Abstract
The vaginal bacterial microbiota of 19 premenopausal women was examined by PCR-denaturing gradient gel electrophoresis (DGGE) and sequencing of the V2-V3 region of the 16S rRNA gene. Ten of the women were studied further to investigate the effect and persistence of vaginally inserted capsules containing viable lactobacilli. PCR-DGGE indicated that most subjects had a microbiota represented by one to three dominant DNA fragments. Analysis of these fragments revealed that 79% of the women possessed sequences with high levels of similarity to Lactobacillus species sequences. Sequences homologous to Lactobacillus iners sequences were the most common and were detected in 42% of the women tested. Alteration of the vaginal microbiota could be detected by PCR-DGGE in several women after the instillation of lactobacilli. Additionally, randomly amplified polymorphic DNA analysis of lactobacilli isolated from selective media demonstrated that the exogenous strains could be detected for up to 21 days in some subjects. This study demonstrates that non-culture-based techniques, such as PCR-DGGE, are useful adjuncts for studies of the vaginal microbiota.
The microbes that inhabit the vagina play a major role in illnesses of the host, including bacterial vaginosis, yeast vaginitis, cancer, and sexually transmitted diseases, such as human immunodeficiency virus infection, as well as in the maintenance of a healthy tract. Our understanding of the nature and functionality of these organisms has progressed in recent years, but it is still far from optimal. For some time the microbiota of so-called normal women of child-bearing age was believed to be dominated by Lactobacillus acidophilus and Lactobacillus fermentum, followed by Lactobacillus brevis, Lactobacillus jensenii, Lactobacillus casei, and other species (12). More recently, molecular methods have shown that Lactobacillus crispatus and Lactobacillus jensenii are the most common isolates (2, 12), and in one study a previously undescribed Lactobacillus species was found in 15% of women (2). The development of denaturing gradient gel electrophoresis (DGGE) has provided an exciting tool to analyze a given population of organisms within a host. To date, this method has been used successfully to examine the intestinal microbiotas of adults and children (5, 18).
Continuous application of certain Lactobacillus strains vaginally and orally has been shown to alter the microbiota from a microbiota indicative of bacterial vaginosis to a microbiota that is dominated by lactobacilli and regarded as normal (10). Instillation of probiotic lactobacilli has the potential to make a significant impact on the health of women, and therefore, it is important to understand how the vaginal microbiota changes and adapts to the presence of these strains. Therefore, the first goal of the present study was to utilize PCR-DGGE and to sequence different 16S DNA fragments to determine which bacterial species were most common among the vaginal samples of premenopausal women. The second goal was to use DGGE to examine the impact of probiotic strains on the vaginal bacterial microbiota and determine the persistence of exogenous lactobacillus strains by using selective medium and randomly amplified polymorphic DNA (RAPD) profiling (7).
MATERIALS AND METHODS
Subjects, probiotic instillation, and sample collection.
Nineteen premenopausal Caucasian women who had no symptoms or signs of vaginal or urinary tract infection and were otherwise healthy were recruited. Each woman signed an informed consent under a protocol approved by the human ethics review board at the University of Western Ontario. None of the recruits was receiving antimicrobial prescribed therapy or using spermicidal products. Deep vaginal samples were collected by rotating swabs throughout the vagina of each of the subjects prior to the start of the study at zero time and at 6 months. For the 10 subjects in whom lactobacilli were vaginally instilled (subjects 260 to 269), one capsule containing 1 × 109 total CFU of Lactobacillus fermentum RC-14 and Lactobacillus rhamnosus GR-1 was inserted daily into the vagina following the initial swabbing for 3 days. Additional swabs were collected on days 3, 7, 14, and 21 from the subjects who received probiotics. Two swabs were collected per subject at each sampling point, one for the culture of lactobacilli for RAPD analysis and the other for direct bacterial DNA extraction for PCR-DGGE. Once taken, the swabs were immediately placed in transport medium (NCS Diagnostics Inc., Etobicoke, Ontario, Canada) and taken to the lab for processing within 3 h.
Culturing and DNA fingerprinting of Lactobacillus strains by RAPD analysis.
