Abstract
We succeeded in isolating several thermostable mutant fructosyl-amino acid oxidase (FAOX; EC 1.5.3) without reduction of productivity by directed evolution that combined an in vivo mutagenesis and membrane assay screening system. Five amino acid substitutions (T60A, A188G, M244L, N257S, and L261M) occurred in the most thermostable mutant obtained by a fourth round of directed evolution. This altered enzyme, FAOX-TE, was stable at 45°C, whereas the wild-type enzyme was not stable above 37°C. The Km values of FAOX-TE for d-fructosyl-l-valine and d-fructosyl-glycine were 1.50 and 0.58 mM, respectively, in contrast with corresponding values of 1.61 and 0.74 mM for the wild-type enzyme. This altered FAOX-TE will be useful in the diagnosis of diabetes.
Fructosyl-amino acid oxidase (FAOX) is a candidate for the enzymatic detection of nonenzymatically glycated proteins. This nonenzymatic reaction is called “glycation” to distinguish it from the enzymatic glycosylation of proteins. Glycation of protein has been implicated in the development of diabetic complications and the aging process (3, 4, 11). Glycation of blood proteins such as hemoglobin and albumin is enhanced in diabetics with high blood glucose. The amounts of these glycated proteins reflect the level of blood glucose in periods corresponding to the half-life of the protein (14 to 20 days for albumin and 1 to 2 months for hemoglobin). Since the glycation of blood proteins is not affected by transient increases in blood glucose, the levels of glycated proteins are good indices for monitoring diabetes mellitus patients during therapy. HbA1c, a good index of the medical condition of patients with diabetes mellitus, is defined as an amino end of the β-subunit (valine residue) of hemoglobin that has been glycated nonenzymatically (1). Glycated albumin is defined as a ɛ-amino group of lysine residue in polypeptide that has been glycated (8). We discovered fructosyl-amino acid oxidase from Corynebacterium sp. strain 2-4-1 (FAOX-C) (7), cloned the FAOX-C gene, and expressed it in Escherichia coli (15). FAOX-C showed high activity toward d-fructosyl-l-valine (FV), but no activity toward Nɛ-fructosyl-lysine (ɛFLys) and Nɛ-fructosyl Nα-formyl-lysine (ɛFfLys). Many other investigators have discovered fungal FAOXs (6, 13, 14, 20, 21, 23-25), but these enzymes (from fungi) showed activity toward ɛFLys and Nɛ-fructosyl Nα-Z-lysine. When the amount of HbA1c in the total blood is measured enzymatically, the activity toward ɛFLys can be a source of contamination and error, and hence, the substrate specificity of FAOX-C is beneficial to the measurement of HbA1c. We wanted to develop an enzymatic method to measure the amount of HbA1c with a combination of FAOX and the enzyme that releases FV from HbA1c. FAOX-C is considered to be most efficient for the measurement of HbA1c, but its thermal sensitivity is a weak point in diagnostic applications. Many enzymes in diagnostic reagents are transported in solution, so more stable enzymes are desirable. In addition, cost efficiency is an important factor for diagnostic reagents. We should therefore consider both the productivity and thermostability of the enzymes.
In this study, we applied directed evolution with in vivo mutagenesis and a membrane assay to increase the thermostability of the enzyme step by step as the amino acid substitutions accumulated. These mutant enzymes also had low Km values for d-fructosyl-glycine (FG) and showed high productivity in recombinant E. coli cells. The mutant strains produced as much FAOX as the wild-type strain. We also purified one enzyme, FAOX-TE, and characterized its properties. The purified FAOX-TE showed high specificity for FV and FG and inactivity toward ɛFLys, like the wild-type enzyme, FAOX-C, did. The thermostable enzyme, FAOX-TE, is therefore considered to be more suitable than FAOX-C for measurement of HbA1c.
MATERIALS AND METHODS
Materials.
FG, FV, and ɛFfLys were prepared as described previously (7). Restriction enzymes and DNA-modifying enzymes were obtained from Daiichi Kagaku (Tokyo, Japan), Takara Shuzo (Kyoto, Japan), and TOYOBO (Osaka, Japan). All other reagents were of analytical grade.
Oligonucleotides.
Oligonucleotides were synthesized on a model 382 synthesizer (ABI). The sequences of the oligonucleotides used in this study are listed in Table 1 and are based on the nucleotide sequence of the FAOX-C gene.
TABLE 1.
Oligonucleotides utilized in this studya
| Oligonucleotide sequence | Utility |
|---|---|
| AGAGATCACATATGTGGTGGACCGCTACCAA | PCR for pFA5 |
| CTTTTGCGCATATGCTAGGAGAACCGGCCCGGCC | PCR for pFA5 |
| GAACGCTCCGCTCCCTATCAC | Mutagenesis of T60A |
| GAACGCTCCNNKCCCTATCAC | Mutagenesis of T60X |
| CGGGCTACCGGCGTCGAAACC | Mutagenesis of A188G |
| CGGGCTACCNNKGTCGAAACC | Mutagenesis of A188X |
| GCGGCAGTGCTGAATACGCCG | Mutagenesis of M244L |
| GCGGCAGTGNNKAATACGCCG | Mutagenesis of M244X |
| AATCCGGGTAGCACTTTTGCC | Mutagenesis of N257S |
| AATCCGGGTNNKACTTTTGCC | Mutagenesis of N257X |
| ACTTTTGCCATGGATCACGAC | Mutagenesis of L261M |
| ACTTTTGCCNNKGATCACGAC | Mutagenesis of L261X |
N indicates a mixture of four nucleotides. X indicates any of 20 amino acids.
