Abstract
mRNA differential display has been used to identify cyclohexanone oxidation genes in a mixed microbial community derived from a wastewater bioreactor. Thirteen DNA fragments randomly amplified from the total RNA of an enrichment subculture exposed to cyclohexanone corresponded to genes predicted to be involved in the degradation of cyclohexanone. Nine of these DNA fragments are part of genes encoding three distinct Baeyer-Villiger cyclohexanone monooxygenases from three different bacterial species present in the enrichment culture. In Arthrobacter sp. strain BP2 and Rhodococcus sp. strain Phi2, the monooxygenase is part of a gene cluster that includes all the genes required for the degradation of cyclohexanone, while in Rhodococcus sp. strain Phi1 the genes surrounding the monooxygenase are not predicted to be involved in this degradation pathway but rather seem to belong to a biosynthetic pathway. Furthermore, in the case of Arthrobacter strain BP2, three other genes flanking the monooxygenase were identified by differential display, demonstrating that the repeated sampling of bacterial operons shown earlier for a pure culture (D. M. Walters, R. Russ, H. Knackmuss, and P. E. Rouvière, Gene 273:305-315, 2001) is also possible for microbial communities. The activity of the three cyclohexanone monooxygenases was confirmed and characterized following their expression in Escherichia coli.
It is now well recognized that the diversity of microbial species and their metabolic capabilities constitute a tremendous source of biocatalysts (6, 10, 39). Only a small fraction of microorganisms in most environments can be readily isolated (1, 58); therefore, gene discovery techniques which overcome the need for strain isolation provide access to the diversity of microbial chemistry. Direct cloning approaches can be very successful (21, 27, 28, 48), but they require a genetic selection or an easy screen as well as the efficient expression of the cloned DNA in an appropriate host (15). Other approaches, based on PCR amplification from environmental DNA, target only highly conserved gene families (50). While these techniques are powerful, they often are not applicable. Differential display (DD) is an alternate technique that can be used for the discovery of bacterial genes, requiring neither a genetic selection or screen nor the presence of highly conserved genes. This technique of DD involves the reproducible amplification of DNA fragments from the mRNA population at arbitrary sites by reverse transcription (RT) followed by PCR (RT-PCR) (36, 37, 57). DD is used to compare the mRNA pools from cells grown under different physiological conditions. Genes expressed at the same level in all cultures will be amplified equally from all cultures, while genes expressed only under a specific condition will give rise to RT-PCR bands only under that condition. DD is a gene discovery technique that can be applied to identify differentially expressed genes. It does not rely on prior knowledge of the genes targeted or on a genomic sequence but only on the fact that the activity that these genes encode is inducible.
DD has been applied extensively to eukaryotic systems and takes advantage of the poly(A) tails of eukaryotic mRNA by using poly(dT) primers to synthesize cDNAs by RT (36, 37, 57). This approach of DD cannot be applied to prokaryotes, which lack stable poly(A) tails. A second variation of DD uses arbitrary oligonucleotide primers to initiate RT of the message at random sites (57) and thus can be applied to archaeal and bacterial species. Application of prokaryotic DD has been limited to fewer than 25 studies, half of them published in the last 2 years (2-4, 9, 16, 25, 26, 44, 46, 47, 52, 56). We have recently shown that a high-throughput approach to DD, using a large set of arbitrary oligonucleotides to initiate RT-PCR, resulted in the repeated identification of an operon responsible for the degradation of 2,4-dinitrophenol (56). We called this high-throughput approach to DD high-density sampling differential display.
Our objective for the present study was to apply high-density sampling DD to identify multiple genes or operons carrying out the dominant physiology of a microbial community. The culture used for this work originated from a wastewater bioreactor and was enriched for growth on cyclohexanone. We show here that DD is a robust technique for gene discovery in prokaryotes and is well suited for isolating genes encoding metabolic enzymes from complex microbial communities.
MATERIALS AND METHODS
Enrichment for cyclohexanone-degrading bacteria.
An enrichment culture degrading cyclohexanone was obtained by the successive transfers of wastewater bioreactor sludge in mineral salt medium [50 mM KHPO4 (pH 7.0), 10 mM (NH4)SO4, 2 mM MgCl2, 0.7 mM CaCl2, 50 μM MnCl2, 1 μM FeCl3, 1 μM ZnCl3, 1.72 μM CuSO4, 2.53 μM CoCl2, 2.42 μM Na2MoO2, and 0.0001% FeSO4] with 0.1% cyclohexanone added as a carbon source.
Community analysis.
