Abstract
A prolyl aminopeptidase (PAP) (EC 3.4.11.5) was isolated from the cell extract of Debaryomyces hansenii CECT12487. The enzyme was purified by selective fractionation with protamine and ammonium sulfate, followed by two chromatography steps, which included gel filtration and anion-exchange chromatography. The PAP was purified 248-fold, with a recovery yield of 1.4%. The enzyme was active in a broad pH range (from 5 to 9.5), with pH and temperature optima at 7.5 and 45°C. The molecular mass was estimated to be around 370 kDa. The presence of inhibitors of serine and aspartic proteases, bestatin, puromycin, reducing agents, chelating agents, and different cations did not have any effect on the enzyme activity. Only iodoacetate, p-chloromercuribenzoic acid, and Hg2+, which are inhibitors of cysteine proteases, markedly reduced the enzyme activity. The Km for proline-7-amido-4-methylcoumarin was 40 μM. The enzyme exclusively hydrolyzed N-terminal-proline-containing substrates. This is the first report on the identification and purification of this type of aminopeptidase in yeast, which may contribute to the scarce knowledge about D. hansenii proteases and their possible roles in meat fermentation.
Yeast are involved in a variety of food fermentation processes, such as baking, brewing, and cheese and sausage making. Thus, knowledge of all biochemical pathways is of great importance relation to yeast physiology and performance in these industrial processes (1). Yeast proteases are involved in numerous biological functions, such as septum formation, sporulation, protein turnover, catabolite inactivation, enzyme secretion, and nutrition (10). The proteolytic system of Saccharomyces cerevisiae is the best characterized so far. This system consists of the cytosolic proteasome, vacuolar and mitochondrial proteases, and proteases of the secretory pathway (11, 20). The major cellular proteases, such as carboxypeptidases Y and S, proteinases A and B, dipeptidyl aminopeptidase B, and aminopeptidases I and Y, are localized in the vacuole (12). These vacuolar hydrolases have been implicated in several processes that can be a consequence of adaptation to changing nutritional conditions, as may occur in the course of food fermentations (18). Among the enzymes identified in S. cerevisiae, aminopeptidases and carboxypeptidases are thought to be specially involved in the utilization of exogenous supplied peptides as nutrients (10).
Debaryomyces hansenii is the most frequent yeast species found in protein-rich fermented products, such as sausages and cheeses (2, 5, 26). The better adaptation of this species to certain ecosystems, compared to Saccharomyces, seems to be related to its high salinity tolerance and ability to grow at low temperatures. Therefore, interest in the physiology and biochemistry of D. hansenii is increasing. This species metabolizes organic acids and amino acids, regulating the acidity of the fermented product, and also provides lipolytic and proteolytic activities contributing to flavor development (2, 3, 23, 34). Proteolysis is a significant process during meat fermentation that leads to the generation of small peptides and free amino acids. These products can be important, physiologically as nutrient compounds and technologically as taste compounds or precursors of aroma compounds. Most of the studies on proteases of meat microorganisms have been carried out with lactobacilli (6) and, especially, with Lactobacillus sakei (29, 30). Nevertheless, a recent study proved the ability of D. hansenii CECT 12487, originally isolated from sausages, to hydrolyze muscle sarcoplasmic proteins (27). Thus, our present goal is to identify the specific proteases involved.
This work focused on the purification of an aminopeptidase from D. hansenii which represents a novel protease in yeasts. The characterization of the enzyme contributes to the knowledge of the proteolytic system in this species and its potential roles in meat fermentation.
MATERIALS AND METHODS
Yeast strain and growth conditions.
D. hansenii CECT 12487 was isolated from the natural microflora of a fermented sausage and selected as a possible starter culture on the basis of its physiological and biochemical properties and its ability to compete in a process of manufacturing of dry fermented sausages (28). It was routinely grown in malt extract agar or broth (Scharlau, Barcelona, Spain) at 27°C for 48 to 72 h and then stored at 4 or −80°C in 15% glycerol. For purification the microorganism was grown in 1.17% (wt/vol) Yeast Carbon Base (Difco, Detroit, Mich.) plus 0.1% (wt/vol) urea as a nitrogen source. A 120-ml portion of this medium was inoculated and incubated at 27°C for 2 days, in an orbital incubator at 110 rpm. This preculture was used to inoculate 400 ml of fresh medium, which was incubated under the same conditions for 5 days and finally used for enzyme purification.
Preparation of cell extract.
