Abstract
Enterococci are one of the major facultative anaerobic bacterial groups that reside in the human gastrointestinal tract. In the present study, the composition of the enterococcal fecal flora in three healthy humans was analyzed before, during, and after the daily consumption of ∼125 g of a raw-milk Cheddar-type cheese containing 3.2 × 104 enterococci/g of cheese. Enterococcal counts ranged between 1.4 × 102 and 2.5 × 108 CFU/g of feces and differed from subject to subject and from week to week. The cheese contained mainly Enterococcus casseliflavus and a small population of Enterococcus faecalis. Clonal relationships were determined by pulsed-field gel electrophoresis. Before and after consumption of the cheese, samples from humans contained mainly Enterococcus faecium, with some of the clones being resident. During consumption of the cheese, one particular transient clone of E. faecalis, clone Fs2, which was present in small numbers in the cheese, largely dominated the feces. Two clones of E. casseliflavus from the cheese were also found in the feces of one of the subjects during cheese consumption. These results suggest that a clone need not be present in a food in high numbers to establish itself in the intestine.
The human gastrointestinal tract (GIT) harbors a complex bacterial ecosystem. Up to 1014 bacteria may be present (1, 8), comprising 400 to 500 species, although generally only a few predominate (22). The major bacterial groups are normally stable (19). Studies in which known strains were ingested showed that certain strains are detected in feces constantly and over a long period, while others are found only occasionally (1, 41). Sears et al. (37) called the persistent strains “resident” and the strains found occasionally “transient.”
The human intestinal flora is a complex network of mutual and/or antagonistic interactions. To establish in the intestine, bacteria must either adhere to the mucosa to avoid being swept away by peristalsis (1) or multiply at a rate exceeding their rate of elimination (22); bacteria must also compete for nutrients, growth factors, and binding sites and confront colonization resistance from already established bacteria (1) which generate an environment that is inhibitory towards potential competitors. Inhibitory environments of this kind can be generated by changes in pH and oxidation-reduction potential and by the production of H2S and volatile fatty acids (8). Recent studies have monitored the passage of lactobacilli and bifidobacteria through the GIT (2, 21, 38).
Enterococci are the predominant gram-positive cocci in human stools at 105 to 108 CFU/g of feces (20, 22), and Enterococcus faecalis is the most common Enterococcus species found (7, 24, 34). Franz et al. (10) and Murray (29) suggest that the presence of Enterococcus faecium and E. faecalis in humans is dependent on geographical location.
Enterococci are the cause of a variety of infections, including endocarditis and neonatal, central nervous system, and respiratory tract infections (18). They may also infect the abdomen biliary tract, burn wounds, soft tissues, paranasal sinuses, and ear, eye, and periodontal tissue (20). Despite these involvements, the study of these organisms at the strain level is limited. Of major concern are the sources of nosocomial infections. Many of these have been identified, but a high percentage remains obscure in origin and some presumably originate from the GIT. It is believed that enterococci exit the epithelial cells or migrate in phagocytes and spread in a hematogenous manner to distant sites (20). That enterococci have become the focus of attention is due also to their increasing resistance to antibiotics. Not only are they resistant to vancomycin, they are also resistant to teicoplanin, penicillins, and aminoglycosides. In addition, vancomycin-dependent enterococci have also been reported (9). The presence of vancomycin-resistant enterococci in hospitals is met with considerable apprehension (10, 14, 27, 28). A major issue of concern is the transfer of antibiotic resistance from enterococci to more-virulent pathogens such as multiple-drug-resistant staphylococci (28). Vancomycin-resistant enterococci have also been isolated from food (10, 14, 17, 32). From this point of view, the statement by Garg and Mital (14)—“a food should be free not only from disease-producing organisms, but also from those that have the potential of causing disease”—seems legitimate.
Nevertheless, enterococci are used as silage inoculants (36), starter cultures (17), and probiotics (10, 12), possess antilisterial activities (6, 31), produce various metabolic compounds that can interfere with the growth of undesirable bacteria (8), and have a beneficial role in ripening and flavor development of cheese (3, 23, 30). Finally, enterococci from dairy products show higher sensitivity to antibiotics and have had a long history of safe use (17).
In a previous study (16), the enterococcal flora of a raw-milk, farmhouse cheese was compared with the microflora of human and bovine feces. The cheese and the human feces contained two dominant strains of Enterococcus casseliflavus and one of E. faecalis and lower numbers of other clones from the two species. The cows were not the source of enterococci in the cheese, but the presence of both E. casseliflavus clones in the milking equipment suggested that contamination of the milk starts there. The presence of identical clones of enterococci in human feces and cheese was presumed to be explained by consumption of milk and cheese by the humans, but this was not proven.
In the present paper we describe the results of a study in which three healthy human subjects consumed cheese containing enterococci (15, 16). The purpose of our investigation was to determine the impact that consumption of cheese containing enterococci had on the enterococcal flora of the feces. Valuable information was also obtained about the regular enterococcal flora.