A terminal-restriction fragment length polymorphism (T-RFLP) analysis was performed to determine the complexity of the community (33, 40). For T-RFLP, DNA was extracted from 1 ml of enrichment culture that was resuspended in 200 μl of buffer P1 from the RNeasy RNA purification kit (Qiagen, Valencia, Calif.). Buffer P2 from the same kit and 0.3 ml of zirconia beads (Biospec Products, Bartlesville, Okla.) were added to the resuspended cells in bead beating tubes. The cells were then disrupted at 2,400 beats per min for 2 min in a bead beater (Biospec Products). DNA was purified by standard phenol extraction and ethanol precipitation protocols (49). 16S ribosomal DNA (rDNA) genes were amplified in a standard PCR by using Taq (Qiagen), a rhodamine-labeled primer (5′-ACGGGCGGTGTGTAC-3) and a second nonlabeled primer (5′-GAGTTTGATCCTGGCTCAG-3′). The PCR conditions included a single 5-min cycle at 94°C, 20 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and one final elongation cycle at 72°C for 7 min. Following amplification, four separate PCRs were purified by using the QIAquick PCR purification kit (Qiagen) and eluted in 80 μl of H2O each. Thirty-eight microliters of this product was used in a 50-μl digestion reaction volume with either AluI, MseI, or NlaIII restriction enzymes used as indicated by the manufacturer (New England Biolabs, Beverly, Mass.). Restriction fragment lengths were determined on an ABI 3700 sequencer in the GeneScan mode. The data were collected by using GeneScan Analysis 3.1 software (Applied Biosystems, Foster City, Calif.). Finally, the results were analyzed internally by using PatScan. The fluorescence threshold was placed at 100 relative fluorescence units. Fragments with sizes smaller than 50 bp were not included in the analysis. Predicted lengths of T-RFLP fragments for identified species were matched with chromatograms within 2 bp (29).
Individual strains were isolated from the community by spreading the enrichment culture on R2A Agar (Difco, Sparks, Md.) at 30°C. Strains were streaked to purity on the same medium and were identified by 16S rDNA sequence analysis. 16S rDNA was amplified from chromosomal DNA by using several primers corresponding to conserved regions of the 16S rDNA gene (32). The following temperature program was used: 95°C for 5 min; 25 cycles of 95°C for 1 min; 55°C for 1 min; 72°C for 1 min, followed by 72°C for 8 min, and then a 4°C hold.
Induction of cyclohexanone oxidation genes.
One milliliter of the culture was suspended in 25 ml of minimal medium (described above) with 0.1% yeast extract, Casamino Acids, and peptone (YECAAP) and incubated overnight at 30°C with agitation. During this incubation residual cyclohexanone was consumed. The next day 10 ml of the overnight culture was resuspended in a total volume of 50 ml of minimal medium with 0.1% YECAAP to an optical density at 600 nm of 0.29. After equilibration at 30°C for 30 min, the culture was split into two separate flasks. Cyclohexanone (0.1%) was added to one of these 25-ml cultures, and both cultures were incubated for an additional 3 h. After that time, the cultures were chilled on ice, harvested by centrifugation in a rotor cooled to −4°C, washed with 2 volumes of ice-cold minimal salts medium, and diluted to an optical density of 1 at 600 nm. Six milliliters of culture were placed in a water-jacketed respirometry cell equipped with an oxygen electrode (Yellow Springs Instruments Co., Yellow Springs, Ohio) at 30°C. After establishing the baseline respiration for each cell suspension, cyclohexanone was added to a final concentration of 0.1% and the rate of O2 consumption was further monitored. To confirm the viability of the control culture, 2 mM potassium acetate was added after the cyclohexanone.
Isolation of total cellular RNA.
RNA isolation was performed with the same cultures that were used for the respirometry experiment. After the 3-h induction period with cyclohexanone that was described above, 2 ml each of the control and induced samples was harvested by centrifugation at 17,000 × g in a rotor cooled to −4°C and resuspended in 900 μl of buffer RLT (Qiagen). A 300-μl volume of zirconia beads (Biospec Products) was added, and cells were disrupted by use of a bead beater (Biospec Products) at 2,400 beats per min for 3 min. Each of these samples was split into six aliquots for nucleic acid isolation by the RNeasy RNA purification kit (Qiagen), and each was eluted with 100 μl of RNase-free distilled water supplied with the kit. DNA was degraded in the samples by using 10 mM MgCl2-60 mM KCl-2 U of RNase-free DNase I (Ambion, Austin, Tex.) at 37°C for 4 h. Following testing for total DNA degradation by PCR with one of the arbitrary oligonucleotides used for RT-PCR, RNA was purified by use of the RNeasy minikit in the same manner as described above. The RNA was eluted from the column in 100 μl of RNase-free H2O.
Generation of randomly amplified polymorphic DNAs from arbitrarily reverse-transcribed total RNA.
A set of 240 primers with the sequence CGGAGCAGATCGAWXYZ, where WXYZ represents all but four of the 244 combinations of the three bases A, G, and C, were used in 480 separate RT-PCRs with RNA from either control or induced cells (56). These 480 reactions were performed in five 96-well PCR plates in which each primer was distributed in two adjacent wells. The four primer variants that were predicted to form the strongest primer dimers were omitted from the experiment.