Vaginal swabs were agitated in 1 ml of sterile phosphate-buffered saline (PBS) (pH 7.5) and serially diluted. To determine the persistence of L. rhamnosus GR-1 and L. fermentum RC-14 within the vagina, aliquots of each dilution were plated onto MRS plates (BBL, Becton Dickinson, Cockeysville, Md.) containing selective agents for each strain (7) (fusidic acid [32 μg/ml; Sigma Chemical Co., St. Louis, Mo.] and tetracycline [8 μg/ml; Sigma], respectively) and incubated anaerobically by using the BBL GasPack system at 37°C for 48 h. Ten colonies from each subject were selected for testing by RAPD analysis by the method of Gardiner et al. (7).
Extraction of bacterial DNA from swabs for PCR.
Swabs were vigorously agitated in 1 ml of PBS to dislodge the cells. The cells were pelleted by centrifugation (10,000 × g, 5 min) and washed once in PBS, and total DNA was extracted by using Instagene matrix (Bio-Rad Laboratories, Hercules, Calif.) according to the manufacturer's instructions. PCRs were carried out in 0.2-ml tubes with a thermocycler (Mastercycler; Eppendorf, Wesseling-Berzdorf, Germany). The HDA eubacterial PCR primers and amplification conditions of Walter et al. were utilized (18).
DGGE, DNA fragment excision from gels, reamplification, and sequencing.
Preparation of DGGE gel gradients and electrophoresis were carried out by using the manufacturer's guidelines for the D-code universal detection system of Bio-Rad. A 100% solution was defined as a mixture of 7 M urea and 40% formamide. The concentrations of polyacrylamide, denaturant, and Tris-acetate buffer (40 mM Tris, 20 mM glacial acetic acid, 1 mM EDTA [pH 8.0]) were 8%, 30 to 50%, and 1×, respectively. Other parameters have been described previously (18). Fragments of interest were excised from DGGE gels with a sterile scalpel, washed once in 1× PCR buffer, and incubated in 20 μl of the same buffer overnight at 4°C. Five microliters of the buffer solution was used as the template for PCR. Reamplification was conducted by using the primers described previously but without the GC clamp (18). Sequences of the reamplified fragments were determined by the dideoxy chain termination method (Sequencing Facility, John P. Robarts Research Institute, London, Ontario, Canada). Analysis of the partial 16S rRNA sequences was conducted by using the GenBank database and the BLAST algorithm (1). Identities of isolates were determined on the basis of the highest score.
RESULTS
DGGE and sequencing of DNA fragments before probiotic use indicated that most of the vaginal samples from the 19 women studied had one to three dominant fragments, as observed within a lane of a DGGE gel (Fig. 1). For subjects 261, 264, and 268 5 to 10 fragments were detected (Fig. 1). When the dominant fragments from every sample were sequenced, the majority of women tested (15 of 19 women) had at least one sequence homologous to a sequence of a species of Lactobacillus (Table 1). A significant discovery was that an organism that was recently found in the vagina (4), Lactobacillus iners, was the most commonly recovered species and was detected in 42% of the women.
FIG. 1.
DGGE of the V2-V3 16S rRNA gene amplicons from vaginal samples: profiles for 19 subjects (zero time, prestudy samples). The arrowheads indicate the DNA fragments sequenced from specific lanes, while in unmarked lanes only the dominant fragment was sequenced. BLAST sequence homologies are shown in Table 1.
TABLE 1.
BLAST analysis of vaginal bacterial V2-V3 16S rRNA sequences of excised fragments from DGGE gels (zero time)
| Subject | Fragment in gel | Most closely related bacterial sequence | % Identity | Accession no. |
|---|---|---|---|---|
| 250 | 1 | Gardnerella vaginalis | 98 | M58744 |
| 252 | 1 | Lactobacillus crispatus | 100 | AF257097 |
| 2 | Gardnerella vaginalis | 98 | M58744 | |
| 253 | 1 | Lactobacillus crispatus | 98 | AF257097 |
| 254 | 1 | Lactobacillus iners | 100 | Y16329 |
| 255 | 1 | Lactobacillus crispatus | 97 | AF257097 |
| 256 | 1 | Lactobacillus crispatus | 100 | AF257097 |
| 2 | Lactobacillus iners | 99 | Y16329 | |
| 257 | 1 | Lactobacillus crispatus | 98 | AF257097 |
| 2 | Lactobacillus iners | 100 | Y16329 | |
| 258 | 1 | Streptococcus agalactiae | 100 | AF015927 |
| 259 | 1 | Lactobacillus gasseri | 100 | AF243165 |
| 260 | 1 | Lactobacillus iners | 100 | Y16329 |
