0.5% triethylenediamine and observed with an Olympus epifluorescence microscope (BX-60) and a Leica confocal laser scanning microscope (TSC-SP).
The previously reported probes used in this study were EUBAC, TT-484V3, and TT-1248V8 (9). EUBAC binds to most eubacterial cells and was labeled at the 5′ end with Texas Red and used as the control for the permeability of the cells. The target sequences of TT-484V3 and TT-1248V8 are conserved for most sequences of the spirochetes in termite Treponema cluster II. These two probes were labeled at the 5′ end with 6-carboxyfluorescein (6-FAM) and were always used simultaneously. A new probe (TTI-732) against the conserved sequence among spirochetes of termite Treponema cluster I was newly designed in this study. Its sequence is 5′-CTCAGCGTCAGTCATTGGCT-3′. The probe was labeled at the 5′ end with either Texas Red or 6-FAM. No difference in binding affinity was observed between the two fluorescent labels. Based on the sequence comparison, fluorescently labeled probes specific for the phylotypes were also designed in this study. Their sequences are as follows: Dev1-486, 5′-TCTCGGTCATTTCCTACCGG-3′; Dev14-486, 5′-ACGGTCATTACCTACCATGC-3′; Dev14-Tp652, 5′-CCTCCTAGACTCGAGCTCA-3′; Oxy8-Tp444, 5′-CCGCTTATTCCTCCGCAATA-3′; and Oxy26-Tp1281, 5′-GCCGGGTTTTTGCGCTTCG-3′. The probes Dev14-486 and Dev14-Tp652 are specific for phylotype Dev14 and were used simultaneously for in situ hybridization. These sequence-specific probes were labeled at the 5′ end with 6-FAM.
Nucleotide sequence accession numbers.
The sequences determined in this study will appear in the nucleotide sequence databases under accession numbers AB084952 to AB085168.
RESULTS
In situ localization of spirochetes with cluster consensus probes.
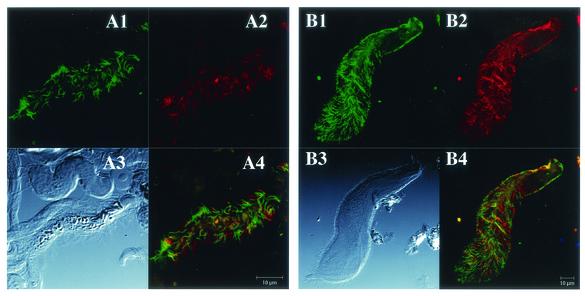
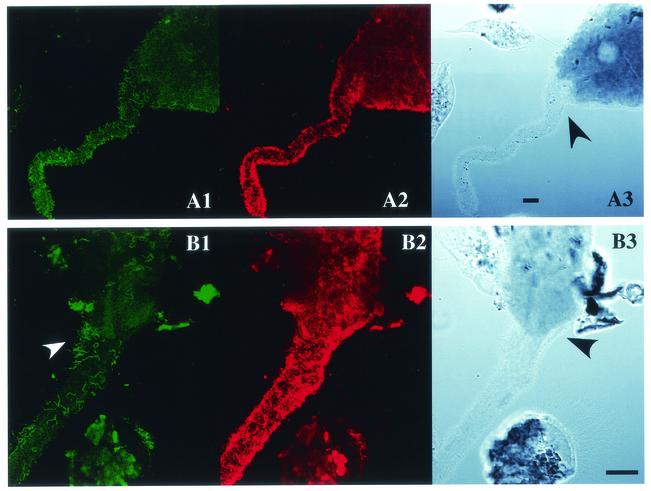
In a previous report, the spirochetes in the termite Treponema cluster II were identified as ectosymbionts attached to the oxymonad protists Dinenympha spp. and Pyrsonympha sp. in the guts of the termites R. speratus and H. sjoestedti by in situ hybridization using a cluster II consensus probe (9). In order to identify the in situ localization of the cluster I spirochetes, we designed a cluster I consensus probe by comparing the 16 rDNA sequences of the termite gut spirochetes reported so far. Figure 1 shows the results of in situ hybridizations labeled simultaneously with the cluster I and cluster II consensus probes. Signals of both probes were observed from the ectosymbiotic spirochetes on Dinenympha porteri in R. speratus (Fig. 1, A1 to A4) and on Pyrsonympha sp. in H. sjoestedti (Fig. 1, B1 to B4). Overlaying images of both signals (Fig. 1, and A4) showed that no cell gave a doubly stained signal, confirming the specificity of both probes. The cluster I consensus probe also detected populations of spirochetes existing freely in the gut fluid of both termite species, whereas the cluster II consensus probe rarely detected free-swimming spirochetes.