The SuperScript one-step RT-PCR system (Life Technologies Gibco BRL, Rockville, Md.) reaction mixture was used with 2 to 5 ng of total RNA per individual 25-μl reaction volume. For each 96-well PCR plate, two 2.5-ml reaction mixtures sufficient for 48 reactions were prepared according to the manufacturer's instructions. Each contained buffer, nucleotides, RNA and DNA polymerase, and one of the two RNA samples (0.1 to 0.2 μg of total RNA). Each mixture was dispensed with a multichannel pipette in the odd or even wells of the 96-well PCR plates containing the prealiquoted oligonucleotide primers.
The following temperature program was used: 4°C (2 min), 5-min ramp to 37°C (1 h), followed by 95°C incubation (3 min), 1 cycle with 94°C (1 min), 40°C (5 min), and 72°C (5 min), 40 cycles with 94°C (1 min), 60°C (1 min), and 72°C (1 min), followed by an incubation at 70°C (5 min) and 4°C. Products of these PCR amplifications were separated by electrophoresis at 1 V/cm in polyacrylamide gels (Amersham Pharmacia Biotech, Piscataway, N.J.). Products resulting from the control mRNA (no cyclohexanone induction) and from the mRNA from induced cells were analyzed side by side and visualized by silver staining by use of an automated gel stainer (Amersham Pharmacia Biotech).
Reamplification of differentially expressed DNA fragments.
A 25-μl volume of DNA elution buffer (10 mg of NaCN/ml, 20 mM Tris-HCl [pH 8.0], 50 mM KCl, and 0.05% NP-40) was incubated with each excised gel band containing a differentially amplified DNA fragment at 95°C for 20 min. Reamplification of this DNA fragment was achieved in a PCR by using 5 μl of the elution mixture in a 25-μl reaction volume with the primer used in the RT-PCR. The temperature program for reamplification was as follows: 94°C (5 min), 20 cycles of 94°C (1 min), 55°C (1 min), and 72°C (1 min), followed by 72°C (7 min). The reamplification products were directly cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, Calif.) and were sequenced by using an ABI 377 DNA sequencer with ABI BigDye terminator sequencing chemistry (Applied Biosystems). To compensate for the possible reamplification of background DNA excised with the RT-PCR bands, eight clones were sequenced for each band reamplified. The nucleotide sequences of the cloned fragments were compared against the nonredundant GenBank database by using the BlastX program (National Center for Biotechnology Information).
Sequencing of cyclohexanone oxidation pathway genes.
Rhodococcus sp. strain Phi2 and Arthrobacter sp. strain BP2 cosmid libraries were constructed with the pWEB cosmid cloning kit (Epicentre Technologies, Madison, Wis.). The Rhodococcus strain Phi1 cosmid library was constructed with the SuperCos 1 cosmid vector kit (Stratagene, La Jolla, Calif.). Cosmids were screened by PCR with primers designed against the differentially amplified fragments with homology to known cyclohexanone degradation genes (Table 1). Recombinant Escherichia coli strains carrying the cosmid clones were used as the template in these PCRs with 1 μl of cell culture added to 24 μl of PCR mixture. Cosmids from recombinant E. coli from any of the three libraries screened, yielding a product of the size corresponding to a monooxygenase gene, were partially digested with Sau3A1. Fragments with sizes between 10 and 15 kb from these partial digests were subcloned into the cloning vector pSU19 (41). These subcloned plasmids were isolated by using Qiagen Turbo96 Miniprep kits and rescreened by PCR as described above. Plasmids carrying the correct sequence were disrupted by in vitro transposition using the GPS-1 genome priming system kit (New England Biolabs, Inc.). Plasmids carrying randomly inserted transposons were sequenced from each end of the transposon to obtain the sequence of kilobase-long DNA fragments. Sequence assembly was performed with the Sequencher program (Gene Codes Corp., Ann Arbor, Mich.).
TABLE 1.
16S rDNA typing of strains isolated from the cyclohexanone degrading enrichmenta
| Isolate (strain)b | Closest relative | Accession no. | % Identity |
|---|---|---|---|
| a (BP2) | Arthrobacter keyseri strain 12B | AF256196 | 99 |
| b (Phi1) | Rhodococcus pyridinovorans | AF173005 | 99 |
| c (Phi2) | Rhodococcus ruber | RR16SR6 | 99 |
| d | Caulobacter intermedius | AB023784 | 96 |
| e | Brevundimonas diminuta | BD16S1635 | 97 |
| f | Stenotrophomonas sp. strain DB1 | AF309081 | 98 |
| g | Rhizobium sp. strain X59 | AF345555 | 95 |
| h | Bacillus sp. | AB017592 | 91 |
Strains were isolated on R2A medium. Arthrobacter strain BP2, Rhodococcus strain Phi1, and Rhodococcus strain Phi2 can grow on cyclohexanone as a sole carbon and energy source.