Cells were harvested at 4,080 × g for 10 min at 4°C, washed with 20 mM sodium phosphate (pH 6.5), and then resuspended in the same buffer. An equivalent volume of glass beads (0.5-mm diameter; Sigma, St. Louis, Mo.) was added to the cell suspension. Cell disruption was carried out in a Bead Beater (Biospec Products, Washington, N.C.) by four shakings for 30 s each with 2-min intervals on ice. Glass beads, nonbroken cells, and debris were separated by centrifugation (27,000 × g, 30 min, 4°C), and the supernatant obtained constituted the cell extract used for enzyme purification.
Enzyme assay.
Prolyl aminopeptidase (PAP) was measured by adding 50 μl of enzyme to 250 μl of McIlvaine buffer (100 mM citric acid, 200 mM disodium phosphate [pH 7.5]) containing 0.12 mM l-proline-7-amido-4-methylcoumarin (Pro-AMC; Sigma). The reaction mixture was incubated at 37°C for 1 min. Fluorescence was measured in a multiscan fluorometer (Fluoroskan II; Labsystem, Oy, Finland), using excitation and emission wavelengths of 355 and 460 nm, respectively. Three replicates were measured for each experimental point. One unit of enzyme activity was defined as the release of 1 μmol of substrate hydrolyzed per h at 37°C.
Enzyme purification. (i) Protamine sulfate fractionation.
Protamine sulfate at concentration of 4 mg/g of protein was slowly added to the cell extract with stirring at 5°C for 20 min. Afterwards, the solution was centrifuged (14,500 × g for 11 min), and then protamine sulfate at 100 mg/g of protein was added to the new supernatant as described above. The solution was centrifuged (27,000 × g for 11 min), and the pellet was finally resuspended in 0.2 M sodium phosphate, pH 7.0. After 5 min of resting, 3.5 μl of 1% (wt/vol) salmon DNA per mg of protein was added. The solution was then centrifuged (27,000 × g for 10 min), and the supernatant was subjected to the following purification steps.
(ii) Ammonium sulfate fractionation.
The supernatant was precipitated with ammonium sulfate at 60% saturation. After centrifugation (27,000 × g at 5°C for 20 min), the resultant pellet was redissolved in a minimum volume of 25 mM Tris-HCl (pH 7.5), containing 0.1 M NaCl.
(iii) Gel filtration chromatography.
The redissolved pellet was injected onto a 70- by 1.6-cm Sephacryl S-300 HR column (Pharmacia, Uppsala, Sweden) previously equilibrated with 25 mM Tris-HCl (pH 7.5) containing 0.1 M NaCl. The column was run at a flow rate of 21 ml/h. Fractions of 4.9 ml were collected and assayed for aminopeptidase activity. The two fractions containing the maximum activity against Pro-AMC were pooled and subjected to the following purification step.
(iv) Anion-exchange chromatography.
The pooled fractions were injected into a Resource Q column (1 ml; Amersham Pharmacia Biotech AB, Uppsala, Sweden). Proteins were eluted with an initial isocratic period in 25 mM Tris-HCl (pH 7.5) containing 280 mM NaCl, followed by a linear gradient of 280 to 350 mM NaCl over 25 min. The flow rate was 1 ml/min, and fractions of 1 ml were collected.
Determination of protein concentration.
The protein concentration was determined by the bicinchoninic acid method (33) with the bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.). Bovine serum albumin was used as a standard. The eluted fractions from the chromatographic separations were also monitored at 280 nm.
Electrophoresis.
The purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), using 10% separating gels (19). Proteins were stained with Coomassie brilliant blue R-250 and silver. Broad-range molecular mass standards (Bio-Rad, Hercules, Calif.) were simultaneously run.
Molecular mass determination.
The molecular mass of the native enzyme was estimated by gel filtration with a Sephacryl S-300 column (Pharmacia) as previously described. The column was calibrated by using the standard proteins ferritin (450 kDa), β-amylase (200 kDa), bovine serum albumin (68 kDa), and cytochrome c (12.4 kDa). Dextran blue was used to estimate the void volume. The molecular mass of the enzyme under denaturing conditions was also determined by SDS-PAGE as described above.
Effect of pH and temperature.
The PAP activity was assayed against Pro-AMC in the pH range from 3 to 11, at intervals of 0.5 pH unit, using the following buffers: from pH 3.0 to 8.0, McIlvaine buffer (100 mM citric acid, 0.2 M disodium phosphate); from pH 8.0 to 10.0, Clark and Lub's borate buffer (100 mM boric acid in 100 mM KCl and 0.1 N NaOH); and from pH 10.0 to 11.0, Sorensen's glycine II buffer (100 mM glycine in 0.1 N NaCl and 0.1 N NaOH). The results were expressed as the percentage of the activity obtained at the optimum pH.