FIG. 1.
In situ hybridization of ectosymbiotic spirochetes of gut protists stained simultaneously with the consensus probes for cluster I and cluster II. The protists D. porteri in R. speratus (A1 to A4) and Pyrsonympha sp. in H. sjoestedti (B1 to B4) are shown. The cluster II consensus probes TT-484V3 and TT-1248V8, labeled with 6-FAM (A1 and B1), and the cluster I consensus probe TTI-732, labeled with Texas Red (A2 and B2), were used. Confocal laser scanning microscopy visualized the fluorescent signals. (A3 and B3) Differential interference contrast micrographs. Bars, 10 μm. (A4) Merged image of panels A1 and A2; (B4) merged image of panels B1 and B2.
We also investigated the in situ localization of gut spirochetes in the termites N. koshunensis and C. formosanus, using the consensus probes for each cluster. Most of the spirochetes in N. koshunensis attached to the devescovinid protist (Devescovina sp.), the calonymphid protist (Stephanonympha sp.), and the oxymonad protist (Oxymonas sp.) and gave signals with the cluster I consensus probe. In C. formosanus, ectosymbiotic spirochetes of a hypermastigote protist, Holomastigotoides mirabile, also gave specific signals with the cluster I consensus probe. In both of these termite species, the cluster I consensus probe detected almost all the spirochete cells that were stained by the eubacterial universal probe, including the free-swimming cells in the gut fluid. In situ hybridization using the cluster II consensus probe failed to detect any spirochetes in either of the termite species, suggesting that cluster II spirochetes were absent in the gut communities.
Spirochetal 16S rDNA clones and their phylogeny.
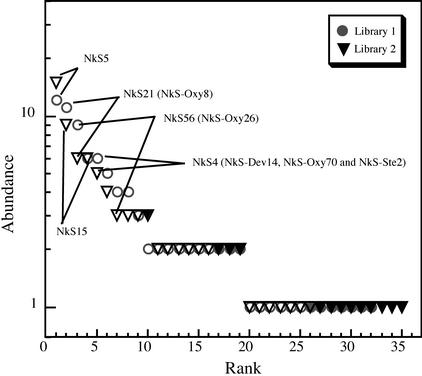
In order to address the phylogenetic diversity of the gut spirochetes in N. koshunensis, 16S rDNAs were PCR amplified from the gut community with a spirochete-specific primer set and cloned. We constructed two clone libraries from independent PCRs. Ninety-three and 91 clones in the two libraries were partially sequenced (500 to 700 bases) and sorted into 32 and 35 phylotypes, respectively, based on a comparison of sequence identity. A phylotype represented a group of clones showing >97% identity, which probably reflected a species level classification. A total of 45 phylotypes were obtained, and 22 phylotypes were common to the two libraries (Fig. 2). These 22 phylotypes included 80 clones (86%) and 73 clones (80%) from the respective libraries, suggesting that they represented the predominant spirochete species in the gut.
FIG. 2.
Rank abundance plots showing the distribution of clones into phylotypes in N. koshunensis. Abundance is plotted in each of the two clone libraries from the whole gut contents. Phylotypes are ranked according to their clone abundances in descending order. Phylotypes common to the two libraries are shown by open symbols; the solid symbols indicate phylotypes found in only one library. The five most abundant phylotypes are indicated, three of which have close relatives in the spirochete phylotypes identified from the isolated protist cells (shown in parentheses) (Table 2).
We specifically identified the 16S rDNA sequences of the ectosymbiotic spirochetes on the three protist species in the gut of N. koshunensis. Each protist species was carefully isolated with the aid of a micromanipulator and used as a template for PCR. The clones of the amplified 16S rDNA were sorted into groups based on the comparison of their restriction fragment patterns, and representatives were selected to determine the nucleotide sequences. Two, three, and six phylotypes were identified from the fractions of Devescovina sp., Stephanonympha sp., and Oxymonas sp., respectively (Table 1).
TABLE 1.