The predicted T-RFLP fragment for each strain is indicated in Fig. 1.
Sequence analysis.
Sequences obtained from the cosmids were compared to the nonredundant GenBank Database at the National Center for Biotechnology Information by using the BlastX program. Multiple sequence alignments were generated by use of the ClustalW program. Phylogenetic trees were calculated with the neighbor-joining tree method.
Biochemical characterization of monooxygenases.
The cyclohexane monooxygenase genes from Arthrobacter strain BP1, Rhodococcus strain Phi1, and Rhodococcus strain Phi2 were cloned into the expression vector pTrcHis-topo (Invitrogen) such that the expressed proteins contained an N-terminal histidine tag. To overexpress each of these proteins, a 1-liter E. coli culture was grown in Luria-Bertani broth with riboflavin (1 μg/ml) at 30°C until the absorbance at 600 nm reached 0.5. At this point, the temperature was shifted to 16°C and the cultures were allowed to equilibrate for 0.5 h, and then IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.1 mM to induce expression. Culture growth was allowed to proceed at 16°C overnight (∼14 h), and cells were harvested and resuspended in 5 ml of buffer A (300 mM NaCl, 5% glycerol, 20 mM Tris-HCl [pH 8.0]) containing 10 mM EDTA and 10 μg of lysozyme/ml. Following a 30-min incubation on ice, cells were disrupted by sonication and the particulate fraction was removed by centrifugation. The supernatant was mixed for 1 h at 4°C with 500 μl of a metal chelation agarose (Ni-nitrilotriacetic acid Superflow; Qiagen). The resin was washed batchwise with a series of 10-ml volumes of buffer A containing 0.15, 0.3, 0.45, 0.6, 1.2, and 60 mM imidazole in order to remove proteins binding nonspecifically. The bound proteins were then eluted with 300 mM imidazole buffer, the eluted proteins were concentrated by ultrafiltration with a Centricon device (cutoff, 10,000 Da; Amicon, Danvers, Mass.), and the buffer was replaced by buffer A such that the final concentration of protein was 1 mg/ml. The monooxygenase activity of each overexpressed enzyme was assayed spectrophotometrically by monitoring the decrease of absorbance at 340 nm which corresponds to the co-oxidation of NADPH. Assays were performed in quartz cuvettes that contained the following in a 400 μl volume: 31.7 mM morpholineethanesulfonic acid (MES)-HEPES-sodium acetate buffer (pH 7.5), 15 mM NADPH, 1.25 mM of each substrate, and 25 ng of enzyme solution/ml.
Nucleotide sequence accession numbers.
The sequences of the three gene clusters have been deposited in GenBank under the accession numbers AY123972, AY123973, and AY123974.
RESULTS
Enrichment for a cyclohexanone-degrading microbial population.
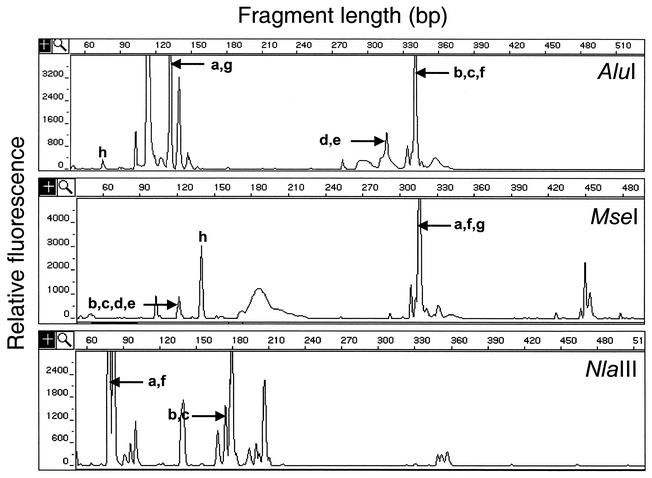
To test the applicability of DD to mixed microbial communities as a method to identify genes involved in cyclohexanone degradation, we established an enrichment culture growing on 0.1% cyclohexanone. The overall complexity of this microbial population was assessed by T-RFLP analysis. T-RFLP identifies “ribotypes,” i.e., groups of species generating DNA fragments of the same length when analyzed with a specific fluorescently labeled primer and restriction enzyme (33). T-RFLP analysis cannot identify species or even genera precisely, since distantly related genera can yield the same T-RFLP fragment of identical length. For our purpose this technique provides an estimation of the complexity of the population and sets a measure of its minimum complexity. The electrophoretic patterns of T-RFLP fragments obtained with three different restriction enzymes are presented in Fig. 1
FIG. 1.