Strains and vector.
The strains and plasmids used in this study are described in Table 2. An expression plasmid, pFA101, was constructed from pUTE500k′. PCR amplification was carried out with the oligonucleotides listed in Table 1, with pFA5 as a template. After digestion by NdeI, the resultant fragment was inserted into pUTE500k′ to obtain pFA101. Antibiotics were routinely used at the following concentrations: ampicillin, 50 μg/ml; and kanamycin, 50 μg/ml.
TABLE 2.
Strains and plasmids utilized in this study
| E. coli strain or plasmid | Genotype | Utility | Source or reference |
|---|---|---|---|
| Strains | |||
| DH5α | F−Φ80 lacZΔM15 Δ(lacZYA-argF) U169 hsdR17 (rk−, mk+) recA1 endA1 deoR thi-1 supE44 gyrA96 relA1 λ− | Enzyme production | TOYOBO |
| XL1-Red | endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10 (Tetr) | In vivo mutagenesis | TOYOBO |
| MV1184 | ara Δ(lac-proAB) rpsL thi (lacZΔM15) Δ(srl-recA) 306::Tn10 (Tetr)/F′ [traD36 proAB+lacIqlacZΔM15] | Site-directed mutagenesis | Takara Shuzo |
| Plasmids | |||
| pUTE500k′ | Expression vector; Apr Kmr | 12 | |
| pFA5 | FAOX-C gene coding; Apr | 15 | |
| pFA101 | Expression of FAOX-C gene and template of mutagenesis; Apr Kmr | This work | |
| pFAH11 | Positive clone of 1st round of directed evolution; Apr Kmr | This work | |
| pFAH21 | Positive clone of 2nd round of directed evolution; Apr Kmr | This work | |
| pFAH22 | Positive clone of 2nd round of directed evolution; Apr Kmr | This work | |
| pFAH31 | Positive clone of 3rd round of directed evolution; Apr Kmr | This work | |
| pFAH41 | Positive clone of 4th round of directed evolution; Apr Kmr | This work | |
| pBluescriptII SK+ | For sequence of mutant FAOX genes; Apr | TOYOBO | |
| pKF19k | For site-directed mutagenesis; Kmr | Takara Shuzo | |
| pFA19k | FAOX-C gene in pKF19k for site-directed mutagenesis; Kmr | This work | |
| pFAT60Ak | Harboring T60A mutation derived from pFA19k; Kmr | This work |
FAOX assay.
FAOX activity was measured at 37°C by the formation of a quinone dye following A555 (ɛ, 39.2 cm2/μmol) with a Hitachi U-2001 spectrophotometer. The reaction mixture contained 100 mM potassium phosphate buffer (pH 8.0), 2.7 purpurogallin units of peroxidase, 0.45 mM 4-aminoantipyrine, 0.5 mM N-ethyl-N-(2-hydroxy-3-sulfopropyl)-m-toluidine (TOOS), and 5.0 mM FG in a total volume of 3 ml. The reaction was linear from 1 to 3 min under these conditions. One unit of enzyme activity was defined as the amount of enzyme that produced 0.5 μmol of quinone dye per min. This amount corresponds to the formation of 1.0 μmol of H2O2 per min.
To test the thermostability, we treated the crude extracts with a warm (47°C) water bath for 10 min before the assay. The kinetic parameters were determined by fitting the data with the Michaelis-Menten equation. To determine the Km values, the substrate (FG or FV) concentration was varied from 0.25 to 10 mM.
Random in vivo mutagenesis and screening.
For diagnostic applications, we should consider productivity as well as thermostability. We tested the productivity of the enzymes by using an assay involving non-heat-treated membranes. Random mutagenesis was performed by introduction of pFA101 into E. coli XL1-Red. E. coli XL1-Red cells transformed with pFA101 were plated on Luria-Bertani (LB) broth agar plates containing ampicillin. These plates were incubated at 30°C for 24 h. Mutated plasmids were retrieved from colonies on the plates by the alkaline sodium dodecyl sulfate (SDS) method (16). The membrane assay was used for the selection of thermostable mutants. E. coli DH5α was transformed with the mutated plasmids retrieved from XL1-Red transformants and plated on LB broth agar plates containing ampicillin. The plates were incubated at 30°C for 24 h, and two replicas of each plate were prepared with velvet cloth when colonies appeared on the plates (16). After 24 h at 30°C, colonies on the LB-ampicillin plates were transferred to nylon membranes (Hybond-N+; Amersham Pharmacia Biotech, Tokyo, Japan). One membrane containing the colonies was placed on agar plates and incubated at 55°C for 1 h at the first screening, and the other one was left at room temperature as a control for thermostability and to check the productivity of the enzyme. The incubation times were 1.5, 2, and 2.5 h at the second, third, and fourth screenings, respectively. Each of the membranes was placed on assay membranes containing 100 mM potassium phosphate buffer (pH 8.0), 5 mM FG, 0.5 U of peroxidase, 1 mM 4-aminoantipyrine, and 1 mM TOOS and then incubated at 37°C for 1 h. The thermostability of enzymes in each colony was checked by comparing the intensities of purple spots on the heat-incubated membranes with those on the non-heat-incubated membranes. The productivity of enzymes in each colony was checked according to the intensity of the purple spots on the non-heat-incubated membranes. The candidates for thermostable mutants were cultured in LB broth containing 100 μg of ampicillin per ml at 30°C for 20 h and disrupted by sonication to obtain the crude extracts, which were tested for thermostability by the above-mentioned FAOX assay, either after heat treatment or after no heat treatment. Screening was performed four times, with the control used at each screening being the parent clone that was obtained by the previous screening.