Sorting of spirochete 16S rDNA clones from isolated protist cells into phylotypes
| Isolated protist | No. of clones analyzed | No. of phylotypes | Representative clonea (no. of clonesb) | Identical phylotype from whole gutc |
|---|---|---|---|---|
| Devescovina sp. | 31 | 2 | NkS-Dev1 (28) | NkS93 |
| NkS-Dev14 (3) | NkS4 | |||
| Stephanonympha sp. | 25 | 3 | NkS-Ste2 (15) | NkS4 |
| NkS-Ste1 (6) | NkS93 | |||
| NkS-Ste9 (4) | (NkS56)d | |||
| Oxymonas sp. | 27 | 6 | NkS-Oxy8 (11) | NkS21 |
| NkS-Oxy26 (7) | NkS56 | |||
| NkS-Oxy1 (4) | −e | |||
| NkS-Oxy3 (2) | NkS97 | |||
| NkS-Oxy25 (2) | NkS9 | |||
| NkS-Oxy70 (1) | NkS4 |
Representative clones were included in the phylogenetic analysis shown in Fig. 3.
Number of clones that were grouped into the same phylotype as the representative clone.
The identical phylotype that was identified in the spirochete 16S rDNA libraries from the whole gut community of N. koshunensis.
The clone NkS-Ste9 had no identical phylotypes in the libraries from the whole gut community but was closely related (96.1% nucleotide identity) to the clone NkS56.
The clone NkS-Oxy1 had no close relatives in the libraries from the whole gut community.
The entire nucleotide sequences of the 22 16S rDNA phylotypes common to the two libraries from the whole gut community and a total of 11 phylotypes from the three protists were determined. No chimerical trait was detected in these sequences even after careful inspection (see Materials and Methods). The authenticity of the sequences was also supported by the sequence analysis of at least one additional clone that belonged to the identical phylotype group but was derived from the other library, because identical chimeras in the independent PCRs were not expected.
Phylotypes NkS-Dev14, NkS-Ste2, and NkS-Oxy70 showed >99% sequence identity to one another. Phylotypes NkS-Dev1 and NkS-Ste1, as well as NkS-Ste9 and NkS-Oxy26, also showed sequence identities of 97.0 and 95.4%, respectively, to each other. These relationships indicated that closely related spirochetes were common in the fractions of different protist species. The phylotypes obtained from the protist ectosymbionts were close relatives of the phylotypes from the whole gut community (Fig. 3). The phylotype NkS-Oxy1, which had no close relatives among the 22 common phylotypes in the two libraries, had a corresponding clone in only one of the two libraries. Among the phylotypes from the whole gut community, the phylotypes that corresponded to NkS-Dev14, NkS-Oxy26, and NkS-Oxy8 (NkS4, NkS56, and NkS21, respectively) consisted of abundant clones (Fig. 2), suggesting that these phylotypes represented the dominant spirochete population in the gut community. However, the PCR usually introduced some biases, so this result is not definitive.
Figure 3 shows the phylogeny of the phylotypes from N. koshunensis. All the phylotypes except one (clone NkS34) were grouped together and affiliated with cluster I. The grouping was supported by a significant bootstrap value of 94%. The one exception, phylotype NkS34, was grouped with the genus Spirochaeta, although the grouping was not supported statistically. The sequence of a clone from a termite gut (Za29, published only in the databases under accession AJ419823) was closely related to that of the clone NkS34. No cluster II phylotypes were obtained from N. koshunensis. Even after including the partial sequences of 45 phylotypes in the phylogenetic analysis, we still could not detect cluster II spirochetes (data not shown). Within cluster I, the phylotypes from N. koshunensis showed remarkable diversity, suggesting the presence of numerous spirochete species in the gut community. Although the branching orders in cluster I were not fully resolved, the phylotypes obtained from the protists formed distinct lineages.
In situ identification of ectosymbiotic spirochetes.
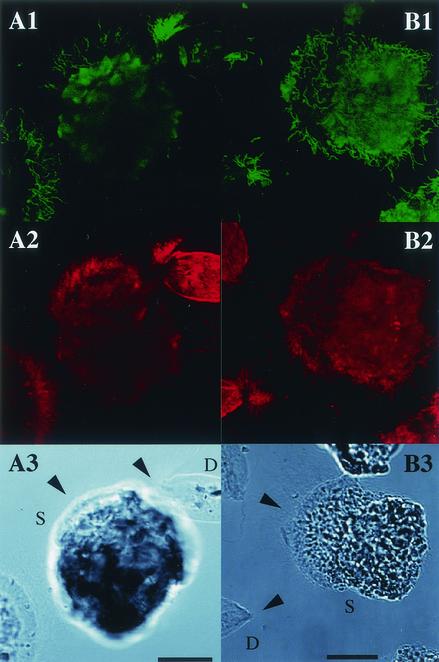
We designed oligonucleotide probes to target the phylotypes NkS-Dev14, NkS-Dev1, NkS-Oxy26, and NkS-Oxy8 for in situ identification of ectosymbiotic spirochetes. As a single probe gave only a weak hybridization signal for the phylotype NkS-Dev14, two kinds of probes were used simultaneously. Figure 4 shows the results of in situ hybridization using the NkS-Dev1-specific probe and the NkS-Dev14-specific probes. The NkS-Dev1-specific probe and the NkS-Dev14-specific probes gave a strong signal in the ectosymbiotic spirochetes on Devescovina sp. and Stephanonympha sp., confirming that the spirochete species represented by these phylotypes were indeed ectosymbionts of the gut protists. The ratios of specific ectosymbiotic spirochete cells were significantly different in the two protist species (Table 2). Approximately twice as many cells were detected by the NkS-Dev1 probe as by the NkS-Dev14 probes for spirochetes on Devescovina sp. Conversely, for Stephanonympha sp., the number of cells detected with the NkS-Dev1 probe was about half of that detected by the NkS-Dev14 probes. The NkS-Oxy26-specific probe, which also recognized the sequence of the NkS-Ste9 phylotype, sometimes detected a small portion of the ectosymbiotic spirochete population on Stephanonympha sp., but this probe did not detect any ectosymbionts of Devescovina sp.