Complexity of the cyclohexanone degradation enrichment. T-RFLP analysis was performed with DNA extracted from the enrichment culture. Three restriction enzymes, AluI, MseI, and NlaIII, were used. Letters indicate the T-RFLP fragment predicted for each of the eight bacterial strains isolated from the enrichment: a, Arthrobacter strain BP2; b, strain Rhodococcus strain Phi1; c, strain Rhodococcus strain Phi2. Other isolates (d to h) are listed in Table 1. Fragments smaller than 50 bp were not recorded. Isolates d, e, g, and h have a predicted T-RFLP fragment 41 bp long with NlaIII.
At the sensitivity and resolution of the ABI3700 sequencing machine, more than 50 peaks, each with individual fluorescence greater than 0.1% of the total fluorescence, were seen for each digest. Each peak corresponds to a different ribotype. These data indicate that at least 50 species were present in the enrichment in an abundance >0.1%. The combined fluorescences of the rare ribotypes (fluorescence between 0.1 and 1% of the total fluorescence) contribute to a significant fraction of the population, accounting for between 6 and 18% of the total fluorescence according to the restriction enzyme used. On the other hand, 7 or 8 ribotypes (according to the restriction enzyme used) account for approximately 70% of the fluorescence. As judged by the complexity of the T-RFLP patterns, such a population is qualitatively as complex as those reported for activated sludges from municipal wastewater treatment plants (29, 38, 40) but not as complex as those in natural environments such as soils (40), aquifers (38,), or open sea waters (43). However, the population was more complex than specialized natural populations such as arid soil populations (20) or those surrounding rice roots (17).
Strain isolation and analysis were carried out in addition to the T-RFLP analysis of this enrichment. Serial dilutions of the enrichment were spread at 30°C on R2A medium, a low-nutrient medium used for environmental isolates. After 72 h, eight strains with different colony morphologies or colors were isolated. These strains were typed by sequencing of their 16S rDNA genes (Table 1). The positions of the T-RFLP fragments predicted from their sequence are shown in Fig. 1. Three of the eight strains isolated, Arthrobacter strain BP2, Rhodococcus strain Phi1, and Rhodococcus strain Phi2, could use cyclohexanone as a sole source of carbon and energy.
The predicted T-RFLP fragments for Arthrobacter strain BP2 correspond to a predominant peak in samples digested with all three restriction enzymes. Assuming that all species yield a T-RFLP fluorescence signal representative of their abundance (despite many caveats relative to cell disruption, PCR amplification bias, and number of RNA operons [29, 33]), the calculated AluI T-RFLP fragment of Arthrobacter strain BP2 (129 bp matching peaks at 131 bp) indicates that that species probably does not account for more than 15% of the enrichment culture. Similarly, the two Rhodococcus species cannot be distinguished by the three restriction enzymes used in our analysis. The calculated MseI T-RFLP fragment (120 bp) indicates that these two Rhodococcus species combined account for no more than 2% of the population.
Induction of cyclohexanone oxidation genes.
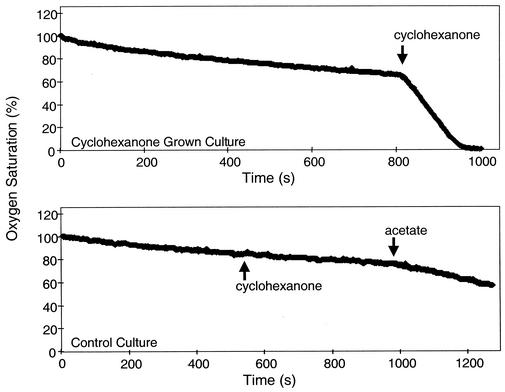
To test for induction, the enrichment culture was grown overnight in minimal medium supplemented with 0.1% YECAAP but lacking cyclohexanone, in order to allow the cells to return to an uninduced state. Subsequently the culture was diluted fivefold in fresh minimal medium, with YECAAP being then split into two separate cultures, one of which received 0.1% cyclohexanone. After 3 h, oxygen consumption in each culture was tested. As shown in Fig. 2, the culture previously exposed to cyclohexanone increased its rate of O2 consumption upon addition of cyclohexanone (Fig. 2, top panel), indicating that the genes responsible for cyclohexanone oxidation were induced. The control culture, not previously exposed to cyclohexanone, showed no increase in O2 consumption upon addition of cyclohexanone (Fig. 2, bottom panel). A decrease in O2 saturation was observed when acetate was added to the control culture, confirming that the cells were metabolically active.
FIG. 2.
Inducibility of cyclohexanone degradation in the enrichment culture. The oxygen consumption of cultures grown on acetate or cyclohexanone was measured before and after addition of acetate or cyclohexanone (indicated by arrows). Cyclohexanone-exposed enrichment culture (top panel) but not control culture (bottom panel) can oxidize cyclohexanone, indicating the inducibility of this pathway in at least some species of the enrichment.