| 261 | 1 | Lactobacillus iners | 99 | Y16329 |
| 2 | Arthrobacter sp. | 100 | AJ243423 | |
| 3 | Gardnerella vaginalis | 99 | M58744 | |
| 262 | 1 | Lactobacillus acidophilus | 97 | AF375937 |
| 2 | Lactobacillus iners | 96 | Y16329 | |
| 263 | 1 | Lactobacillus delbrueckii | 97 | AF375917 |
| 264 | 1 | Lactobacillus iners | 92 | Y16329 |
| 2 | Gardnerella vaginalis | 98 | M58744 | |
| 265 | 1 | Lactobacillus crispatus | 98 | AF257097 |
| 266 | 1 | Lactobacillus iners | 96 | Y16329 |
| 267 | 1 | Caulobacter sp. | 98 | M83799 |
| 2 | Gardnerella vaginalis | 97 | M58744 | |
| 268 | 1 | Butyrivibrio fibrisolvens | 95 | AF125217 |
| 2 | Gardnerella vaginalis | 97 | M58744 | |
| 269 | 1 | Lactobacillus crispatus | 99 | AF257097 |
Sequence analysis indicated that Gardnerella vaginalis was present in six of the study participants at zero time; three of these women (subjects 250, 267, and 268) would have been characterized as having asymptomatic bacterial vaginosis by the Nugent criteria (9). In three of the subjects with G. vaginalis, other microorganisms not commonly found in the vagina, including Arthrobacter sp., Caulobacter sp., and Butyrivibrio fibrisolvens, were detected. G. vaginalis and Lactobacillus species were simultaneously detected in three subjects at the first sampling time (Table 1).
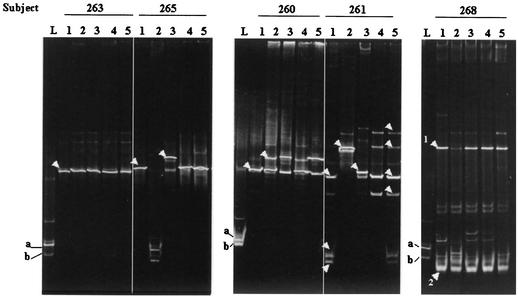
After probiotic instillation, DGGE and sequencing results showed that in five patients there was no apparent major alteration in the existing vaginal microbiota, regardless of whether one fragment (subject 263) (Fig. 2) or more DNA fragments (subjects 262, 264, 267, and 269) (data not shown) were initially detected. No changes were observed in the DGGE profile of subject 266, other than detection of the exogenous lactobacilli in the first sample after instillation. Subject 260 acquired an L. crispatus strain (100% homology with accession no. AF257097 sequence) in addition to the original L. iners strain 3 days after probiotic instillation was begun (Fig. 2).
FIG. 2.
DGGE profiles of the vaginal microbiota from five women during the study. Lanes L contained known isolates L. fermentum RC-14 (a) and L. rhamnosus GR-1 (b). Lanes 1 to 5 contained amplicons from samples taken at zero time (prestudy) and at 3, 7, 14, and 21 days after instillation of capsules containing lactobacilli, respectively. The arrowheads indicate DNA fragments that were sequenced. Presumptive identities based on closest BLAST homologies are as follows: for subject 263, lane 1, L. delbrueckii; for subject 265, lane 1, L. crispatus; for subject 265, lane 3, Pseudomonas sp.; for subject 260, lane 1, L. iners; for subject 260, lane 2, L. crispatus; for subject 261, lane 1 (from top to bottom), L. iners, Arthrobacter sp., and G. vaginalis; for subject 261, lane 2, Pseudomonas sp.; for subject 261, lane 3, S. agalactiae; for subject 261, lane 5, L. iners (all fragments); for subject 268, lane 1, B. fibrisolvens and G. vaginalis (fragments 1 and 2, respectively).
The G. vaginalis DNA fragment present in subject 261 disappeared immediately following lactobacillus treatment and was detected again only at day 21. This subject and subject 265 retained their indigenous lactobacilli (excluding day 3 data for subject 261) but also acquired a Pseudomonas strain (on days 3 and 7, respectively); subject 261 acquired a Streptococcus agalactiae strain on day 7. When other DNA fragments observed in the last two samples in the DGGE gel from subject 261 were sequenced, they were found to be homologous to L. iners and were likely to be spurious PCR artifacts (17). Therefore, if the spurious DNA fragments in subject 261 were ignored, the day 21 microbiota was the same as the microbiota prior to treatment in both subjects. In subject 268 a DNA fragment of B. fibrisolvens was present at zero time, and although the intensity of the fragment significantly decreased at day 3, the intensity was similar to the intensity in the zero-time microbiota in subsequent day 7, 14, and 21 samples tested (Fig. 2). The follow-up samples obtained from the women after 6 months showed that most women (10 of 18 women, with one woman noncompliant) had altered DGGE profiles, indicating that their bacterial microbiota had changed compared to the microbiota in the prestudy samples.