FIG. 4.
Detection by in situ hybridization of spirochetes attached to the gut protists Devescovina sp. and Stephanonympha sp. (A1 and B1) Epifluorescent images with probes specific for phylotypes NkS-Dev1 (Dev1-486) and NkS-Dev14 (Dev14-486 and Dev14-Tp654), respectively. (A2 and B2) Epifluorescent images with the eubacterial universal probe (EUBAC). Note that rod-shaped (nonspiral) bacteria are also attached to the cells of Devescovina sp. (A3 and B3) Differential interference contrast micrographs. The protist species are labeled as D (Devescovina sp.) and S (Stephanonympha sp.). Bars, 20 μm. The arrowheads indicate the anterior parts of the protist cells.
TABLE 2.
Percentages of populations of ectosymbiotic spirochete cells of Devescovina sp. and Stephanonympha sp. represented by phylotypes NkS-Dev1 and NkS-Dev14
| Protist | Percentagea
|
||||
|---|---|---|---|---|---|
| Cluster I/eubacteria | NkS-Dev1/eubacteria | NkS-Dev14/eubacteria | NkS-Dev1/cluster I | NkS-Dev14/cluster I | |
| Devescovina sp. | 85 ± 13 | 58 ± 18 | 33 ± 16 | 65 ± 17 | 32 ± 12 |
| Stephanonympha sp. | 77 ± 8.6 | 30 ± 13 | 55 ± 9.0 | 34 ± 14 | 68 ± 10 |
Values are percentages (mean ± standard deviation [SD]) of numbers of cells detected by the probes for cluster I of spirochetes and for the phylotypes NkS-Dev1 and NkS-Dev14 per number of cells detected by the probes for either eubacteria or cluster I of spirochetes. In the case of detection by the probe for eubacteria, only the cells showing spirochete-like morphology were counted. Five to 22 images from confocal laser scanning microscopy were counted in each case. The mean numbers (±SD) of cells in an image that were detected by the probes for eubacteria and cluster I of spirochetes were 28 ± 15 and 31 ± 20 cells, respectively, in the case of Devescovina sp. and 154 ± 74 and 83 ± 47 cells, respectively, in the case of Stephanonympha sp. Note that these cell numbers did not correspond to the total number of ectosymbiotic spirochete cells of each protist species, since only a section of the protist cell imaged by confocal laser scanning microscopy was used for counting.
Of the six phylotypes detected from the Oxymonas sp. fraction in the clone analysis, two were confirmed as ectosymbiotic spirochetes by in situ hybridization (Fig. 5). The probe specific for the NkS-Oxy8 phylotype, which represented the most abundant clones from Oxymonas sp., detected ectosymbionts distributed over the entire surface of the cell. The NkS-Oxy26 probe also detected ectosymbiotic spirochetes, but many of them were located at the base of the rostellum, a stalk-like structure at the tip of which the cell was connected to the gut epithelium. Neither the NkS-Dev1 nor the NkS-Dev14 probe could detect ectosymbionts on Oxymonas sp., although the clone (NkS-Oxy70) of the NkS-Dev1 phylotype was obtained from this protist.
FIG. 5.
In situ identification of the ectosymbiotic spirochetes on Oxymonas sp. from N. koshunensis. (A1 and B1) Epifluorescent images with probes specific for phylotypes NkS-Oxy8 (Oxy8-Tp444) and NkS-Oxy26 (Oxy26-Tp1281), respectively. The arrowhead in panel B1 indicates the presence of a dense population of the stained spirochetes. (A2 and B2) Epifluorescent images labeled with the consensus probe for termite Treponema cluster I (TTI-732). (A3 and B3) Differential interference contrast micrographs. The arrowheads indicate cells of Oxymonas sp. Bars, 20 μm.