DD analysis of the enrichment culture.
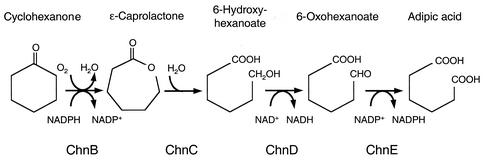
Following the demonstration that the degradation of cyclohexanone was inducible in at least some members of the enrichment community, we performed the DD analysis of the genes induced by cyclohexanone. Randomly amplified DNA fragments were generated from total RNA of the cyclohexanone-induced and control enrichment cultures in 240 parallel RT-PCR experiments. Each pair of RT-PCRs was directed with a different primer (56). From approximately 10,000 RT-PCR DNA fragments visualized on polyacrylamide gels, we chose to analyze 59 bands that were amplified from RNA of the cyclohexanone-induced culture but not from RNA of the control culture. After excision from the gel, these differentially amplified fragments (100 to 400 bp) were reamplified by PCR and cloned. The sequences from eight isolates of each cloning experiment were obtained. Twelve fragments among the 59 analyzed were recognized as putative homologues of genes necessary for biochemical conversion of cyclohexanone to adipic acid as described for Acinetobacter sp. strain SE19 (Fig. 3). Nine of the 12 fragments encoded protein sequences similar to that of the ChnB cyclohexanone monooxygenase from Acinetobacter. One of them overlapped two open reading frames (ORFs) (Fig. 4). The upstream region encoded a homologue of the C terminus of a cyclohexanone monooxygenase, while the downstream region encoded the N terminus of an aldehyde dehydrogenase similar to the 6-oxohexanoate dehydrogenase from Acinetobacter (12, 30). Of the other four fragments, two fragments were homologues of the hydrolases similar to the ɛ-caprolactone hydrolase, and two were homologues of a hydroxy-acid dehydrogenase like the 6-hydroxycaproate dehydrogenase. Several of these fragments had sequence overlaps as depicted in Fig. 4. Homologues identified for the remaining 46 differentially amplified fragments not predicted to be involved in cyclohexanone oxidation included metabolic genes as well as genes coding for core physiological functions such as ribosomal proteins or DNA polymerase. With the exception of stable RNA genes that were differentially amplified in eight RT-PCR DNA fragments, no gene outside those proposed to be involved in cyclohexanone oxidation was sampled more than once. These genes were not studied further.
FIG. 3.
Oxidation pathway of cyclohexanone into adipic acid. The nomenclature of the cyclohexanone genes is derived from that of Acinetobacter (12, 30).
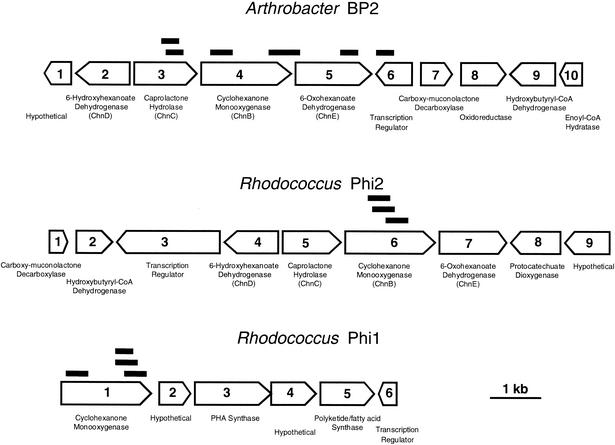
FIG. 4.
Organization of the gene clusters identified though DD in three bacteria. Black bars correspond to RT-PCR bands specifically amplified from the RNA of a cyclohexanone-induced culture. Names of genes follow the nomenclature chosen for Acinetobacter genes (12, 30).
Identification and sequencing of genes encoding cyclohexanone oxidation enzymes.
Following the DD experiment, we tried to determine whether the differentially expressed gene fragments that potentially belong to cyclohexanone oxidation genes originated from one of the isolated strains. Primers based on the sequences of the differentially amplified monooxygenase gene fragments were used to screen by PCR for the presence of the various gene fragments in the eight strains isolated. DNA extracted from the whole enrichment culture was used as a positive control template. These fragments could be specifically amplified from either the Arthrobacter sp. strain BP2, Rhodococcus sp. strain Phi1, or Rhodococcus sp. strain Phi2 but not from the other isolated strains (data not shown). Products of these amplifications were sequenced to confirm their identity.