The plasmids pFAH11, pFAH21, pFAH22, pFAH31, and pFAH41 were prepared from selected mutants and sequenced.
Site-directed mutagenesis.
Mutan-Super Express Km (Takara) was used for site-directed mutagenesis. The plasmid pFA19k used as the template for site-directed mutagenesis was constructed from pFA101 and pKF19k. The plasmid pFA101 was digested by NdeI. Its resultant smaller fragment was inserted into pKF19k, digested with NdeI, and dephosphorylated with alkaline phosphatase (Takara). The oligonucleotides used for site-directed mutagenesis are listed in Table 1. A plasmid harboring the T60A mutation derived from pFA19k, pFAT60Ak, was used to construct the double mutant containing T60A and M244L. E. coli MV1184 transformed with mutated plasmids was selected on LB broth agar plates containing kanamycin and sequenced.
DNA sequencing.
Various restriction fragments derived from the mutant FAOX-C gene were subcloned into pBluescriptII SK+ and sequenced with a DNA model 373A sequencer (ABI).
Purification of the thermostable mutant FAOX-TE.
E. coli DH5α cells harboring pFAH41 were cultured in 6.0 liter of LB broth containing ampicillin at 30°C for 24 h. Seventy grams (wet weight) of the cells was harvested by centrifugation and resuspended in 1.0 liter of 20 mM potassium phosphate buffer (pH 8.0) containing 50 mM KCl. The suspended cells were sonicated, and the insoluble extracts were removed by centrifugation. The enzyme solution was added to 1.0 liter of QAE-Sephadex A-50 resin. The resin was washed with 5.0 liters of 20 mM potassium phosphate buffer (pH 8.0) containing 50 mM KCl. The enzyme was eluted with 1.5 liter of 20 mM potassium phosphate buffer (pH 8.0) containing 600 mM KCl. The eluate was dialyzed with 20 mM potassium phosphate buffer (pH 8.0) containing 200 mM KCl and then applied to a Q-Sepharose Fast Flow column (diameter, 8 cm; height, 20 cm), followed by elution in a gradient from 5.0 liters of 200 mM KCl to 5.0 liters of 400 mM KCl.
Protein determination.
The protein concentration was determined by the Bradford assay (2) with a protein assay kit (Bio-Rad) by using bovine serum albumin as the standard. Protein analysis by SDS-polyacrylamide gel electrophoresis was carried out according to the method of Laemmli (9).
RESULTS
Isolation of the thermostable mutant FAOX in directed evolution.
In order to investigate the rate of mutation, we sequenced 10 clones selected randomly from approximately 10,000 bacterial colonies. One of the clones included two mutations in the FAOX-C coding region, and both mutations had substituted amino acids. Four of the clones included one mutation that had substituted amino acid. Five of the clones included one mutation that did not have substituted amino acid. The productivity of half of the colonies was indistinguishable from that of the wild-type strain, DH5α(pFA101), as evaluated by the intensity of purple spots. From these colonies carrying random in vivo-mutagenized pFA101, five thermostable mutants were selected with the membrane assay as described in Materials and Methods. We then confirmed the thermostability of the crude extracts from the five positive clones. The wild-type FAOX-C decreased its activity to only 1% of the initial activity by heat treatment at 47°C for 10 min. In contrast, about 30% of the initial activity still remained in the crude extracts from the five mutants under the same conditions as the wild type (Table 3). We isolated the plasmids from the selected mutants and determined the nucleotide sequences. Two of the five mutants carried base changes of C to G at base 563, G to C at base 564, and A to C at base 730; the other three mutants carried the base changes of C to G at base 563, G to C at base 564, and A to T at base 730. These two types of base changes resulted in the substitutions of Gly (GGC) for Ala (GCG) at position 188 and Leu (CTG or TTG) for Met (ATG) at position 244 in the amino acid sequence (Table 3). We named the former mutant plasmid “pFAH11.”
TABLE 3.