The in situ hybridization using the phylotype-specific probes for NkS-Dev1, NkS-Dev14, NkS-Oxy8, and NkS-Oxy26 gave no signal in the free-swimming spirochetes in the gut of the termite N. koshunensis.
DISCUSSION
In situ hybridizations using consensus probes for cluster I or cluster II revealed localization of the gut spirochetes at the large-cluster level. Identification of the spirochetes possessing cluster II 16S rDNA sequences as ectosymbionts of gut protists had been reported previously in the termites R. speratus and H. sjoestedti (9), whereas free-swimming spirochetes in cluster II were rare. Conversely, cluster I spirochetes in all four termite species examined here included both ectosymbionts on protists and free-swimming spirochetes in the gut fluid. This dual localization of cluster I spirochetes is not because each species is present at both localizations but because individual species inhabit only one of the two distinct locations. It has been reported that gut methanogenic archaea harbored by gut protists are phylogenetically distinct from those attached to the gut epithelium (31). The gut dwellers are not randomly dispersed over ecological niches in the termite gut.
The cluster II consensus probe failed to detect any spirochetes in the guts of N. koshunensis and C. formosanus. The absence of cluster II spirochetes in the gut community was confirmed by intensive analysis of the spirochete 16S rDNA clones in N. koshunensis. Only the spirochete 16S rDNA sequences belonging to cluster I have been reported from the gut community of C. formosanus (14). Members of cluster I have been reported from all the termites examined so far (14, 17, 24), while those of cluster II are restricted to Reticulitermes flavipes (14), R. speratus, and H. sjoestedti. Only one sequence, sp40-12 from the termite Mastotermes darwiniensis (3), is somewhat related to cluster II. Since both Reticulitermes and C. formosanus belong to the termite family Rhinotermitidae, but H. sjoestedti (Termopsidae) does not, the preference for the cluster II spirochetes seems to be unrelated to termite evolution. Although in situ localization of the gut spirochetes has not yet been investigated in R. flavipes, the cluster II spirochetes are found attached to the oxymonad in the genera Dinenympha and Pyrsonympha. Since these genera occur in Reticulitermes and H. sjoestedti but not in the other termite species, the presence of cluster II spirochetes may be related to the presence of these protists in the gut.
The ectosymbiotic spirochetes are considered to be dominant species in the gut spirochete community. In particular, the three ectosymbiont phylotypes, NkS-Dev14, NkS-Oxy26, and NkS-Oxy8, corresponded to the abundant phylotypes in the clone libraries from the whole gut community (NkS4, NkS56, and NkS21, respectively) (Fig. 2). In fact, the three protist species harboring these ectosymbiotic spirochetes are the dominant protists in the gut, and individual protist cells of Stephanonympha sp. and Oxymonas sp. frequently harbor several hundred or more spirochete cells. The other protist species that harbor ectosymbiotic spirochetes in N. koshunensis are scarce. The surfaces of the protist cells are considered to harbor a large proportion of the gut prokaryote cells, as described previously (1). The two phylotypes represented by the clones NkS5 and NkS15 were predominant in the clone libraries, but they did not correspond to the phylotypes identified from the gut protists. This is probably because they represent spirochete populations existing freely in the gut fluid.
Great phylogenetic diversity of the gut spirochetes in N. koshunensis is revealed in the 16S rDNA clone analysis. A total of 45 phylotypes are recognized in this termite species alone, although we determined only a partial sequence for about half of these phylotypes. An extension of the phylogenetic analysis, including most of the sequences reported as gut spirochetes of termites, reveals that the phylotypes identified from N. koshunensis are unique (data not shown). Most of the phylotypes from N. koshunensis do not show >97% sequence identity to those from the other termites. The only exception is the phylotype NkS37, which is closely related to the phylotype Gf35 from the termite Glyptotermes fuscus (Kalotermitidae) (17). The results confirm that termite guts are indeed an enormous reservoir of novel spirochetes, as proposed previously (14).