The DNA regions flanking each monooxygenase were cloned and sequenced (Fig. 4). Contigs assembled from Arthrobacter strain BP2 and Rhodococcus strain Phi2 carried the four genes required for oxidation of cyclohexanone to adipic acid as determined for Acinetobacter sp. strain SE19 (12) (Fig. 3, Table 2). The organization of the gene clusters in Arthrobacter strain BP2 is identical to that of Rhodococcus strain Phi2 with respect to the sequence and position of the metabolic genes. However, the organization differs from that of the cyclic ketone degradation gene clusters in Acinetobacter strain SE19 (12), Brevibacterium strain HCU (8), Arthrobacter (13), and Rhodococcus strain SC1 (34). Both Arthrobacter strain BP2 and Rhodococcus strain Phi2 gene clusters also lack a short-chain Zn-independent alcohol dehydrogenase homologue of the cyclohexanol dehydrogenases found in Acinetobacter strain SE19 (12), Arthrobacter (13), and Brevibacterium strain HCU (8).
TABLE 2.
Sequence similarity of ORFs involved in cyclohexanone degradationa
| ORF | Closest homologues involved in cyclohexanone degradation (organism) | Accession no. or reference | Identity (%) | Similarity (%) | E valueb |
|---|---|---|---|---|---|
| Arthrobacter strain BP2 ChnD | 6-Hydroxycaproate dehydrogenase (Arthrobacter oxydans) | 13 | 90 | 93 | e-121 |
| Arthrobacter strain BP2 ChnC | Caprolactone hydrolase (Arthrobacter oxydans) | 13 | 85 | 90 | e-153 |
| Arthrobacter strain BP2 ChnC | Cyclohexanone monooxygenase (Xanthobacter flavus) | CAD10801 | 60 | 74 | 0.0 |
| ChnB Cyclohexanone monooxygenase (Acinetobacter sp. strain SE19) | AAG10026 | 57 | 72 | e-180 | |
| Arthrobacter strain BP2 ChnE | Succinic semialdehyde dehydrogenase (Deinococcus radiodurans) | BAA21377 | 51 | 67 | e-131 |
| ChnE 6-oxohexanoic dehydrogenase (Acinetobacter) | AAG10022 | 33 | 52 | 4e-66 | |
| Arthrobacter strain BP2 ChnR | TetR/AcrR family transcriptional regulator (Bacillus subtilis) | BAA19366 | 33 | 55 | e-20 |
| Rhodococcus strain Phi2 ChnR | ChnR2 transcriptional regulator (Brevibacterium sp. strain HCU) | AAK73166 | 31 | 44 | 8e-54 |
| Rhodococcus strain Phi2 ChnD | ChnD 6-hydroxyhexanoate dehydrogenase (Brevibacterium sp. strain HCU) | AAK73165 | 48 | 71 | 2e-99 |
| Rhodococcus strain Phi2 ChnC | ChnC 6-hexanolactone hydrolase (Acinetobacter sp. strain NCIMB9871) | BAB61745 | 59 | 75 | 2e-96 |
| Rhodococcus strain Phi2 ChnB | Cyclohexanone monooxygenase (Xanthobacter flavus) | CAD10801 | 59 | 72 | 0.0 |
| Rhodococcus strain Phi2 ChnE | Succinic semialdehyde dehydrogenase (Deinococcus radiodurans) | BAA21377 | 53 | 68 | e-137 |
| ChnE 6-oxohexanoic dehydrogenase (Acinetobacter) | AAG10022 | 32 | 50 | e-62 | |
| Rhodococcus strain Phi1 ChnB | Cyclohexanone monooxygenase (Xanthobacter flavus) | CAD10801 | 58 | 71 | 0.0 |
The predicted functions of surrounding genes not involved in cyclohexanone degradation pathways are shown in Fig. 4.
Probability that sequence similarity is due to chance.
Once the sequences of the three gene clusters were determined, we compared them to those of all the differentially expressed bands identified in the DD experiment. As represented in Fig. 3, each of the 12 fragments predicted to be involved in cyclohexanone oxidation were included in one of the three gene clusters sequenced. Four of the five cyclohexanone oxidation genes from Arthrobacter strain BP2 were represented in the differentially expressed fragments, but only the cyclohexanone monooxygenase was sampled from each of the Rhodococcus species. A 13th differentially amplified fragment with homology to the gene of a TetR family transcriptional regulator was found later to be also located on the Arthrobacter gene cluster (Fig. 4).
The regions surrounding the cyclohexanone degradation genes in both Arthrobacter strain BP2 and Rhodococcus strain Phi2 include genes characteristic of the degradation pathways of aromatic compounds. ORF7 in Arthrobacter strain BP2 as well as the partial ORF1 in Rhodococcus strain Phi2 encode carboxy-muconolactone decarboxylases. The fragment of a carboxy-muconolactone decarboxylase gene was similarly found upstream of one of the two cyclohexanone gene clusters in Brevibacterium strain HCU (8). ORF8 in Rhodococcus strain Phi2 encodes a protocatechuate dioxygenase homologue.