MATERIALS AND METHODS
Collection of samples.
Four healthy Belgian subjects (two males and two females) who were between 26 and 52 years old participated in this study. Fecal samples were collected weekly in sterile containers by the volunteers over a 10- to 12-week period except in week 4. The control period was the first 3 weeks, after which three of the four volunteers began to consume between 100 and 150 g of an Irish farmhouse raw-milk Cheddar-type cheese (15) daily, giving a daily dose of 3.2 × 106 to 4.8 × 106 enterococci for 4 weeks. The feces were not sampled during the first week of cheese consumption. Weekly fecal samples were also collected for 3 weeks after cheese consumption had stopped. The fourth volunteer was used as a control. During this period the volunteers maintained their lifestyles and their usual diets, which included other cheeses in some cases. None of the subjects was administered antibiotics during the study period. When a weekly fecal sample was not obtained for any reason, the experiment was prolonged by 1 week.
Isolation of strains.
The cheese was emulsified at a 1:10 dilution in a 2% (wt/vol) trisodium citrate solution (pH 8.75), homogenized with the aid of a stomacher apparatus, and plated on kanamycin esculin azide (KAA; Merck, Darmstadt, Germany) agar.
Fecal samples (3 to 6 g) were immediately diluted 1:10 in sterile peptone saline solution (0.1% peptone [Oxoid, Basingstoke, England], 8.5% NaCl [Merck]), emulsified in a stomacher apparatus, diluted in peptone saline solution, and plated on KAA agar.
All plates were incubated overnight at 37°C under microaerophilic conditions (5% O2, 10% CO2, 85% N2). A total of 480 colonies were picked from the plates of cheese showing over 100 colonies. Ten colonies were randomly picked from the highest dilution of each fecal sample. All colonies were purified twice on KAA and once on BM agar (2% tryptose [Oxoid], 0.5% NaCl [Merck], 0.5% yeast extract [Merck], 0.5% glucose [Merck], pH 6.85). Cultures were maintained at −20°C in a 1:2 glycerol-BM broth mixture. Isolates from stock were streaked on BM agar plates.
Strain typing by pulsed-field gel electrophoresis (PFGE).
As the running conditions for PFGE are different for the yellow-pigmented E. casseliflavus from those for other enterococci, all isolates were first distinguished by colony color when grown on BM agar. The yellow-pigmented colonies were assumed to represent E. casseliflavus, as determined in previous studies (15, 16), while the white colonies represented E. faecalis, E. faecium, Enterococcus hirae, and other enterococci commonly present in feces.
The strains were grown overnight at 37°C on BM agar (2% tryptose [Oxoid], 0.5% NaCl [Merck], 0.5% yeast extract [Merck], 0.5% glucose [Merck], pH 6.85). All reagents were obtained from Sigma (St. Louis, Mo.) unless otherwise stated. One loopful of cells from an overnight culture was washed three times in 1 ml of EET buffer (100 mM EDTA, 10 mM EGTA, 10 mM Tris-HCl [pH 8.0]). After centrifugation, the pellet was resuspended in EC buffer (6 mM Tris-HCl [pH 7.6], 1 M NaCl [Merck], 100 mM EDTA [pH 8.0], 0.5% polyoxyethylene 20 cetyl ether [Brij 58], 0.2% deoxycholate, 0.5% N-laurylsarcosyl) and mixed with an equal volume of 1.6% (wt/vol) low-melting-point agarose (Bio-Rad, Richmond, Calif.) in EC buffer and pipetted into plug molds. The solidified plugs were incubated overnight at 37°C in 1 ml of EC buffer-lysozyme solution (2.88 mg of lysozyme per ml of EC buffer). The lysis buffer was replaced with 1 ml of protein digestion solution (3.3 mg pronase E in 1 ml of EET buffer containing 1.6% [wt/vol] sodium dodecyl sulfate [SDS]), and the plugs were incubated again overnight at 37°C. The agarose plugs were washed three times for 1 h in EET buffer, twice for 1 h in Milli-Q water, and once for 1 h in the appropriate restriction buffer (Buffer Y+/Tango; MBI Fermentas, St. Leon-Rot, Germany) at room temperature. The restriction was carried out overnight at 27.5°C in 300 μl of restriction buffer containing 30 U of SmaI (MBI Fermentas). The digestion was stopped by adding 0.5 ml of 0.5 M EDTA (pH 8.0), and the plugs were stored at 4°C. The restriction fragments were separated by PFGE in a contour-clamped homogeneous electric field MAPPER system (Bio-Rad) by loading pieces of the plugs in 1% (wt/vol) pulsed-field-certified agarose (Bio-Rad) gel prepared with a 0.5× TBE buffer (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA). Electrophoresis of E. casseliflavus was performed in 2 liters of 0.5× TBE buffer at 14°C for 22 h at 6 V/cm and an angle of 120°, with pulse times ramping linearly from 0.41 to 15.11 s. For all the other Enterococcus strains, pulse times ramping linearly from 5 to 30 s were chosen.