The presence of the instilled exogenous Lactobacillus species could not always be detected within the vaginal samples by PCR-DGGE. However, RAPD profiling (Table 2) detected the exogenous lactobacillus strains in 80% of the women after 1 week and in 20% of the women after 3 weeks (L. rhamnosus GR-1 only). The detection of instilled Lactobacillus strains by RAPD analysis inversely correlated with detection of G. vaginalis by DGGE and sequence analysis in samples from subject 261 (data not shown).
TABLE 2.
Detection of Lactobacillus strains by selective culturing and subsequent RAPD analysis in a group of 10 women
| Lactobacillus strain | No. of women positive on the following days after instillation:
|
|||
|---|---|---|---|---|
| 3 | 7 | 14 | 21 | |
| GR-1 | 10 | 8 | 6 | 2 |
| RC-14 | 5 | 4 | 2 | 0 |
DISCUSSION
A number of interesting findings emerged from this study. L. iners, which was not detected in other studies of the vaginal microbiota (2, 12), is clearly a common constituent of the women sampled in this study. This species does not grow on the major selective media used for isolation of Lactobacillus, including MRS and Rogosa-Sharp medium (4). This might explain the failure to detect this organism, or the organism may have been confused with members of the L. acidophilus complex (4). The potential importance of L. iners in protecting the vagina from disease and its possible use as a probiotic remain to be determined. Since most of the urogenital bacterial microbiota originates from the gastrointestinal tract (14) and while species of vaginal lactobacilli have also been detected in feces (14, 15, 18), we can only assume that this is the origin of L. iners. However, because there has been no selective medium or species-specific primers described for L. iners, this cannot be confirmed at present.
The discovery of three strains not commonly detected in the vagina is also intriguing. Arthrobacter spp. are gram-positive organisms typically isolated from soil, although some are now regarded as opportunistic pathogens, having been recovered from blood and urine (6). Caulobacter spp. are freshwater organisms, and B. fibrisolvens is a fecal organism. Although we cannot be certain of the precise origin of these organisms in the three subjects in which they were found, the findings suggest that the vaginal microbiota may also be influenced by environmental organisms, perhaps acquired through bathing and exposure to the soil.
The correlation between a healthy vaginal tract, as defined by lack of symptoms and signs of disease, and dominance of lactobacilli (9) supports the belief that these commensals play a major role in preventing certain types of vaginal infections. In the zero-time samples of three of six subjects G. vaginalis was detected in conjunction with a species of Lactobacillus. Thus, the presence of lactobacilli does not necessarily exclude potential pathogens from the vagina. The question becomes, what virulent properties or other factors result in an infection? The balance between an infectious state and a healthy state is likely a constant battle, and we speculate that this battle involves interactions between bacteria and interactions between bacteria and host defenses (11).
The immediate detection of changes in the DGGE profiles of four subjects (subjects 260, 261, 265, and 268) following lactobacillus instillation and the subsequent reversion of the profiles to the profiles of the prestudy state in three of the subjects over the course of the study suggest that these changes were probably not attributable to temporal variation of the microbiota. Pseudomonas species can be a cause of urinary tract infections (3, 13). The detection of Pseudomonas sp. in samples from subjects 261 and 265 following instillation of the probiotic might have been due to emergence of endogenous and potentially opportunistic microorganisms within the vagina at levels below the detection limit of PCR-DGGE (8, 17). Such microorganisms may become increasingly prevalent upon minor alteration of the vaginal microenvironment. Persistence of microorganisms at levels below the detection threshold of PCR-DGGE was demonstrated by culturing vaginal swabs on selective antibiotic media preferential for the supplanted Lactobacillus strains and typing isolates by RAPD analysis. For up to 21 days after the initial instillation, the exogenous strains could be detected in the samples from some women by RAPD analysis but not by PCR-DGGE. Whether probiotic microorganisms create a slight perturbation of the microbiota following which other persistent endogenous microorganisms, including lactobacilli (such as L. crispatus in subject 260), take advantage to replenish their populations has yet to be determined. However, the instillation of two probiotic strains showed that non-hydrogen-peroxide-producing L. rhamnosus GR-1 persisted longer than the L. fermentum RC-14 strain, a known H2O2 producer, emphasizing that expression of this factor alone is probably insufficient for restoration of a lactobacillus-dominant microbiota, as previously proposed (16).