The effect of temperature was determined in the range from 5 to 55°C. The substrate solution (250 μl) was previously equilibrated at each temperature, and then the reaction was initiated by the addition of the purified enzyme (50 μl). After incubation, the reaction was stopped by addition of 100 μl of 0.6 M acetic acid. The results were expressed as the percentage of the activity obtained at the optimum temperature.
Analysis of potential enzymatic inhibitors.
The activity of the purified enzyme was assayed in the presence of different chemical agents to identify possible inhibitors or activators by the standard procedure. Leupeptin, puromycin, bestatin, trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane (E-64), and pepstatin A were assayed at 50 to 500 μM; iodoacetate, 3,4-dichloroisocoumarin (3,4-DCI), phenylmethylsulfonyl fluoride (PMSF), 4-(2-amino-ethyl)-benzosulfonylfluoride hydrochloride (Pefabloc SC), and p-chloromercuribenzoic acid were assayed at 0.1 to 1 mM; the chelating agents EDTA, EGTA, and phenanthroline were assayed at 1 to 5 mM, 1 to 5 mM, and 0.1 to 1 mM, respectively; and the reducing agents dithiothreitol and 2-mercaptoethanol were assayed at 1 to 5 mM. The effects of the divalent cations CaCl2, MnCl2, CoCl2, CuCl2, CdCl2, HgCl2, and MgCl2 were determined at 50 to 500 μM.
All reagents were purchased from Sigma, except for Pefabloc SC, which was from Merck (Darmstadt, Germany), and metal salts, which were from Panreac (Barcelona, Spain).
Determination of kinetic parameters.
The kinetic parameters of the purified enzyme were estimated for Pro-AMC, using concentrations ranging from 5 to 200 μM. Activity was continuously measured at 37°C as described above, and kinetic parameters were calculated from Lineweaver-Burk plots.
Substrate specificity.
The activity towards different fluorimetric substrates at 100 μM was tested by the standard activity assay.
The activity was also assayed against different peptides. The reaction mixture consisted of 50 μl McIlvaine's buffer (100 mM citric acid, 200 mM disodium phosphate [pH 7.5]), 25 μl of 10 mM peptide solution, and 25 μl of purified enzyme (samples) or water (control). Two independent test and control samples were assayed for each peptide. The reaction was stopped by adding 35 μl of 0.6 M acetic acid after 45 min of incubation at 37°C. The relative activity was determined by measuring the disappearance of substrate by high-performance liquid chromatography (HPLC) on a Hewlett-Packard 1050 HPLC system (Agilent Technologies, Palo Alto, Calif.). The hydrophobic peptides were analyzed by reverse-phase HPLC in a Symmetry C18 (4.6 by 250 mm) column (Waters Corporation, Milford, Mass.). The following solvents were used: 0.1% (vol/vol) trifluoroacetic acid in MilliQ water (solvent A) and acetonitrile-MilliQ water-trifluoroacetic acid (60:40:0.085, vol/vol) (solvent B). The hydrophobic peptides were eluted at a flow rate of 0.9 ml/min and different concentrations of solvent B: Val-Val at 20%; Pro-Leu, Leu-Pro, and Pro-Phe-Gly-Lys at 25%; Pro-Pro-Gly-Phe-Ser-Pro (Bradykinin, fragment 2-7) at 30%; Pro-Phe at 32%; and Leu-Leu at 38%. The separations were performed at 40°C. The hydrolysis of hydrophilic peptides was analyzed by cation-exchange HPLC in a Spherisorb SCX (25- by 0.46-cm) column (Tracer analítica; Teknokroma, Barcelona, Spain). The following solvents were used: 20% acetonitrile in 6 mM HCl (solvent A) and 20% acetonitrile-1 M NaCl in 6 mM HCl (solvent B). Lys-Lys was eluted at 55% solvent B. The rest of the hydrophilic peptides, i.e., Ala-Ala-Ala, Glu-Glu, Pro-Gly, and Gly-Pro, were eluted by applying a linear gradient of 10 to 45% of solvent B for 10 min, followed by an isocratic period at 45% solvent B for 2 min. The separations were carried out at flow rate of 0.9 ml/min and 40°C.
RESULTS
Purification of the PAP.