A Staphylococcus aureus strain (R-6314; Department for Medical Microbiology, University of Antwerp, Antwerp, Belgium) was used as a molecular weight marker. The genome was prepared as described above, with the exception that 500 U of mutanolysin was added to the lysozyme solution. The gels were stained with ethidium bromide.
Species identification by SDS-polyacrylamide gel electrophoresis (PAGE).
Cells were grown for 24 h on BM agar at 37°C under microaerophilic conditions. Whole-cell protein extracts and protein separation were carried out as described by Gelsomino et al. (16) on one to three isolates from each clone clustered by PFGE.
RESULTS
Total counts of enterococci in cheese and human feces.
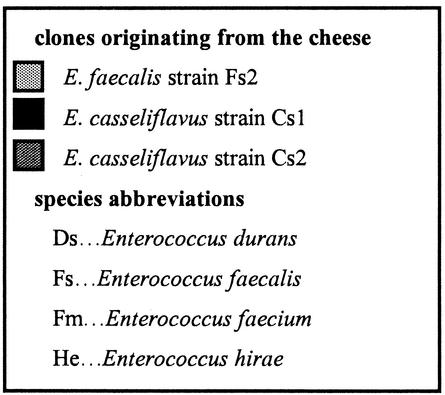
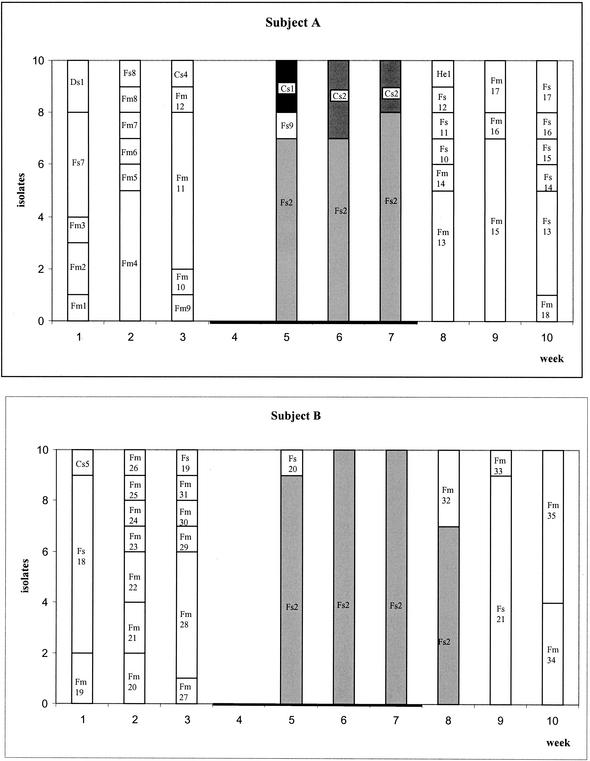
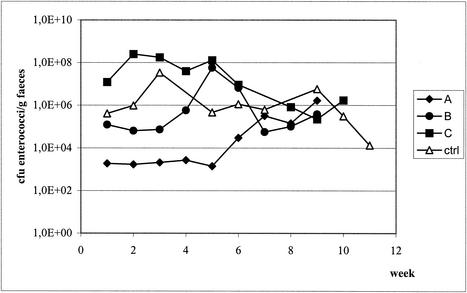
The cheese used contained an average of 3.2 × 104 enterococci/g (standard deviation = 5.6 × 103; data not shown). Figure 1 shows the number of enterococci in the fecal samples as well as the clonal diversity at each sampling point. The subjects started eating the cheese in week 4, during which no samples were taken. Recovery of enterococci in the feces differed from subject to subject and from week to week. In subject A, the level of enterococci remained stable during the first 6 weeks at 1.9 × 103 CFU/g of feces, after which it increased to 1.6 × 106 CFU/g. Subject B had stable numbers of enterococci in the first 3 weeks (8.7 × 104 CFU/g of feces) and in the last 3 weeks (1.8 × 105 CFU/g), but the level peaked at 5.7 × 107 CFU/g during cheese consumption. Subject C had a level of ∼1 × 108 CFU/g of feces during the first 5 weeks and a lower number during the last 3 weeks (9.1 × 105 CFU/g). The number of enterococci from the control subject increased during the first 3 weeks from 4.1 × 105 CFU/g of feces to 3.4 × 107 CFU/g, remained stable at around 7.3 × 105 CFU/g between weeks 6 and 8, and decreased from 5.8 × 106 to 1.3 × 104 CFU/g in the last 3 weeks (Fig. 2).
FIG. 1.
Diagrams showing the numbers of enterococci and the species and strains recovered from the feces of single subjects before, during, and after cheese consumption. Numbers refer to different clones. The black horizontal bars show the periods of cheese consumption. Column colors in the key refer to the strains that were recovered from both humans and cheese.
FIG. 2.