The detection of instilled lactobacillus strains by RAPD analysis of cultured organisms at low levels but not by DGGE in certain samples may have been the result of the ability to plate out the entire contents of a vaginal sample on agar. PCRs for DGGE, however, rely on efficient DNA extraction and multiple cells to be present to ensure that a representative DNA molecule from each bacterial type is present in each aliquot used for a reaction. Other factors that may also influence amplification strength may include dominant DNA templates outcompeting lesser species, PCR primer bias, and rRNA operon copy numbers that are different in different microorganisms (8, 17). However, previous culture studies have failed to identify the presence of certain species, including L. iners. Our data suggest that PCR-DGGE may be a superior technique for detecting the dominant microbiota that may not be detectable by standard culture techniques. Furthermore, PCR-DGGE was a useful tool for detecting changes in the vaginal microbiota after the addition of lactobacillus strains. We suggest that the DGGE technique is a very useful adjunct for clinical studies of the vaginal tract.
Acknowledgments
We thank Dee Beuerman and Ivo Braunstein for recruiting subjects and performing bacterial culturing and DNA extraction.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Antonio, M. A., S. E. Hawes, and S. L. Hillier. 1999. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180:1950-1956. [DOI] [PubMed] [Google Scholar]
- 3.Bonadio, M., M. Meini, P. Spitaleri, and C. Gigli. 2001. Current microbiological and clinical aspects of urinary tract infections. Eur. Urol. 40:439-444. (Discussion, 40:445.) [DOI] [PubMed]
- 4.Falsen, E., C. Pascual, B. Sjoden, M. Ohlen, and M. D. Collins. 1999. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int. J. Syst. Bacteriol. 49:217-221. [DOI] [PubMed] [Google Scholar]
- 5.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funke, G., R. A. Hutson, K. A. Bernard, G. E. Pfyffer, G. Wauters, and M. D. Collins. 1996. Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J. Clin. Microbiol. 34:2356-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardiner, G., C. Heinemann, M. Baroja, A. W. Bruce, D. Beuerman, J. Madrenas, and G. Reid. 2002. Oral administration of the probiotic combination Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 for human intestinal applications. Int. Dairy J. 12:191-196. [Google Scholar]
- 8.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 9.Nugent, R. P., M. A. Krohn, and S. L. Hillier. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reid, G., D. Beuerman, C. Heinemann, and A. W. Bruce. 2001. Probiotic Lactobacillus dose required to restore and maintain a normal vaginal flora. FEMS Immunol. Med. Microbiol. 1351:1-5. [DOI] [PubMed] [Google Scholar]
- 11.Reid, G., J. Howard, and B. S. Gan. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424-428. [DOI] [PubMed] [Google Scholar]
- 12.Sobel, J. D. 1999. Biotherapeutic agents as therapy for vaginitis, p. 221-244. In G. Elmer, L. V. McFarland, and C. Surawicz (ed.), Biotherapeutic agents and infectious diseases. Humana Press, Totowa, N.J.
- 13.Takeyama, K., Y. Kunishima, M. Matsukawa, S. Takahashi, T. Hirose, N. Kobayashi, I. Kobayashi, and T. Tsukamoto. 2002. Multidrug-resistant Pseudomonas aeruginosa isolated from the urine of patients with urinary tract infection. J. Infect. Chemother. 8:59-63. [DOI] [PubMed] [Google Scholar]
- 14.Tannock, G. W. 1999. The bowel microflora: an important source of urinary tract pathogens. World J. Urol. 17:339-344. [DOI] [PubMed] [Google Scholar]
- 15.Tannock, G. W. 1995. Normal microflora: an introduction to microbes inhabiting the human body, 1st ed. Chapman & Hall, London, United Kingdom.
- 16.Vallor, A. C., M. A. Antonio, S. E. Hawes, and S. L. Hillier. 2001. Factors associated with acquisition of, or persistent colonization by, vaginal lactobacilli: role of hydrogen peroxide production. J. Infect. Dis. 184:1431-1436. [DOI] [PubMed] [Google Scholar]
- 17.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 18.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]