The four phylotypes of the ectosymbiotic spirochetes NkS-Dev1, NkS-Dev14, NkS-Oxy26, and NkS-Oxy8 were dispersed in cluster I of the phylogeny, and they formed completely distinct lineages. The phylotypes identified from the protist ectosymbionts but not yet confirmed by an in situ hybridization experiment also formed lineages distinct from these four phylotypes and from each other (except for phylotype NkS-Ste9, which is related to NkS-Oxy26). The phylotypes of cluster I that have been identified from the isolated protists of Dinenympha spp. and Pyrsonympha sp., RsDiSp8, HsDiSp319, HsDiSp320, and HsPySp4, in R. speratus and H. sjoestedti were also distantly related to the phylotypes of the ectosymbiotic spirochetes in N. koshunensis. This indicates that the ectosymbiotic spirochetes belonging to cluster I are a polyphyletic group. For the cluster II ectosymbiotic spirochetes, there are two distinct lineages, although the two are monophyletic among the known Treponema species (supported by a 93% bootstrap value). There seem to be multiple independent origins of spirochete attachment to gut protists.
Each protist cell was found to harbor multiple ectosymbiotic spirochete species. At least two spirochete phylotypes were identified on the three protist species in N. koshunensis. Spirochetes belonging to both clusters I and II were attached to cells of Dinenympha spp. and Pyrsonympha sp. in the guts of R. speratus and H. sjoestedti. Two ultrastructurally distinct morphotypes of the attached spirochetes have been reported in a hypermastigote protist, Joenia annectens (26). The attachment of multiple species seems to be a common feature among the spirochete-harboring protists.
It has been noted that common phylotypes are often shared among the protist species. For Devescovina sp. and Stephanonympha sp., the two phylotypes NkS-Dev1 and NkS-Dev14 are common ectosymbionts. The NkS-Oxy26 phylotype, which is abundant on Oxymonas sp., is also occasionally found on Stephanonympha sp. A common phylotype in cluster I, represented by either HsPySp4 or HsDiSp320, has been identified from Pyrsonympha sp. and Dinenympha sp. in H. sjoestedti (9). One can presume that, among free-swimming spirochetes in cluster I, some species that are able to attach to protist cells have emerged, probably several times during their evolution, and within the gut microbial community they have established ectosymbiotic relationships with multiple protist species. However, the NkS-Dev1 and NkS-Dev14 phylotypes were never found on Oxymonas sp., and the NkS-Oxy8 phylotype attaching to Oxymonas sp. did not occur on Devescovina sp. or Stephanonympha sp. Hence, at least some specificity to the host protist is present. Furthermore, for D. porteri and Pyrsonympha sp., the ectosymbiotic phylotypes are somewhat related (RsDiSp8 and HsPySp4, RsDiSp9 and HsPySp1, and RsDiSp1 and HsPySp15, respectively), even though these two protists inhabit evolutionarily distant termites, R. speratus and H. sjoestedti. The phylogenetic relationships of these ectosymbiotic spirochetes suggest that they have not coevolved with their host termites but rather with their host protists. Although the protist genera Oxymonas, Dinenympha, and Pyrsonympha belong to the order Oxymonadida, the last two are more closely related to each other than to Oxymonas, since they are in the family Pyrsonymphidae while Oxymonas is in the family Oxymonadidae (33). The apparent restriction of cluster II spirochetes to pyrsonymphids is consistent with the evolutionary divergence of that family from the other oxymonads. Nevertheless, the establishment and evolution of ectosymbiosis on the cell surface of gut protists are considered to be complex.
Most of the spirochetes represented by the NkS-Oxy26 phylotype are found on a restricted portion of the surfaces of Oxymonas sp. cells. D. porteri protists in R. speratus often harbor a dense population of cluster I spirochetes on the posterior end of the cell, which was revealed by in situ hybridization (data not shown). For the protist H. mirabile in the termite C. formosanus, ectosymbiotic spirochetes in cluster I occur on only the posterior surface, although their corresponding phylotypes have not yet been identified. These features evoke a specific association between the cellular organization of the protists and the function of the spirochetes. These apparent preferences of some spirochetes for attachment sites are in contrast to the lack of site specificity of the NkS-Oxy8 phylotype, which is distributed over the entire surface of its host, Oxymonas sp. Spirochetes on Pyrsonympha sp. in H. sjoestedti attach to the entire cell surface, and both clusters I and II of the spirochetes occur randomly. While most of the ectosymbiotic spirochetes occur on the anterior surfaces of Devescovina sp. and Stephanonympha sp., each detected phylotype seemed to disperse randomly. For the latter species, the spirochetes were interspersed with the flagella of the protist, and they were also found on the posterior surfaces of several cells. No relationship was found between the phylogenetic positions of the phylotypes and their preference for attachment sites. For example, two phylogenetically different phylotypes, NkS-Oxy26 and RsDiSp8 (a unique cluster I phylotype identified from D. porteri), mainly localize in restricted regions of the protists' cell surfaces. The phylotypes NkS-Oxy8 and HsPySp4 are also distantly related, but they are distributed randomly on the surfaces of the protists.