Total counts of enterococci found in subjects A, B, and C and the control subject.
Species identification by SDS-PAGE.
A total of 480 colonies were isolated from plates of the cheese showing >102 colonies. These were divided into yellow-pigmented, sticky colonies (n = 443) and white, thick, creamy colonies (n = 37) and counted. Forty randomly selected yellow-pigmented colonies were identified as E. casseliflavus, and the 37 white colonies were identified as E. faecalis by SDS-PAGE. After PFGE had been performed on the fecal isolates and the band patterns had been clustered, one to three isolates from each clone were also identified by SDS-PAGE as E. faecium, E. faecalis, E. casseliflavus, Enterococcus durans, or E. hirae.
During the preconsumption period (weeks 1 to 3), the presence of high numbers of E. faecium was detected in the feces of subjects A, B, and C (73, 70, and 100%, respectively). Other species were detected in lower numbers, namely, E. faecalis (16.5%), E. durans (6.5%), and E. casseliflavus (3.5%) in subject A and E. faecalis (26.5%) and E. casseliflavus (3.5%) in subject B. In the control subject the distribution of enterococcal species remained fairly stable during the entire experiment. E. faecium (84.5%) was the most frequently encountered species; much lower numbers of E. faecalis (11.0%) and E. hirae (4.5%) were detected.
During cheese consumption the detection frequency of E. faecalis was much higher in subjects A, B, and C. Subject A showed high numbers of E. faecalis (76.5% of isolates) and lower numbers of E. casseliflavus (23.5% of isolates), while E. faecalis was the only species detected in the samples from subject B. In subject C, 33% of the isolates were still E. faecium and they were all found in week 6. E. faecalis was found in weeks 5 and 7 (67% of isolates).
In the postconsumption period, E. faecium appeared again in subjects A, B, and C (56.5, 46.5, and 56.5% of isolates, respectively) while E. faecalis was present in 40, 53.5, and 40% of the isolates, respectively. E. hirae was present in low numbers in subjects A (3.5%) and C (3.5%).
Strain typing by PFGE.
A total of 437 isolates were typed by PFGE (360 human isolates [i.e., 10 isolates picked from four human fecal samples over nine sampling points] and 77 isolates from the cheese [i.e., 40 yellow-pigmented and 37 white colonies]). In the present study, we labeled the E. faecalis clones Fs1 to Fs32, the E. faecium clones Fm1 to Fm69, the E. casseliflavus clones Cs1 to Cs5, the E. durans clone Ds1, and the E. hirae clones He1 to He5. In a previous study (16), the clones were called F1 to F7 (E. faecalis) and C1 to C3 (E. casseliflavus).
A total of 39 E. casseliflavus isolates were identical to clone Cs2 and 1 isolate was identical to clone Cs1 of E. casseliflavus from the previous study (16), while 33 E. faecalis isolates were identical to clone Fs1 and 4 were identical to clone Fs2 (called C2, C1, F1, and F2, respectively, in Gelsomino et al. [16]). It was presumed that the other 403 yellow-pigmented isolates from the cheese were either clone Cs1 or Cs2. All these clones, except Fs2, were also found in the cheese in the previous study, proving that the flora of this particular cheese remains constant over a period of at least 3 years. Clone Fs2 was found in the milk and the human feces in the previous study. Taking into consideration that the average enterococcal content in the cheese was 3.2 × 104 CFU/g of cheese, we conclude that each gram of cheese contained at least 2.7 × 102 CFU of clone Fs2, 7.4 × 102 CFU of clone Cs1, 2.2 × 103 CFU of clone Fs1, and ∼2.9 × 104 CFU of clone Cs2.
None of the clones detected in the cheese were found in the human feces in the 3-week preconsumption period. The clones found in this period, especially in the control and in subjects A and B, were very diverse. Subject C showed different dominant clones in each week: Fm37 in week 1, Fm38 in week 2, and Fm39 in week 3 (Fig. 1).
During consumption of the cheese, E. faecalis clone Fs2, which was a minor component of the cheese, dominated the feces of all cheese-consuming subjects, especially subjects A (73.5% of isolates) and B (96.5% of isolates). In addition, the feces of subject A contained E. casseliflavus clones Cs1 (16.5%) and Cs2 (6.5%), both of which were also found in the cheese. The latter was the dominant clone in the cheese. Subject C's feces contained clone Fs2 but only in weeks 5 and 7; in week 6, E. faecium clone Fm38, which had also been found in the preconsumption period, was the only clone detected (Fig. 1). During the first week of cheese consumption, a number of other E. faecalis clones were isolated, one of which (Fs20) was isolated from the feces of two different subjects (B and C).