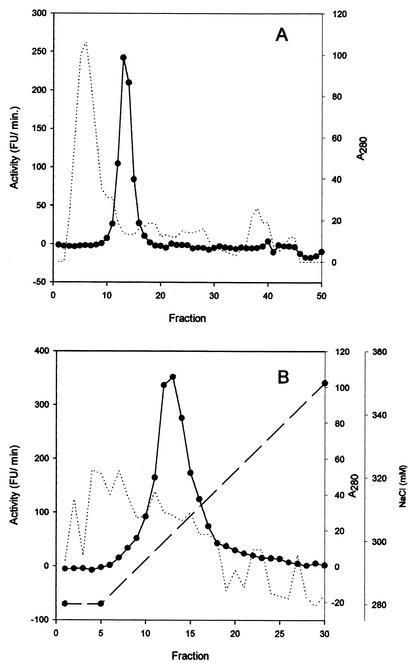
The results of the purification of the PAP from the cell extract of D. hansenii are summarized in the Table 1. The first protamine precipitation resulted in a moderate protein reduction and elimination of DNA (data not shown). In the second step, the addition of a higher concentration of protamine to the obtained supernatant allowed the precipitation of PAP and an increase in specific activity. The pellet was resuspended, and the protamine was eliminated due to the binding of this compound to the added DNA. The sample obtained was further concentrated by ammonium sulfate precipitation. The gel filtration chromatography step resulted in an important specific activity enrichment (Table 1). The final step was carried out by strong anion-exchange chromatography using a narrow gradient (from 280 to 350 mM in 25 min). The enzyme eluted at 300 mM (Fig. 1). The yield from the whole purification process was 1.4% and resulted in an increase in specific activity of 247.8-fold. Once purified, PAP was stable for at least 3 months at 4°C.
TABLE 1.
Purification of PAP from D. hansenii CECT 12487
| Purification step | Protein (mg) | Total activity (U)a | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 416.250 | 106.5 | 0.3 | 100.4 | 1 |
| Supernatant from first protamine precipitation | 345.736 | 115.5 | 0.3 | 108.4 | 1.3 |
| Resuspended pellet from second protamine precipitation | 24.616 | 95.8 | 3.9 | 90.0 | 15.2 |
| Gel filtration | 0.426 | 9.2 | 21.7 | 8.6 | 84.6 |
| Strong anion exchange | 0.034 | 1.5 | 63.4 | 1.4 | 247.8 |
1 U = 1 μmol of released AMC per h at 37°C.
FIG. 1.
Chromatograms from different steps in the purification of PAP from D. hansenii. (A) Gel filtration in a Sephacryl S-300 column. (B) Strong anion-exchange chromatography in a Resource-Q column. Protein was detected by measuring the absorbance at 280 nm (dotted lines), aminopeptidase activity is expressed in fluorescence units (FU) per minute (solid lines), and the NaCl gradient is indicated (dashed line).
Molecular mass and purity.
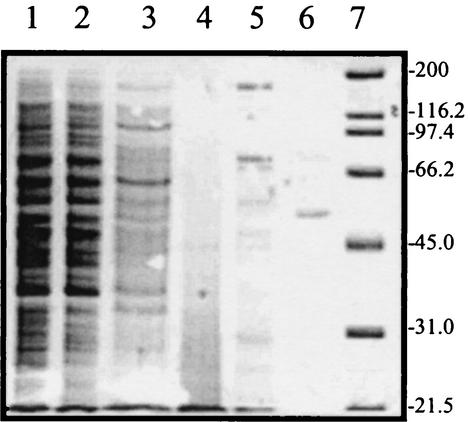
The molecular mass of native PAP calculated by gel filtration was 370 kDa. The SDS-PAGE analysis showed a single band of approximately 53.5 kDa, indicating that it is a multimeric enzyme that seems to consist of seven similar subunits.
Enzymatic characterization of PAP.
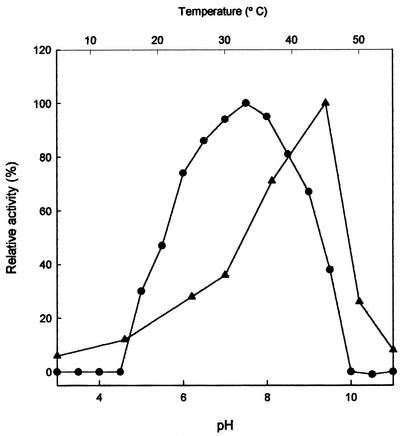
The enzyme was active in a broad range from pH 5 to 9.5, with an optimum at pH 7.5 (Fig. 2). The maximum activity was found to be at 45°C. The activity sharply decreased above the optimum (Fig. 2).
FIG. 2.
Effects of pH (•) and temperature (▴) on PAP activity from D. hansenii.