Rounds of thermostabilization of FAOXa
| Round | Productivity (U/ml) | % of activity at 47°C for 10 min | Km value (mM) | Mutation sites |
|---|---|---|---|---|
| Parent (pFA101, FAOX-C) | 0.10 ± 0.01 | 1.0 ± 0.02 | 0.74 | |
| 1st (pFAH11) | 0.11 ± 0.01 | 30.0 ± 0.55 | 0.57 | A188G M244L |
| 2nd | ||||
| pFAH21 | 0.10 ± 0.01 | 35.5 ± 0.59 | 0.42 | A188G M244L L261M |
| pFAH22 | 0.11 ± 0.01 | 70.0 ± 1.25 | 0.50 | T60A A188G M244L |
| 3rd (pFAH31) | 0.10 ± 0.01 | 80.0 ± 1.52 | 0.50 | T60A A188G M244L L261M |
| 4th (pFAH41 FAOX-TE) | 0.10 ± 0.01 | 90.0 ± 1.61 | 0.50 | T60A A188G M244L N257S L261M |
Two thermostable mutants were obtained in the second screening round. The Km value was measured for FG. The data represent the means ± standard deviations of three measurements.
We performed a second screening from approximately 10,000 bacterial colonies carrying random in vivo-mutagenized pFAH11, from which seven thermostable mutants were selected. These mutants could be grouped into distinct types by the degree of thermostability. In the extracts from three of the seven mutants, 35.5% of the FAOX activity remained after heat treatment at 47°C for 10 min; in the extracts from the other four, 70% of the FAOX activity remained (Table 3). We isolated the plasmid from the two types of mutants and determined the nucleotide sequence. In the former group (35.5% of remaining activity), the change of base from T to A was found at base 781. This base change resulted in the substitution of Met (ATG) for Leu (TTG) at position 261 in the amino acid sequence (Table 3). We named this mutant plasmid “pFAH21.” In the latter group (70% of remaining activity), the change of base from A to G was found at base 178. This base transition resulted in the substitution of Ala (GCT) for Thr (ACT) at position 60 in the amino acid sequence (Table 3). We named this mutant plasmid “pFAH22.”
We performed a third screening from approximately 10,000 bacterial colonies carrying random in vivo-mutagenized pFAH22. We isolated five mutants in which 80% of the FAOX activity remained in the crude extract after heat treatment at 47°C for 10 min. The change of base from T to A at base 781 was found in all five of the mutants, the plasmid of which was named “pFAH31.” This base change resulted in the substitution of Met (ATG) for Leu (TTG) at position 261 in the amino acid sequence (Table 3). A fourth screening from approximately 10,000 bacterial colonies carrying random in vivo-mutagenized pFAH31 resulted in the isolation of four mutants in which 90% of the FAOX activity remained in the crude extract. The change of base from A to G at base 770 was found in all four of the mutants, the plasmid of which was named “pFAH41.” This base change resulted in the substitution of Ser (AGC) for Asn (AAC) at position 257 in the amino acid sequence (Table 3). In every screening round of directed evolution, we confirmed that the productivity of the mutants was indistinguishable from that of the wild type (Table 3).
Comparison of the wild-type and mutant FAOXs.
The isolated mutants, E. coli DH5α(pFAH11, pFAH21, pFAH22, pFAH31 and pFAH41), were cultured at 30°C for 24 h. These activities are presented in Table 3 and are the same as those for the wild-type FAOX-C produced by DH5α(pFA101). The Km values for the FG substrate of the mutant FAOXs were lower than that of the wild-type FAOX-C (Table 3).
Site-directed mutagenesis.
From the random in vivo mutagenesis and membrane assay screening, we found five mutation sites (T60A, A188G, M244L, N257S, and L261M) that were predicted to be effective for the thermostabilization of FAOX-C. In order to investigate the individual effects of the amino acid residues at each of the five sites, we independently replaced each position of FAOX-C with one of the other possible amino acid residues by site-directed mutagenesis. The thermostability of the mutants was tested with a nylon membrane assay. The nucleotide sequences of the mutants were determined. The most thermostable mutations without associated reductions in productivity were the substitutions of Ala for Thr at position 60 and Leu for Met at position 244 in the amino acid sequence (Fig. 1 and Table 4). On the other hand, any amino acid substitutions at positions 188, 257, and 261 did not give independent thermostability. The substitutions of Gly for Ala at position 188, Ser for Asn at position 257, and Met for Leu at position 261 of FAOX-C did not give independent thermostability (Table 4).
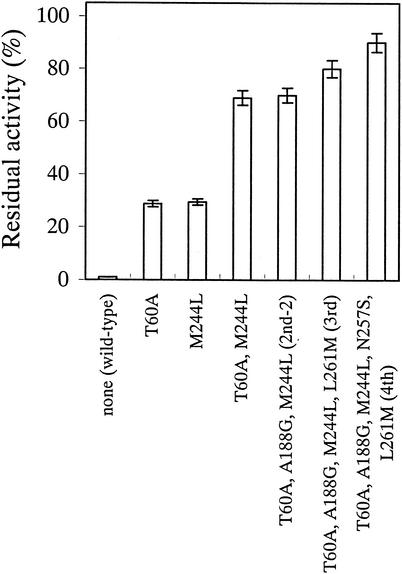
FIG. 1.