This study highlights the complexities of diversity, host specificity, preference for attachment site, and evolution of the ectosymbiotic spirochetes associated with gut protists in termites. In order to understand the real nature of this ectosymbiosis, further research is needed to investigate the functions or roles of individual spirochetes toward their hosts. Further studies of the phylogenetic and in situ identification of the ectosymbiotic spirochetes on diverse protist species are necessary to more fully understand their evolution. The culture-independent approaches described in this study would be advantageous not only for this purpose but also to investigate the structure of the microbial community in the termite guts, since a wide variety of protist-prokaryote associations have been observed in the gut community.
Acknowledgments
We thank O. Kitade for identification of the flagellated protists. We also thank G. Tokuda, T. Agusa, and A. Takase for collection of the termites and K. Nakamura for assistance with nucleotide sequencing.
This work was partially supported by grants for the Bioarchitect Research Program and the Eco Molecular Science Research Program from RIKEN. One of us (S.N.) is a recipient of support from the Special Postdoctoral Researcher Program of RIKEN, and A.Y. is supported by a grant from the Junior Research Associate Program of RIKEN.
REFERENCES
- 1.Berchtold, M., A. Chatzinotas, W. Schönhuber, A. Brune, R. Amann, D. Hahn, and H. König. 1999. Differential enumeration and in situ localization of microorganisms in the hindgut of the lower termite Mastotermes darwiniensis by hybridization with rRNA-targeted probes. Arch. Microbiol. 172:407-416. [DOI] [PubMed] [Google Scholar]
- 2.Berchtold, M., and H. König. 1996. Phylogenetic analysis and in situ identification of uncultivated spirochetes from the hindgut of the termite Mastotermes darwiniensis. Syst. Appl. Microbiol. 19:66-73. [Google Scholar]
- 3.Berchtold, M., W. Ludwig, and H. König. 1994. 16S rDNA sequence and phylogenetic position of an uncultivated spirochete from the hindgut of the termite Mastotermes darwiniensis Froggatt. FEMS Microbiol. Lett. 123:269-273. [DOI] [PubMed] [Google Scholar]
- 4.Bloodgood, R. A., and T. P. Fitzharris. 1976. Specific associations of prokaryotes with symbiotic flagellate protozoa from the hindgut of the termite Reticulitermes and the wood-eating roach Cryptocercus. Cytobios 17:103-122. [PubMed] [Google Scholar]
- 5.Breznak, J. A. 1984. Hindgut spirochetes of termites and Cryptocercus punctulatus, p. 67-70. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 6.Breznak, J. A. 2000. Ecology of prokaryotic microbes in the guts of wood- and litter-feeding termites, p. 209-234. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termite: evolution, sociality, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Cleveland, L. R., and A. V. Grimstone. 1964. The fine structure of the flagellate Mixotricha paradoxa and its associated micro-organisms. Proc. R. Soc. Lond. B 157:668-683. [Google Scholar]
- 8.Friedrich, M. W., D. Schmitt-Wagner, T. Lueders, and A. Brune. 2001. Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl. Environ. Microbiol. 67:4880-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iida, T., M. Ohkuma, K. Ohtoko, and T. Kudo. 2000. Symbiotic spirochetes in the termite hindgut: phylogenetic identification of ectosymbiotic spirochetes of oxymonad protists. FEMS Microbiol. Ecol. 34:17-26. [DOI] [PubMed] [Google Scholar]
- 10.Kirby, H. 1941. Devescovinid flagellates of termites. I. The genus Devescovina. Univ. Calif. Pub. Zool. 45:1-91. [Google Scholar]
- 11.Kirby, H. 1945. Devescovinid flagellates. IV. The genera Metadevescovina and Pseudodevescovina. Univ. Calif. Pub. Zool. 45:247-318.
- 12.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 13.Lilburn, T., G. K. Byzek, R. K. Kim, and J. A. Breznak. 2001. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495-2498. [DOI] [PubMed] [Google Scholar]
- 14.Lilburn, T. G., T. M. Schmidt, and J. A. Breznak. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331-345. [DOI] [PubMed] [Google Scholar]
- 15.Maidak, B. L., J. R. Cole, C. T. Parker, G. M. Garrity, N. Larsen, and B. Li. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margulis, L., and G. Hinkle. 1992. Large symbiotic spirochetes: Clerelandina, Cristispira, Diplocalyx, Hollandina and Pillotina, p. 3965-3978. In A. Balows, H. G. Trupper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer, New York, N.Y.