The effects of potential inhibitors on PAP activity are shown in Table 2. Puromycin and bestatin, which are typical inhibitors of aminopeptidases, did not cause any effect on PAP activity. PAP activity also was not affected by cysteine protease inhibitors (leupeptin and E-64) or serine proteases inhibitors (3,4-DCI, PMSF, and Pentabloc SC) (Table 2) or by chelating agents (EDTA, EGTA, and 1,10-phenanthroline) or reducing agents (dithiothreitol and β-mercaptoethanol) (data not shown). The aspartic protease inhibitor pesptatin A exerted a slight inhibition (15%) at the highest concentration. Only sulfhydryl group reagents, i.e., iodoacetate and p-chloromercuribenzoic acid, caused a significant effect, reducing the optimal activity to 2.6 and 61%, respectively (Table 2). In relation to the effects of different divalent cations, only concentrations of the assayed metal salts of 500 μM resulted in a weak increase (10%) in activity (data not shown). The exception was Hg2+, which dramatically reduced the activity to less than 10% (data not shown).
TABLE 2.
Effects of different inhibitors on the purified PAP
| Chemical | Relative activitya at the following concn (mM):
|
|||
|---|---|---|---|---|
| 0.05 | 0.5 | 0.1 | 1 | |
| Leupeptin | 101 | 116 | —b | — |
| Puromycin | 115 | 109 | — | — |
| Bestatin | 113 | 116 | — | — |
| E-64 | 118 | 111 | — | — |
| Pepstatin A | 101 | 84 | — | — |
| Iodoacetate | — | — | 95 | 2.6 |
| 3,4-DCI | — | — | 103 | 111 |
| PMSF | — | — | 99 | 100 |
| Pentabloc SC | — | — | 105 | 105 |
| p-Chloromercuribenzoic acid | — | — | 101 | 61 |
Expressed as a percentage of the activity obtained in the absence of any added chemical agent, which was given a value of 100%.
—, not determined.
The Vmax and Km values for Pro-AMC were 1 μmol min−1 mg−1 and 40 μM, respectively. The relative activity of PAP was assayed against synthetic substrates and peptides (Table 3). PAP displayed aminopeptidase activity exclusively on Pro-AMC, while it did not hydrolyze other substrates of amino-, dipeptidyl-, or tripeptidylpeptidases. The specificity against natural peptides was in accordance with that observed against synthetic substrates. Only peptides containing proline at the N terminus were hydrolyzed. PAP did not show carboxypeptidase activity, since it could hydrolyze Pro-Gly and Pro-Leu but not Gly-Pro and Leu-Pro (Table 3). The enzyme was able to hydrolyze Pro-Pro bonds and oligopeptides of at least six amino acid residues in length (Pro-Pro-Gly-Phe-Ser-Pro). Presumably, PAP showed a preference for peptides longer than two residues, since Pro-Phe-Gly-Lys was hydrolyzed at a higher rate than Pro-Phe (Table 3). The nature of the amino acid located at the second position had also an effect on hydrolysis rates. The hydrolysis of Pro-Leu was considerably higher than that of Pro-Phe and Pro-Gly (Table 3).
TABLE 3.
Relative activities of the purified PAP on different synthetic AMCs and peptides as substrates
| Substrate | Relative activitya |
|---|---|
| Synthetic AMCs | |
| Pro-AMC | 100 |
| pGlu-AMC | 0 |
| Ala-AMC | 0 |
| Arg-AMC | 0 |
| Leu-AMC | 0 |
| Met-AMC | 0 |
| Tyr-AMC | 0 |
| Gly-AMC | 0 |
| Val-AMC | 0 |
| Phe-AMC | 0 |
| Ser-AMC | 0 |
| Glu-AMC | 0 |
| Pro-Arg-AMC | 0 |
| Gly-Ala-AMC | 0 |
| Ala-Arg-AMC | 0 |
| Gly-Arg-AMC | 0 |
| Lys-Ala-AMC | 0 |
| Arg-Arg-AMC | 0 |
| Ala-Ala-Phe-AMC | 0 |
| Peptides | |
| Pro-Phe-Gly-Lys | 100 |
| Pro-Leu | 97 |
| Pro-Phe | 66 |
| Pro-Gly | 62 |
| Leu-Pro | 0 |
| Leu-Leu | 0 |
| Val-Val | 0 |
| Gly-Pro | 0 |
| Lys-Lys | 0 |
| Glu-Glu | 0 |
| Ala-Ala-Ala | 0 |
Expressed as a percentage of the activity against Pro-AMC or Pro-Phe-Gly-Lys, which was given a value of 100%.