Step-by-step increase in enzyme thermostability as amino acid substitutions accumulated. Shown are the residual activities of the wild-type and mutant FAOX enzymes after heat treatment at 47°C for 10 min. The data represent the means ± standard deviations of three measurements.
TABLE 4.
Thermostabilization of FAOX by site-directed mutagenesisa
| Mutation site(s) | % of activity at 47°C for 10 min | Km (mM) |
|---|---|---|
| None (wild type) | 1.0 ± 0.02 | 0.74 |
| T60A | 28.8 ± 0.56 | 0.50 |
| A188G | 1.0 ± 0.02 | 0.74 |
| M244L | 29.5 ± 0.60 | 0.57 |
| N257S | 1.5 ± 0.04 | 0.70 |
| L261M | 2.5 ± 0.05 | 0.70 |
| T60A M244L | 69.0 ± 1.29 | 0.45 |
| T60A A188G M244Lb | 70.0 ± 1.25 | 0.50 |
The Km value is measured for FG. The data represent the means ± standard deviations of three measurements.
Second mutant in the second screening round.
To investigate the effect of the combination of mutation sites, we obtained a double (T60A M244L) mutant by site-directed mutagenesis. The double mutant showed a higher thermostability than two single (either T60A or M244L) mutants and thermostability as high as that of the second mutant in directed evolution (Fig. 1 and Table 3).
Thermostability of FAOX-TE.
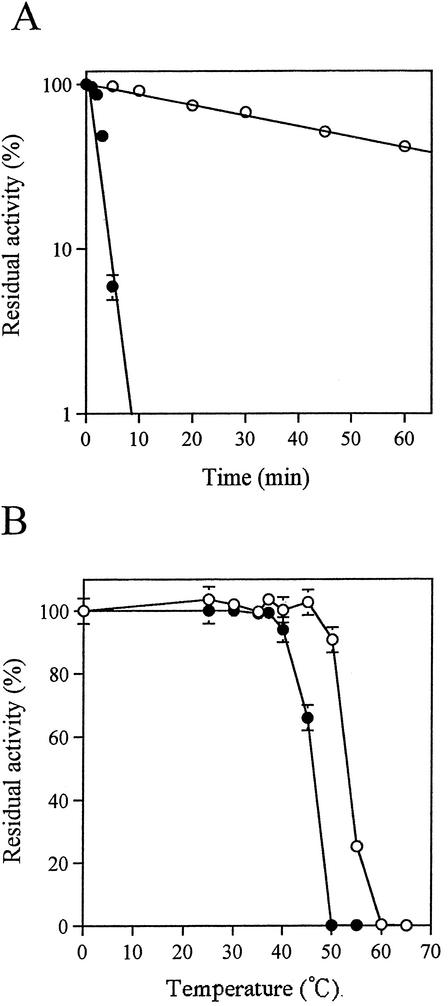
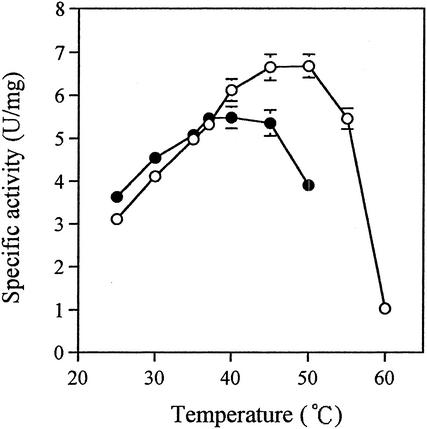
The most thermostable enzyme, FAOX-TE, which had five mutations (T60A, A188G, M244L, N257S, and L261M), was obtained in the fourth round of screening in directed evolution. The wild type (FAOX-C) and FAOX-TE were purified to homogeneity as described in Materials and Methods. We performed two tests to compare the thermostabilities of these enzymes. First, purified enzymes were incubated at 50°C at pH 8.0, and residual FAOX activity was measured at various times. The mutant FAOX-TE showed a greater thermostability than the wild-type FAOX-C (Fig. 2A). The half-lives of the activities of FAOX-C and FAOX-TE were estimated as generally 2.9 and 45.0 min, respectively. Second, each enzyme was kept at various temperatures for 10 min, and the residual FAOX activity was measured. FAOX-TE was stable at 45°C, whereas FAOX-C was not stable above 37°C (Fig. 2B). The optimum temperature of FAOX-TE was about 10°C higher than that of FAOX-C (Fig. 3).
FIG. 2.
Thermostability of the purified wild-type FAOX-C (solid circles) and the mutant FAOX-TE (open circles). (A) Time course of thermoinactivation of FAOX enzymes at 50°C. The enzymes (180 mU) were incubated at 50°C in 100 mM phosphate buffer (pH 8.0). The remaining activities were determined and are shown as percentages of the activity at 0 min. (B) The enzyme (180 mU) in 100 mM phosphate buffer (pH 8.0) was kept at various temperatures for 10 min, and then the remaining activities were determined, which are shown here as percentages of the activity when incubated at 0°C. The data represent the means ± standard deviations of three measurements.