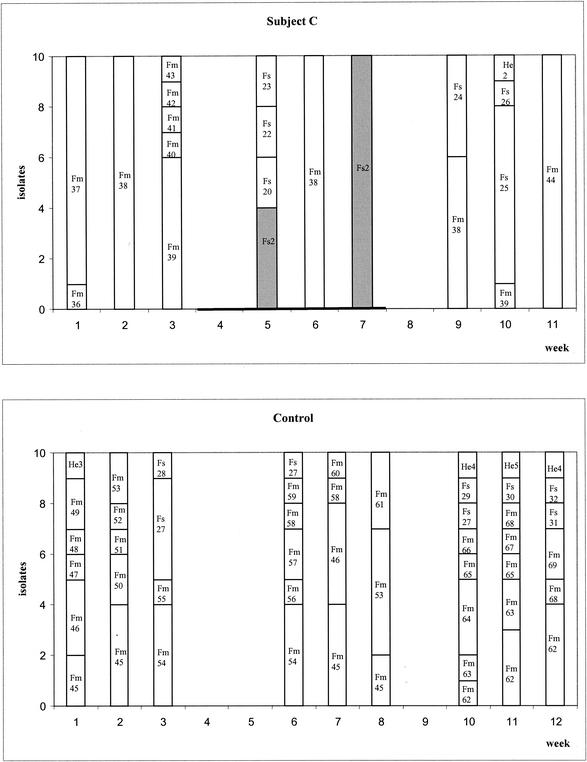
In the postconsumption period, E. faecalis clone Fs2 disappeared from the feces of all subjects except that of subject B, where it was detectable for one more week. New clones of E. faecalis appeared in all subjects, and E. faecium appeared again in subjects A, B, and C. However, all these clones were different from the ones recovered in the cheese. As in the preconsumption period, the feces of subjects A, B, and C showed dominant clones (e.g., Fm44 and Fs25 in weeks 10 and 11 in subject C). All the clones found in the feces of subject C are shown in Fig. 3. Clone Fm38 was recovered in the pre- and postconsumption periods as well as during the cheese consumption period.
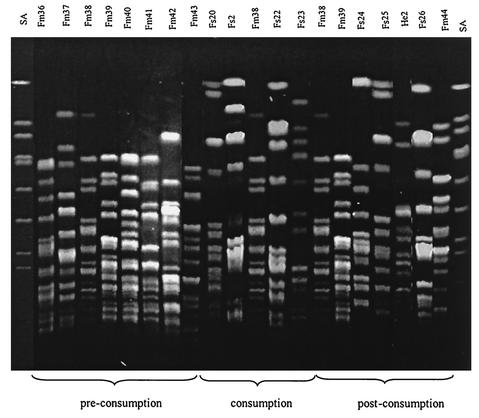
FIG. 3.
PFGE agarose gel showing the strains recovered from subject C. An S. aureus strain was used as molecular weight marker (SA). Clone Fm38 was found before, during, and after the cheese consumption period.
Comparison of the PFGE band patterns of all fecal isolates was performed using a GelCompar apparatus and visual inspection. All clones (except clone Fs20) present in the feces of one subject were unique to that subject and were not present in another subject (data not shown).
DISCUSSION
The aim of this study was to determine the common clones of enterococci in human feces and the effect that daily consumption of a raw-milk cheese, containing known clones of enterococci, has on the fecal enterococcal microflora. Like Tannock et al. (39), we assumed that the bacteria in feces reflect the bacteria in the distal large bowel, although reports by Savage (35) and Tuomola et al. (40) include a claim that fecal samples alone cannot reveal the composition and localization of bacterial communities in the colonic lumen. However, fecal sample analysis is the common method used to study the flora of the GIT (2, 21, 37, 38).
The common enterococcal flora in humans was studied during 3 weeks (preconsumption period) in subjects A to C and during 12 weeks in the control subject. Only the clones isolated from the feces during the first 3 weeks were considered to represent the regular flora, as the effect of cheese consumption can last through the postconsumption period. Total enterococcal counts differed in each of the subjects from week to week. In the preconsumption period, E. faecium was the dominant organism in all subjects (70 to 100%), followed by E. faecalis. These findings confirm the statements of Devriese et al. (4) and Murray (29) that E. faecalis and E. faecium are the dominant species in the human intestine. Generally, each sample of feces contained two to seven different clones. Very often the subjects showed dominant clones or clones that are recovered over several weeks, such as Fm38 in subject C or Fm45 in the control (Fig. 1 and 3). According to the definition of Sears et al. (37), these clones are resident whereas the clones that appear only once are transient. In the case of the only cheese-consuming subject in whose samples a resident clone was detected (Fm 38 in subject C), it is precisely this clone that came back first after the cheese consumption. No resident clone was found in subjects A and B. This may be due to the low numbers of isolates taken per sample. E. hirae clone He4 in the control subject seems to have been resident too, although it was found only twice. According to Sears et al. (37), though, resident strains eventually disappear and are replaced by other resident strains.
During cheese consumption, the subjects showed a drastic change in their fecal flora. The cheese contained an average enterococcal count of 3.2 × 104 CFU/g, which is within the range (103 to 107 CFU/g) of enterococcal counts found in cheese by Fryer (11). Four clones were detected in the cheese (Fs1, Fs2, Cs1, and Cs2). Three of them (Fs1, Cs1, and Cs2) were clones of the same strains as those recovered from the same cheese in the previous study (16). The incidence of these clones differs when compared to the previous trials.