DISCUSSION
A PAP has not been previously described for yeast. Thus, this work constitutes the first evidence of the presence of such a specific enzyme in this microbial group, and it is the only protease purified from D. hansenii. The purification was initiated by protamine fractionation, which resulted in a considerable increase (13-fold) in specific activity due to a drastic reduction in the contaminant proteins (Fig. 3). This purification step was based on the reversible binding of protamine sulfate to PAP. Typical protamines are polypeptides of 5,000 to 6,000 Da that are very rich in arginine residues, which can represent about 60 to 70 mol% of the molecule (31). Further, the binding between the protamine and PAP is released by addition of a high-ionic-strength buffer, and then the protamine can be eliminated by addition of salmon DNA, for which protamine shows higher affinity. PAP eluted at the tail of the large peak that appears at the beginning of the chromatogram of the gel filtration step (Fig. 1), together with other proteins and possibly DNA fragments. This step also resulted in a high purification which was completed after a strong anion-exchange chromatography.
FIG. 3.
SDS-PAGE of different purification steps of PAP from D. hansenii. Lanes: 1, cell extract; 2, supernatant from first protamine precipitation; 3, resuspended pellet from second protamine precipitation; 4, protamine; 5, active samples from gel filtration; 6, purified protein from anion-exchange separation; 7, standard proteins. Numbers on the right are molecular masses in kilodaltons.
The estimated molecular mass (370 kDa) of the native enzyme of D. hansenii is close to those (400 to 270 kDa) determined for enzymes of eukaryotes (7, 14, 21, 35). The enzymes from prokaryotes have lower molecular mass, varying between 150 and 30 kDa (8). The multimeric structure of the enzyme appeared to be a common characteristic of these aminopeptidases in most eukaryote and prokaryote cells. The molecular masses of other PAPs in the denatured state have been found to be 55 to 56 kDa (14, 21, 22), and these are described as hexamers or tetramers. On the other hand, the molecular masses of the denatured PAPs of prokaryotes have been found to be from 34 to 63 kDa, and these have been described as trimers (8), dimers (4), or monomers (36).
The activity of PAP was optimal at 45°C and pH 7.5. Most of the purified PAPs have optimal pHs of between 7 and 8 (7, 14, 16, 32, 35). The optimum temperature of the majority of PAPs is between 37 and 55°C (17, 32). The PAP of Penicillium camemberti has optimal activity at 45°C, as is the case for the PAP of D. hansenii (7).
On the basis of studies with various inhibitors, PAP of D. hansenii seems to be a cysteine protease, as initially described for this enzyme from other origins (14, 35, 36). PAP of P. camemberti is inhibited by thiol reagents but also exhibits inhibition by di-isopropylfluorophosphate, indicating that serine residues are important for the catalytic activity (7). In contrast, PAPs from lactic acid bacteria such as Propionibacterium shermanii and Lactobacillus delbrueckii and from Arthrobacter nicotianae and Hafnia alvei are considered serine proteases on the basis of 3,4-DCI inhibition (8, 16, 25, 32). Studies by site-directed mutagenesis in Bacillus coagulans and Aeromonas sobria also demonstrated that the enzyme from prokaryotes is indeed a serine protease (15).
The unique structure of proline in polypeptide chains restricts their susceptibility to the action of most proteases, and the activity of specific enzymes is required to avoid accumulation of proline-containing polypeptides. There are two major groups of specialized enzymes that are able to cleave peptide bounds involving proline: (i) aminopeptidase P and prolidase, which cleave X-Pro bonds from oligopeptides and dipeptides, respectively, and are both metalloenzymes, and (ii) prolyl oligopeptidase, prolinase, prolyl carboxypeptidase, and prolyl aminopeptidase, which cleave Pro-X bonds and are either serine (8, 15, 17, 25, 32) or cysteine (7, 14, 35, 36) proteases. The PAP of D. hansenii releases proline from the N-terminal position of peptides of at least six amino acid residues (i.e., Pro-Pro-Gly-Phe-Ser-Pro) and shows higher preference for oligopeptides than for dipeptides. Therefore, it is not a prolinase, because can hydrolyze peptides with more than two residues. The Km for Pro-AMC was estimated to be 40 μM. The Km values found in the literature for the substrate Pro-2-naphthylamide vary from 10 to 152 μM (35, 36), and those for Pro-p-nitroanilide vary from 250 to 320 μM (7, 36).
Despite the fact that D. hansenii can generally synthesize proline from arginine, the newly purified PAP could be involved in the release of this free amino acid, which could be used as nutrient, by acting at the N termini of peptides resulting from muscle protein hydrolysis (27). The activity of this specialized enzyme can also be important by allowing the subsequent action of other peptidases and the progress of the proteolytic chain.