FIG. 3.
Optimal temperature of wild-type FAOX-C (solid circles) and mutant FAOX-TE (open circles). Enzyme reactions were performed with the standard assay mixture at various temperatures. The enzyme (180 mU) assayed at 37°C was used in the assay mixtures. The data represent the means ± standard deviations of three measurements.
Properties of FAOX-TE.
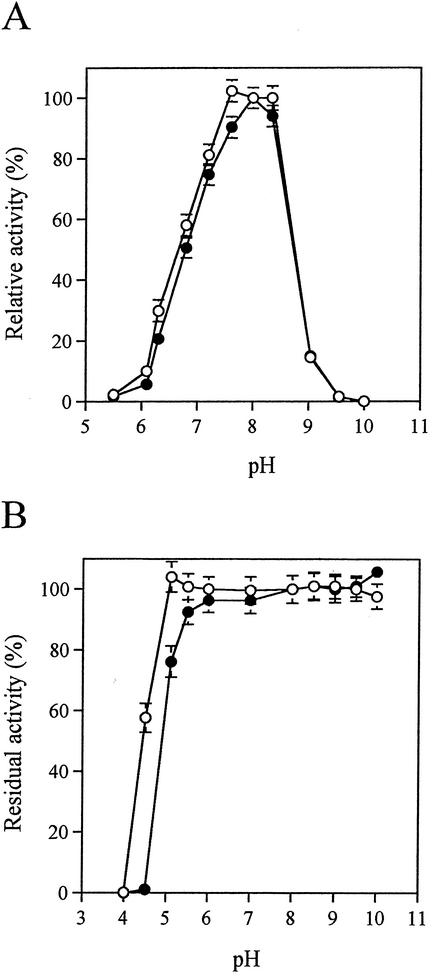
The purified FAOX-C and FAOX-TE were characterized. The specific activity toward FG of the purified FAOX-C was the same as that of purified FAOX-TE (Table 5). FAOX-C and FAOX-TE showed high enzyme activity toward FV and FG, but no activity toward ɛFfLys. Inactivity toward ɛFfLys was conserved in all thermostable mutants (data not shown). The kinetic parameters and pIs of FAOX-C and FAOX-TE are presented in Table 5. The Km and kcat values of FAOX-TE for FV and FG were slightly lower than those of FAOX-C. The catalytic efficiency (kcat/Km) of FAOX-TE was 1.4-fold larger than that of FAOX-C for FG and the same as that of FAOX-C for FV. FAOX-TE exhibited 100% of its optimal activity at pH values of 7.6 to 8.3 and 100% stability for pH values over 5.1. Both the optimum pH values and the pH stability of FAOX-TE make it slightly more suitable for lower-pH conditions than FAOX-C (Fig. 4).
TABLE 5.
Comparison of wild-type FAOX-C and thermostable mutant FAOX-TE by using purified enzymesa
| Enzyme | Sp act for FG (U/mg) | Specificity (%)
|
Km (mM)
|
kcat (s−1)
|
kcat/Km (mM−1 s−1)
|
pI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FG | FV | ɛFf Lys | FG | FV | FG | FV | FG | FV | |||
| FAOX-C (wild type) | 5.46 ± 0.11 | 100 ± 2.1 | 132 ± 2.5 | 0 | 0.74 | 1.61 | 8.01 ± 0.16 | 10.60 ± 0.21 | 10.80 ± 0.22 | 6.57 ± 0.13 | 4.6 |
| FAOX-TE (mutant) | 5.32 ± 0.11 | 100 ± 2.0 | 124 ± 2.3 | 0 | 0.58 | 1.50 | 7.80 ± 0.16 | 9.68 ± 0.19 | 15.60 ± 0.31 | 6.45 ± 0.13 | 4.2 |
The data represent the means ± standard deviations of three measurements.
FIG. 4.
Comparisons of the effects of pH on the activities and stabilities of the wild-type FAOX-C (solid circles) and FAOX-TE (open circles). (A) Optimum pH. Relative activity was measured with a reaction mixture containing 100 mM buffer at various pH values by the assay method with 180 mU of enzyme. Phosphate buffer was used at pH 5.5 to 8.3, and NaHCO3-Na2CO3 buffer was used at pH 9.0 to 10.0. (B) pH stability. Enzyme (180 mU) was kept at 40°C for 10 min at various pH values, and the remaining activity was then measured by the assay method. The following buffers (100 mM each) were used: McIlvaine buffer at pH 4.0 to 6.0, phosphate buffer at pH 6.0 to 8.0, and NaHCO3-Na2CO3 buffer at pH 8.5 to 10.0. The data represent the means ± standard deviations of three measurements.
DISCUSSION
In the present study, we repeatedly applied random mutagenesis and screening to improve thermostability without reducing productivity in diagnostic applications. This process is an application of directed evolution. We already have an efficient FAOX production system in E. coli (15). Our subjects for directed evolution were the construction of mutagenesis and screening systems. We adopted an XL1-Red random in vivo mutagenesis system and membrane assay screening system. The XL1-Red system was more convenient and rapid than error-prone PCR for the free optimization of the protocol. We also performed error-prone PCR and obtained the same result as that of the XL1-Red system at the first screening (data not shown). The membrane assay that was useful for a high-throughput screening with high sensitivity could remedy the fault of the XL1-Red system that could cause unnecessary mutation in other than the coding region.