In addition, during cheese consumption no correlation seemed to exist between the amount of enterococci/g of cheese ingested and the number of enterococci/g in the feces, as decreases and increases in the numbers of enterococci occurred all the time. The majority of the strains isolated from the feces during the consumption period belonged to the same clone strains as those detected in the cheese. Although the cheese contained primarily E. casseliflavus clone Cs2, almost all strains detected in the feces of subjects A, B, and C belonged to that of E. faecalis clone Fs2 (Fig. 1). Why the finding of clone Fs2 was interrupted by that of clone Fm38 in subject C is not clear, but the reasons might be found in dietary habits or in the low number of isolates taken. Clone Fs2, although a minor component of the enterococcal clones in the cheese, apparently thrived best in the human intestine as soon as cheese consumption began and disappeared as soon as the cheese consumption came to an end. This clone most likely found optimal conditions in the bowel and proliferated as long as there was a continuous supplementation of that particular clone. It is possible that other enterococcal strains were brought into the human GIT by consumption of the cheese but were missed because they were present in low numbers. An example of this happening is that of E. faecalis clone Fs20, which was found twice in subject C and once in subject B. Sørensen et al. (38) proved in a similar experiment that enterococci in a suspension pass through the GIT, but in contrast, in this study we were able to prove the effect of a food (in this case, cheese) as a carrier. Similar studies with other organisms (Lactobacillus rhamnosus, Lactobacillus paracasei, and Bifidobacterium lactis) confirm the present finding (2, 25, 38).
A similar result was found in a previous study (16) in which isolates from raw milk, cheese, and milking equipment were compared with isolates from fecal samples from four cheesemakers. In that study, three clones, two of E. casseliflavus (Cs1 and Cs2) and one of E. faecalis (Fs1), largely dominated the milk, cheese, and human fecal samples. Clone Fs2 was never isolated from the cheese in that study; however, it was found in the raw milk and in the human feces. In the present study, this clone was present in very low numbers in the cheese but was the dominant clone isolated from the feces during the cheese consumption period. Why different clones dominated the feces of the Irish cheesemakers and the Belgian consumers is not known. The difference might be due to the fact that the two groups live in different geographical areas, as stated by Franz et al. (10) and Murray (29), or to the fact that the dominant clones of the cheese also dominated the GIT of the cheesemaking family due to consumption over several years. These findings support the idea that the enterococcal population of the fecal flora of the cheesemaking family represents consumption of cheese rather than human fecal contamination of milk (16).
According to Franz et al. (10), enterococci are able to colonize the GIT as they are part of the normal intestinal flora. The allochthonous clone Fs2 (found in a place other than where it originated), however, is not able to colonize the intestine, as it disappears quickly after cheese consumption ends. In subjects A and C, clone Fs2 disappears when cheese consumption stops, while in subject B, it remains for another week (Fig. 1). This finding may be subject dependent. Tannock et al. (39) obtained similar results with a strain of Lactobacillus rhamnosus. The clones probably undergo a reduction in numbers due to the variable conditions in the stomach, the changing nature of the intestinal contents, the rate of movement, the competition for nutrients or binding sites, etc. There may also be an antagonistic effect between the enterococci or other bacteria in the GIT and the new enterococci (such as those of clone Fs2) from the cheese which can only be detected in the feces when relatively large amounts of the new enterococci are consumed.
One of the first reports of a similar experiment was that of Sears et al. (37). Bacteria of E. coli strains were swallowed deliberately in large numbers, but they were recovered for limited periods only and were not established as residents. The present findings corroborate the results presented in that report. Enterococci do not seem to adhere to the intestinal mucosa. Although adhesion is regarded by numerous authors as an essential feature for probiotics (5, 26, 39), there is concern over the use of probiotic bacteria that contain antibiotic resistance genes as these might be transmissible to other bacteria, especially when adhering to the mucosa. Another recommendation addresses the levels of viable cells. According to Ziemer and Gibson (42), probiotics should establish and flourish in the intestine, while others (13, 33) recommend a daily intake of at least 107 CFU per gram or per milliliter. E. faecalis clone Fs1 and E. casseliflavus clone Cs2 were present in the cheese in relatively high numbers (2.2 × 103 and 3.0 × 104 CFU/g, respectively). But it was E. faecalis clone Fs2, which was present in the cheese at only 267 CFU/g (resulting in a daily consumption of 2.6 × 104 to 4.0 × 104 CFU), which proliferated and colonized the intestine during cheese consumption. These results prove that a clone present in very low numbers in cheese can establish itself in the intestine.
Acknowledgments
This work was funded by a Walsh Fellowship from Teagasc, Dublin, Ireland.
We thank Dick and Anne Keating for supplying the cheese and the four volunteers who ate the cheese.