The bitterness of peptides appears to be closely related to the content of certain hydrophobic amino acids, such as leucine, isoleucine, and proline, which were high in some bitter peptide fractions isolated from cheese (13) and sausage (9). Thus, the activity of PAP of D. hansenii could contribute to reduce the bitter taste by degrading proline-containing peptides once they have been transported inside the cell or after the release of the intracellular enzymes to the meat matrix by cell lysis, as occurs in beer (24).
In summary, this study provides valuable biochemical data about the properties of the PAP of D. hansenii that could constitute the basis for further studies focused on its genetic and functional characterization and which will complete the present classification of yeast proteases.
Acknowledgments
This work has been supported by grant AGL2001-1141 from CICYT (Spain). An FPU/MEC scholarship to Tomás Bolumar is fully acknowledged.
REFERENCES
- 1.Brejning, J., and L. Jespersen. 2002. Protein expression during lag phase and growth initiation in Saccharomyces cerevisiae. Int. J. Food Microbiol. 75:27-38. [DOI] [PubMed] [Google Scholar]
- 2.Cook, P. E. 1995. Fungal ripened meats and meat products, p. 110-129. In G. Campbell-Platt and P. E. Cook (ed.), Fermented meats. Chapman & Hall, London, United Kingdom.
- 3.Durá, A., M. Flores, and F. Toldrá. 2002. Purification and characterization of a glutaminase from Debaryomyces spp. Int. J. Food Microbiol. 76:117-126. [DOI] [PubMed] [Google Scholar]
- 4.Ehrenfreud, P., C. Mollay, and G. Kreil. 1992. Purification and properties of an iminopeptidase from culture media of Streptomyces plicatus. Biochem. Biophys. Res. Commun. 184:1250-1255. [DOI] [PubMed] [Google Scholar]
- 5.Encinas, J.-P., T.-M. Lopez-Díaz, M.-L. García-Lopez, A. Otero, and B. Moreno. 2000. Yeast populations on Spanish fermented sausages. Meat Sci. 54:203-208. [DOI] [PubMed] [Google Scholar]
- 6.Fadda, S., Y. Sanz, G. Vignolo, M-C. Aristoy, G. Oliver, and F. Toldrá. 1999. Hydrolysis of pork muscle sarcoplasmic proteins by Lactobacillus curvatus and Lactobacillus sake. Appl. Environ. Microbiol. 65:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuke, Y., and H. Matsuoka. 1993. The purification and characterization of prolyl aminopeptidase from Penicillium camemberti. J. Dairy Sci. 76:2478-2484. [Google Scholar]
- 8.Gilbert, C., D. Atlan, B. Blanc, and R. Portalier. 1994. Proline iminopeptidase from Lactobacillus delbrueckii subsp. Bulgaricus CNRZ 397: purification and characterization. Microbiology 140:537-542. [DOI] [PubMed] [Google Scholar]
- 9.Henriksen, A. P., and L. H. Stahnke. 1997. Sensory and chromatographic evaluations of water soluble fractions from dried sausages. J. Agric. Food Chem. 45:2679-2684. [Google Scholar]
- 10.Jones, E. W. 1983. Genetic approaches to the study of protease function and proteolysis in Saccharomyces cerevisiae, p. 165-203. In J. F. T. Spencer, D. M. Spencer, and A. R. W. Smith (ed.), Yeast genetics. Fundamental and applied aspects, Springer-Verlag, New York, N.Y.