We obtained mutant FAOXs with the remaining activity increasing step by step in every screening round (Table 3). This result indicates that our system functioned effectively and achieved the effectiveness of directed evolution in terms of thermostability. Our aim in this study was to improve thermostability while maintaining productivity. For this, we checked the productivity of each colony on non-heat-treated membranes and obtained thermostable FAOXs without associated reductions in productivity. With four sequential cycles of directed evolution, we observed accumulation of base substitutions in the gene and stepwise improvement in the thermostability of the enzymes. The application of directed evolution to thermostabilization of an enzyme has been reported previously (10, 19). These results showed that the directed evolution system was successful and indicate that we can distinguish a 10% difference in thermostability with the membrane assay. We consider our system for directed evolution useful for the thermostabilization of other oxidases.
The analysis of the thermostable mutation site by site-directed mutagenesis indicates that at some positions, T60A and M244L were effective on only single mutations, and at other positions, A188G, N257S, and L261M were ineffective only on single mutations. The double mutant (containing T60A and M244L) exhibited a quantitative increase in thermostability (Fig. 1 and Table 4). Tables 3 and 4 suggest that the other mutation sites (N257S and L261M) produced an increase in thermostability by a combination of mutation sites (second, third, and fourth mutants in directed evolution). These mutation sites, N257S and L261M, could not be found by a single mutation. Therefore, second, third, and fourth mutants (Table 3) could be obtained by directed evolution, but not by a combination of single mutation sites that increased thermostability. Only the A188G mutation did not produce an increase in thermostability, even in combination with the other mutation sites that were tested in this study (Table 4). We believe the A188G mutation was accumulated in the first cycle of directed evolution along with M244L and that this did not confer improved thermostability.
We performed site-directed mutagenesis to examine the possibility that the other 18 amino acids contribute to thermostabilization at each of the mutation sites. The results suggested that every substitution selected by directed evolution was the best for thermostabilization without a concomitant reduction in productivity. Although we cannot discount the possibility of another mutation site for implementing thermostabilization, we consider that our final mutant, FAOX-TE, satisfied the original objectives of our study. We saw that the thermostability of FAOX increased step by step as the amino acid substitutions accumulated in directed evolution (Fig. 1).
The optimal temperature of the thermostable mutant FAOX-TE was 10°C higher than that of the wild-type FAOX-C, and the activity at 50°C was ∼20% greater than that at 37°C (Fig. 3). These results indicate that FAOX-TE and FAOX-C were inactivated at different temperatures and that higher activity accompanied a temperature increase.
The Km values of the mutant FAOXs were slightly lower than those for the wild type. These results might be attributable to mutation site T60A or M244L (Table 4). The other mutation sites did not appear to affect the Km value in this study. The specific activity and substrate specificity of the mutant FAOX-TE were indistinguishable from those of the wild-type FAOX-C (Table 5). The catalytic efficiency (kcat/Km) of FAOX-TE indicates that FAOX-TE has higher specificity for FG than FAOX-C and specificity as high for FV as that of FAOX-C. The inactivity toward ɛFfLys that is a feature of both FAOX-C and FAOX-TE was beneficial to the development of an enzymatic method for measuring the amount of HbA1c.
The thermostability mechanism of FAOX-TE in this study is unclear, but the coinciding substrate specificities and specific activities between the wild-type FAOX-C and FAOX-TE led us to the hypothesis that the mutation sites found in this study did not affect the substrate recognition of FAOX. The stabilization of proteins through site-directed mutations often leads to decreased enzymatic activity (10, 17, 19), and an inverse correlation between protein stability and activity has been suspected. However, in this study, the amino acid substitutions introduced into the thermostable mutant enzymes did not appear to affect their enzymatic activity. Together with other cases in which the stabilization of proteins was achieved without the loss of their functions (5, 18, 22), our results strongly support the implication that the structure of a protein has a higher tolerance to changes in structural amino acids than in functional ones (17). The analysis of the other mutation sites with either differences in substrate specificity or reduction in productivity, X-ray analysis, and substrate-analog studies will help determine the mechanism of catalysis or stability of FAOX. We speculate that the thermostabilization of FAOX-C accelerates the development of the enzymatic measurement of HbA1c.
Acknowledgments
We thank Y. Koyama and N. Yamaji for continuous support, encouragement, and valuable and fruitful discussions and F. Tatsumi for technical assistance.