- 17.Ohkuma, M., T. Iida, and T. Kudo. 1999. Phylogenetic relationships of symbiotic spirochetes in the gut of diverse termites. FEMS Microbiol. Lett. 181:123-129. [DOI] [PubMed] [Google Scholar]
- 18.Ohkuma, M., and T. Kudo. 1998. Phylogenetic analysis of the symbiotic intestinal microflora of the termite Cryptotermes domesticus. FEMS Microbiol. Lett. 164:389-395. [Google Scholar]
- 19.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohkuma, M., S. Noda, Y. Hongoh, and T. Kudo. 2002. Diverse bacteria related to the bacteroides subgroup of the CFB phylum within the gut symbiotic communities of various termites. Biosci. Biotech. Biochem. 66:78-84. [DOI] [PubMed] [Google Scholar]
- 21.Ohkuma, M., S. Noda, K. Horikoshi, and T. Kudo. 1995. Phylogeny of symbiotic methanogen in the gut of the termite Reticulitermes speratus. FEMS Microbiol. Lett. 134:45-50. [DOI] [PubMed] [Google Scholar]
- 22.Ohkuma, M., S. Noda, and T. Kudo. 1999. Phylogenetic relationships of symbiotic methanogens in diverse termites. FEMS Microbiol. Lett. 171:147-153. [DOI] [PubMed] [Google Scholar]
- 23.Ohkuma, M., K. Ohtoko, C. Grunau, S. Moriya, and T. Kudo. 1998. Phylogenetic identification of the symbiotic hypermastigote Trichonympha agilis in the hindgut of the termite Reticulitermes speratus based on small-subunit rRNA sequence. J. Eukaryot. Microbiol. 45:439-444. [DOI] [PubMed] [Google Scholar]
- 24.Paster, B. J., F. E. Dewhirst, S. M. Cooke, V. Fussing, L. K. Poulsen, and J. A. Breznak. 1996. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl. Environ. Microbiol. 62:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paster, B. J., F. E. Dewhirst, W. G. Weisburg, L. A. Tordoff, G. J. Franser, R. B. Hespell, T. B. Stanton, L. Zablen, L. Mandelco, and C. R. Woese. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radek, R., K. Hausmann, and A. Breunig. 1992. Ectobiotic and endocytobiotic bacteria associated with the termite flagellate Joenia annectens. Acta Protozool. 31:93-107. [Google Scholar]
- 27.Shinzato, N., T. Matsumoto, I. Yamaoka, T. Oshima, and A. Yamagishi. 1999. Phylogenetic diversity of symbiotic methanogens living in the hindgut of the lower termite Reticulitermes speratus analyzed by PCR and in situ hybridization. Appl. Environ. Microbiol. 65:837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, H. E., and H. J. Arnott. 1974. Epi- and endobiotic bacteria associated with Pyrsonympha vertens, a symbiotic protozoon of the termite Reticulitermes flavipes. Trans. Am. Microsc. Soc. 93:180-194. [PubMed] [Google Scholar]
- 29.Smith, H. E., H. E. Buhse, Jr., and S. J. Stamler. 1975. Possible formation and development of spirochete attachment site found on the surface of symbiotic polymastigote flagellates of the termite Reticulitermes flavipes. Biosystems 7:374-379. [DOI] [PubMed] [Google Scholar]
- 30.Smith, H. E., S. J. Stamler, and H. E. Buhse. 1975. A scanning electron microscope survey of the surface features of polymastigote flagellates from Reticulitermes flavipes. Trans. Am. Microsc. Soc. 94:401-410. [Google Scholar]
- 31.Tokura, M., M. Ohkuma, and T. Kudo. 2000. Molecular phylogeny of methanogens associated with flagellated protists in the gut and with the gut epithelium of termites. FEMS Microbiol. Ecol. 33:233-240. [DOI] [PubMed] [Google Scholar]
- 32.Trager, W. 1934. The cultivation of a cellulose-digesting flagellate Trichomonas termopsidis and of certain other termite protozoa. Biol. Bull. 66:182-190. [Google Scholar]
- 33.Yamin, M. A. 1979. Flagellates of the orders Trichomonadida Kirby, Oxymonadida Grassé and Hypermastigida Grassi and Foà reported from lower termites (Isoptera families Mastotermitidae, Kalotermitidae, Hodotermitidae, Termopsidae, Rhinotemitidae, and Serritermitidae) and from the wood-feeding roach Cryptocercus (Dictyoptera: Cryptocercidae). Sociobiology 4:5-119. [Google Scholar]