REFERENCES
- 1.Adlerberth, I., M. Cerquetti, I. Poilane, A. Wold, and A. Collignon. 2000. Mechanisms of colonisation and colonization resistance of the digestive tract. Microb. Ecol. Health Dis. 2(Suppl.):223-239. [Google Scholar]
- 2.Bunte, C., C. Hertel, and W. P. Hammes. 2000. Monitoring and survival of Lactobacillus paracasei LTH 2579 in food and the human intestinal tract. Syst. Appl. Microbiol. 23:260-266. [DOI] [PubMed] [Google Scholar]
- 3.Clark, W. S., and G. W. Reinbold. 1966. Enterococci in young cheddar cheese. J. Dairy Sci. 49:1214-1218. [Google Scholar]
- 4.Devriese, L. A., B. Pot, L. van Damme, K. Kersters, and F. Haesebrouck. 1995. Identification of Enterococcus species isolated from foods of animal origin. Int. J. Food Microbiol. 26:187-197. [DOI] [PubMed] [Google Scholar]
- 5.Dunne, C., L. O'Mahony, L. Murphy, G. Thornton, D. Morrisey, S. O'Halloran, M. Feeney, S. Flynn, G. Fitzgerald, C. Daly, B. Kiely, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 2001. In vitro selection criteria for probiotics bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73(Suppl.):386S-392S. [DOI] [PubMed]
- 6.Elotmani, F., A. M. Revol-Junelles, O. Assobhei, and J. B. Millière. 2002. Characterization of anti-Listeria monocytogenes bacteriocins from Enterococcus faecalis, Enterococcus faecium, and Lactococcus lactis strains isolated from Raïb, a Moroccan traditional fermented milk. Curr. Microbiol. 44:10-17. [DOI] [PubMed] [Google Scholar]
- 7.Facklam, R. R., and L. M. Teixeira. 1998. Enterococcus, p. 669-682. In A. Balows and B. I. Duerden (ed.), Topley & Wilson's microbiology and microbial infections, 9th ed., vol. 2. Arnold, London, United Kingdom.
- 8.Fons, M., A. Gomez, and T. Karjalainen. 2000. Mechanisms of colonisation and colonisation resistance of the digestive tract. Microb. Ecol. Health Dis. 2(Suppl.):240-246. [Google Scholar]
- 9.Fraimow, H. S., D. L. Jungkind, D. W. Lander, D. R. Delso, and J. L. Dean. 1994. Urinary tract infection with an Enterococcus faecalis isolate that requires vancomycin for growth. Ann. Intern. Med. 121:22-26. [DOI] [PubMed] [Google Scholar]
- 10.Franz, C. M. A. P., W. H. Holzapfel, and M. E. Stiles. 1999. Enterococci at the crossroads of food safety? Int. J. Food Microbiol. 47:1-24. [DOI] [PubMed] [Google Scholar]
- 11.Fryer, T. F. 1969. Microflora of cheddar cheese and its influence on cheese flavour. Dairy Sci. Abstr. 31:471-490. [Google Scholar]
- 12.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 13.Gardiner, G. E., E. O'Sullivan, J. Kelly, M. A. E. Auty, G. F. Fitzgerald, J. K. Collins, R. P. Ross, and C. Stanton. 2000. Comparative survival rates of human-derived probiotics Lactobacillus paracasei and L. salivarius strains during heat treatment and spray drying. Appl. Environ. Microbiol. 66:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg, S. K., and B. K. Mital. 1991. Enterococci in milk and milk products. Crit. Rev. Microbiol. 18:15-45. [DOI] [PubMed] [Google Scholar]
- 15.Gelsomino, R., M. Vancanneyt, S. Condon, J. Swings, and T. Cogan. 2001. Enterococcal diversity in the environment of an Irish Cheddar-type cheesemaking factory. Int. J. Food Microbiol. 71:177-188. [DOI] [PubMed] [Google Scholar]
- 16.Gelsomino, R., M. Vancanneyt, T. M. Cogan, S. Condon, and J. Swings. 2002. The source of enterococci in a farmhouse raw-milk cheese. Appl. Environ. Microbiol. 68:3560-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraffa, G., C. Domenico, and E. Neviani. 1997. Enterococci isolated from dairy products: a review of risks and potential technological use. J. Food Prot. 60:732-738. [DOI] [PubMed] [Google Scholar]
- 18.Hardie, J. M., and R. A. Whiley. 1997. Classification and overview of the genera Streptococcus and Enterococcus. Soc. Appl. Bacteriol. Symp. Ser. 26(Suppl.):1S-11S. [PubMed]
- 19.Holdeman, L. V., I. J. Good, and W. E. C. Moore. 1976. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl. Environ. Microbiol. 31:359-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura, K., A. L. McCartney, M. A. McConnell, and G. W. Tannock. 1997. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl. Environ. Microbiol. 63:3394-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleessen, B., E. Bezirtzoglou, and J. Mättö. 2000. Culture-based knowledge on biodiversity, development and stability of human gastrointestinal microflora. Microb. Ecol. Health Dis. 2(Suppl.):53-63. [Google Scholar]