REFERENCES
- 1.Bookchin, R. M., and P. M. Gallop. 1968. Structure of hemoglobin A1c: nature of the N-terminal β-chain blocking group. Biochem. Biophys. Res. Commun. 32:86-93. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bunn, H. F., D. N. Haney, D. S. Kamin, K. H. Gabbay, and P. M. Gallop. 1976. The biosynthesis of human hemoglobin A1c: slow glycosylation of hemoglobin in vivo. J. Clin. Investig. 57:1652-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou, S. H., L. T. Chylack, W. H. Tung, Jr., and H. F. Bunn. 1981. Nonenzymatic glycosylation of bovine lens crystallins. J. Biol. Chem. 256:5176-5180. [PubMed] [Google Scholar]
- 5.Giver, L., A. Gershenson, P. O. Frekgard, and F. H. Arnold. 1998. Directed evolution of a thermostable esterase. Proc. Natl. Acad. Sci. USA 95:12809-12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horiuchi, T., and T. Kurokawa. 1991. Purification and properties of fructosylamine oxidase from Aspergillus sp. 1005. Agric. Biol. Chem. 55:333-338. [Google Scholar]
- 7.Horiuchi, T., T. Kurokawa, and N. Saito. 1989. Purification and properties of fructosyl-amino acid oxidase from Corynebacterium sp. 2-4-1. Agric. Biol. Chem. 53:103-110. [Google Scholar]
- 8.Iberg, N., and J. Fluckiger. 1986. Nonenzymatic glycosylation of albumin in vivo. Identification of multiple glycosylated sites. J. Biol. Chem. 261:13542-13545. [PubMed] [Google Scholar]
- 9.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 10.Meiering, E. M., L. Serrano, and A. R. Fersht. 1992. Effect of active site residues in barnase on activity and stability. J. Mol. Biol. 225:585-589. [DOI] [PubMed] [Google Scholar]
- 11.Monnier, H. F., and A. Cerami. 1982. Non-enzymatic glycosylation and browning of proteins in diabetes. Clin. Endocrinol. Metab. 11:431-452. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura, I., K. Okada, and Y. Koyama. 1996. Cloning and expression of pyranose oxidase cDNA from Coriolus versicolor in Escherichia coli. J. Biotechnol. 52:11-20. [DOI] [PubMed] [Google Scholar]
- 13.Sakai, Y., N. Yoshida, A. Isogai, Y. Tani, and N. Kato. 1996. Purification and properties of fructosyl amino acid oxidase from Fusarium oxysporum S-1F4. Biosci. Biotechnol. Biochem. 59:487-491. [DOI] [PubMed] [Google Scholar]
- 14.Sakai, Y., N. Yoshida, Y. Tani, and N. Kato. 1996. Production of fructosyl lysine oxidase from Fusarium oxysporum S-1F4 on autoclave-browned medium. Biosci. Biotechnol. Biochem. 60:150-151. [DOI] [PubMed] [Google Scholar]
- 15.Sakaue, R., M. Hiruma, N. Kajiyama, and Y. Koyama. 2002. Cloning and expression of fructosyl-amino acid oxidase gene from Corynebacterium sp. 2-4-1 in Escherichia coli. Biosci. Biotechnol. Biochem. 66:1256-1261. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Schreiber, G., A. M. Buckle, and A. R. Fersht. 1994. Stability and function: two constraints in the evolution of barstar and other proteins. Structure 2:945-951. [DOI] [PubMed] [Google Scholar]
- 18.Serrano, L., A. G. Day, and A. R. Fersht. 1993. Step-wise mutation of barnase to binase. A procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J. Mol. Biol. 233:305-312. [DOI] [PubMed] [Google Scholar]
- 19.Shoichet, B. K., W. A. Baase, R. Kuroki, and B. W. Matthew. 1995. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. USA 92:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi, M., M. Pischetsrieder, and V. M. Monnier. 1997. Isolation, purification, and characterization of amadoriase isoenzymes (fructosyl amine:oxygen oxidoreductase, EC 1.5.3) from Aspergillus sp. J. Biol. Chem. 272:3437-3443. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi, M., M. Pischetsrieder, and V. M. Monnier. 1997. Molecular cloning and expression of amadoriase isoenzyme (fructosyl amine:oxygen oxidoreductase, EC 1.5.3) from Aspergillus fumigatus. J. Biol. Chem. 272:12505-12507. [DOI] [PubMed] [Google Scholar]
- 22.Uchiyama, H., T. Inaoka, T. Ohkuma-Soyejima, H. Togame, Y. Shibanaka, T. Yoshimoto, and T. Kokubo. 2000. Directed evolution to improve the thermostability of prolyl endopeptidase. J. Biochem. 128:441-447. [DOI] [PubMed] [Google Scholar]
- 23.Wu, X., M. Takahashi, S. G. Chen, and V. M. Monnier. 2000. Cloning of amadoriase I isoenzyme from Aspergillus sp.: evidence of FAD covalently linked to Cys342. Biochemistry 39:1515-1521. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida, N., Y. Sakai, A. Isogai, H. Fukuya, M. Yagi, Y. Tani, and N. Kato. 1996. Primary structures of fungal fructosyl amino acid oxidases and their application to the measurement of glycated proteins. Eur. J. Biochem. 242:499-505. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida, N., Y. Sakai, M. Serata, Y. Tani, and N. Kato. 1995. Distribution and properties of fructosyl amino acid oxidase in fungi. Appl. Environ. Microbiol. 61:4487-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]