- 11.Jones, E. W. 1991. Three proteolytic systems of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 266:7963-7966. [PubMed] [Google Scholar]
- 12.Jones, E. W., G. C. Webb, and M. A. Hiller. 1997. Biogenesis and function of the yeast vacuole, p. 363-470. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), Molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Kai-Ping, D. L., and J. J. Warthesen. 1996. Preparative methods of isolating peptides from Cheddar cheese. J. Agric. Food Chem. 44:1058-1063. [Google Scholar]
- 14.Khilji, M. A., and G. S. Bailey. 1978. The purification of a bovine kidney enzyme which cleaves melanocyte-stimulating hormone-release inhibiting factor. Biochim. Biophys Acta 527:282-288. [DOI] [PubMed] [Google Scholar]
- 15.Kitazono, A., K. Ito, and T. Yoshimoto. 1994. Prolyl aminopeptidase is not a sulphydryl enzyme: identification of the active serine residue by site-directed mutagenesis. J. Biochem. 116:943-945. [DOI] [PubMed] [Google Scholar]
- 16.Kitazono, A., A. Kitano, T. Kabashima, K. Ito, and T. Yoshimoto. 1996. Prolyl aminopeptidase is also present in Enterobacteriaceae: cloning and sequencing of the Hafnia alvei enzyme-gene and characterization of the expressed enzyme. J. Biochem. 119:468-474. [DOI] [PubMed] [Google Scholar]
- 17.Kitazono, A., T. Kabashima, H.-S. Huang, K. Ito, and T. Yoshimoto. 1996. Prolyl aminopeptidase gene from Flavobacterium meningosepticum: cloning, purification of the expressed enzyme, and analysis of its sequence. Arch. Biochem. Biophys. 336:35-41. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Leonhard, K., B. Guiard, G. Pallecchia, A. Tzagoloff, W. Neupert, and T. Langer. 2000. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5:629-638. [DOI] [PubMed] [Google Scholar]
- 21.Matsushima, M., T. Takahashi, M. Ichinose, K. Miki, K. Kurokawa, and K. Takahashi. 1991. Prolyl aminopeptidases from pig intestinal mucosa and human liver: purification, characterization and possible identity with leucyl aminopeptidase. Biomed. Res. 12:323-333. [Google Scholar]
- 22.Ninomiya, K., K. Kawatani, S. Tanaka, S. Kawata, and S. Makusumi. 1982. Purification and properties of a proline iminopeptidase from apricot seeds. J. Biochem. 92:413-421. [DOI] [PubMed] [Google Scholar]
- 23.Olensen, P.-T., and L.-H. Stahnke. 2000. The influence of Debaryomyces hansenii and Candida utilis on the aroma formation in garlic spiced fermented sausages and model minces. Meat Sci. 56:357-368. [DOI] [PubMed] [Google Scholar]
- 24.Ormrod, I. H. L., E. F. Lalor, and F. R. Sharpe. 1991. Release of yeast proteolytic enzymes into beer. J. Inst. Brew. 97:441-443. [Google Scholar]
- 25.Panon, G. 1990. Purification and characterization of a proline iminopeptidase from Propionibacterium shermanii 13673. Lait 70:439-452. [Google Scholar]
- 26.Petersen, K. M., S. Westall, and L. Jespersen. 2002. Microbial succession of Debaryomyces hansenii strains during the production of Danish surface-ripened cheeses. J. Dairy Sci. 85:478-486. [DOI] [PubMed] [Google Scholar]
- 27.Santos, N., R.-C. Santos-Mendoça, Y. Sanz, T. Bolumar, M.-C. Aristoy, and F. Toldrá. 2001. Hydrolysis of pork muscle sarcoplasmic proteins by Debaryomyces hansenii. Int. J. Food Microbiol. 68:199-206. [DOI] [PubMed] [Google Scholar]
- 28.Santos-Mendoza, R. C. 2000. Ph.D. thesis. Aislamiento, selección y caracterización de levaduras de embutidos con vistas a su utilización como coadyuvante en el proceso de curado. Universidad de Valencia, Valencia, Spain.
- 29.Sanz, Y., and F. Toldrá. 2001. Purification and characterization of an X-prolyl-dipeptidyl peptidase from Lactobacillus sakei. Appl. Environ. Microbiol. 67:1815-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanz, Y., and F. Toldrá. 2002. Purification and characterization of an arginine aminopeptidase from Lactobacillus sakei. Appl. Environ. Microbiol. 68:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saperas, N., C. Buesa, J. Abián, J. Vanderkerckhove, H. E. Kasinsky, and M. Chiva. 1996. The primary structure of cholrichthyan protamine: a new apparent contradiction in protamine evolution. J. Mol. Evol. 43:528-535. [DOI] [PubMed] [Google Scholar]
- 32.Smacchi, E., M. Gobbetti, R. Lanciatti, and P. F. Fox. 1999. Purification and characterization of an extracellular proline iminopeptidase from Arthrobacter nicotianae 9458. FEMS Microbiol. Lett. 178:191-197. [DOI] [PubMed] [Google Scholar]
- 33.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Garthner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 34.Sorensen, B. B., and H. Samuelsen. 1996. The combined effects of environmental conditions on lipolysis of pork fat of the meat starter culture organism Staphylococcus xylosus and Debaryomyces hansenii. Int. J. Food Microbiol. 32:59-71. [DOI] [PubMed] [Google Scholar]
- 35.Waters, S. P., and M. J. Dalling. 1983. Purification and characterization of an iminopeptidase from primary leaf of wheat (Triticum aestivum L.). Plant Physiol. 73:1048-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimoto, T., T. Saeki, and D. Tsuru. 1983. Proline iminopeptidase from Bacillus megaterium: purification and characterization. J. Biochem. 93:469-477. [DOI] [PubMed] [Google Scholar]