- 23.Kurmann, J. A. 1968. Ueber die Ursachen der qualitätsfördernde Wirkung auf Emmentaler und Greyerzerkäse bei einer Einimpfung von Streptokokkus faecalis in keimarmer Rohmilch. Milchwissenschaft 23:193-197. [Google Scholar]
- 24.Leclerc, H., L. A. Devriese, and D. A. A. Mossel. 1996. Taxonomical changes in intestinal (faecal) enterococci and streptococci: consequences on their use as indicators of faecal contamination in drinking water. J. Appl. Bacteriol. 81:459-466. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto, M., T. Tadenuma, K. Nakamura, H. Kume, T. Imai, R. Kihara, M. Watanabe, and Y. Benno. 2000. Effect of Bifidobacterium lactis LKM 512 yogurt on fecal microflora in middle to old aged persons. Microb. Ecol. Health Dis. 12:77-80. [Google Scholar]
- 26.Morelli, L. 2001. Taxonomy and physiology of lactic acid bacteria, effects and functions on nutrition. Report of a joint FAO/W. H. O. expert consultation on evaluation on health and nutritional properties of probiotics in food including powder milk with lactic acid bacteria. [Online.] Food and Agricultural Organization of the United Nations, New York, N.Y. ftp://ftp.fao.org/es/esn/food/Morelli.pdf.
- 27.Morris, J. G., Jr., D. K. Shay, J. N. Hebden, R. J. McCarter, Jr., B. E. Perdue, W. Jarvis, J. A. Johnson, T. C. Dowling, L. B. Polish, and R. S. Schwalbe. 1995. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Ann. Intern. Med. 123:250-259. [DOI] [PubMed] [Google Scholar]
- 28.Morrison, D., N. Woodford, and B. Cookson. 1997. Enterococci as emerging pathogens of humans. Soc. Appl. Bacteriol. Symp. Ser. 26(Suppl.):89S-99S. [PubMed]
- 29.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neviani, E., G. Mucchetti, G. Contarini, and S. Carini. 1982. Ruolo degli enterococci nei formaggi italiani I. Il Latte 7:722-728. [Google Scholar]
- 31.Nuñez, M., J. L. Rodríguez, E. Garcia, P. Gaya, and M. Medina. 1997. Inhibition of Listeria monocytogenes by enterocin 4 during the manufacture and ripening of Manchego cheese. J. Appl. Microbiol. 83:671-677. [DOI] [PubMed] [Google Scholar]
- 32.Perreten, V., B. Kollöffel, and M. Teuber. 1997. Conjugal transfer of the Tn916-like transposon TnFO1 from Enterococcus faecalis isolated from cheese to other Gram-positive bacteria. Syst. Appl. Microbiol. 20:27-38. [Google Scholar]
- 33.Reid, G. 2001. Regulatory and clinical aspects of dairy probiotics. Report of a joint FAO/W. H. O. expert consultation on evaluation on health and nutritional properties of probiotics in food including powder milk with lactic acid bacteria. [Online.] Food and Agricultural Organization of the United Nations, New York, N.Y. ftp://ftp.fao.org/es/esn/food/Reid.pdf.
- 34.Saramago Stern, C., M. G. Squeira Carvalho, and L. Martins Teixeira. 1994. Characterization of enterococci isolated from human and nonhuman sources in Brazil. Diagn. Microbiol. Infect. Dis. 20:61-67. [DOI] [PubMed] [Google Scholar]
- 35.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 36.Seale, D. R. 1986. Bacterial inoculants as silage additives. J. Appl. Bacteriol. 61(Suppl.):9S-26S.
- 37.Sears, H. J., I. Brownlee, and J. K. Uchiyama. 1949. Persistence of individual strains of Escherichia coli in the intestinal tract of man. J. Bacteriol. 59:293-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sørensen, T. L., M. D. M. Blom, D. L. Monnet, N. Frimodt-Møller, R. L. Poulsen, and F. Espersen. 2001. Transient intestinal carriage after ingestion of antibiotic-resistant Enterococcus faecium from chicken and pork. N. Engl. J. Med. 345:1161-1166. [DOI] [PubMed] [Google Scholar]
- 39.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuomola, E., R. Crittenden, M. Playne, E. Isolauri, and S. Salminen. 2001. Quality assurance criteria for probiotics bacteria. Am. J. Clin. Nutr. 73(Suppl.):393S-398S. [DOI] [PubMed]
- 41.Wallick, H., and C. A. Stuart. 1943. Antigenic relationships of Escherichia coli isolated from one individual. J. Bacteriol. 45:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziemer, C. J., and G. R. Gibson. 1998. An overview of probiotics, probiotics and synbiotics in the functional food concept: perspectives and future strategies. Int. Dairy J. 8:473-479. [Google Scholar]