The genes upstream of the Rhodococcus strain Phi1 cyclohexanone monooxygenase gene have not been sequenced and may also be involved in the degradation cyclohexanone. However, the downstream genes code for conserved hypothetical proteins, a phytohemagglutinin synthase, and a polyketide and/or fatty acid synthase, suggesting that the monooxygenase of Rhodococcus strain Phi1 could be part of a biosynthetic pathway. This is the case for the Emericella nidulans Baeyer-Villiger monooxygenase StcW gene that is part of the sterigmatocystin biosynthetic gene cluster (7).
Relationships of the newly identified cyclohexanone monooxygenases to other Baeyer-Villiger flavin monooxygenases.
Sequence comparison using the BLAST programs against the nonredundant GenBank database showed that these newly identified cyclohexanone monooxygenases, along with the previously identified Brevibacterium sp. strain HCU and Acinetobacter sp. strain SE19 cyclohexanone monooxygenases, are part of the large family of flavin-dependent monooxygenases (23, 24). The monooxygenases (ChnB) from the two Rhodococcus species are relatively similar and share 90% amino acid identity and 89% nucleotide identity. The Arthrobacter enzyme is more distantly related, with 84% amino acid identity to both the Rhodococcus strain Phi1 and Rhodococcus strain Phi2 enzymes. All three enzymes cluster together in the family of BV monooxygenases. The other three genes involved in the degradation of cyclic ketones (chnC, chnD, and chnE) and their corresponding proteins show the same sequence divergence between the two species, between 78 and 84% of nucleotide identity and between 80 and 87% of amino acid identity.
Biochemical characterization of the cyclohexanone monooxygenases.
To confirm that the genes identified by DD were those of the targeted pathway, cultures of E. coli cells carrying the cosmids encoding the cyclohexanone oxidation operons from Arthrobacter strain BP2 or Rhodococcus strain Phi2 were grown in the presence of mineral medium containing glucose and 0.1% cyclohexanone as described previously (8). Complete oxidation of cyclohexanone was not observed, but traces of adipic acid (∼5% conversion of the substrate added) were detected by gas chromatography-mass spectrometry. No adipic acid was seen in the control culture lacking the cosmids (data not shown). The low levels of conversion most likely result from inefficient expression of high G+C gram-positive genes and operons in E. coli (8, 35).
Previous work with the cyclohexanone degradation genes of Brevibacterium had shown that the flavin cyclohexanone monooxygenases can be easily expressed in E. coli in an active form, unlike the other genes of the pathway (8, 9). We therefore cloned and expressed in E. coli the three putative cyclohexanone monooxygenase genes to produce His6 tag fusion proteins. The purified proteins all oxidized cyclohexanone as well as a large variety of cyclic and linear ketones. The specific activities (Table 3) are in the range of those previously reported for the monooxygenases of Acinetobacter, Brevibacterium, Rhodococcus, Nocardia, and Pseudomonas (9, 11, 18, 31, 34, 53-55). While following the overall similar patterns of activity for most substrates, the enzymes exhibited different specific activity signatures for some of the substrates. For example, the Rhodococcus strain Phi2 enzyme readily oxidized the linear 2-tridecanone while no activity was detected with the Arthrobacter strain BP2 enzyme (Table 3). We anticipate that these three enzymes will find useful applications in biocatalysis. Investigations are under way to characterize further their substrate specificity as well as the enantiomeric specificity of their products.
TABLE 3.
Substrate specificity of the cyclohexanone monooxygenases identifieda
| Substrate | Activity (μmol of substrate/min/mg of protein)
|
||
|---|---|---|---|
| Arthrobacter strain BP2 | Rhodococcus strain Phi1 | Rhodococcus strain Phi2 | |
| Cyclobutanone | 0.13 | 0.10 | 0.15 |
| Cyclopentanone | 1.49 | 1.37 | 2.45 |
| 2-Methylcyclopentanone | 3.51 | 3.39 | 6.45 |
| Cyclohexanone | 3.57 | 3.68 | 3.75 |
| 2-Methylcyclohexanone | 4.21 | 4.77 | 5.95 |
| Cyclohex-2-ene-1-one | 2.74 | 2.69 | 3.09 |
| 1,2-Cyclohexanedione | 0.24 | 0.08 | ND |
| 1,3-Cyclohexanedione | 0.40 | ND | 0.14 |
| 1,4-Cyclohexanedione | 3.99 | 3.30 | 6.15 |
| Cycloheptanone | 3.85 | 3.62 | 6.23 |
| Cyclooctanone | 0.65 | 0.63 | 0.14 |
| Cyclodecanone | 0.17 | 0.08 | 0.21 |
| 2-Tridecanone | ND | 0.16 | 1.69 |
| Dihexyl ketone | ND | 0.16 | ND |
| 2-Phenylcyclohexanone | ND | 1.05 | 0.73 |
| Dimethyl-2-piperidone | 4.15 | 3.54 | 6.51 |
| Phenylboronic acid | 0.19 | ND | 0.11 |
| β-Ionone | 1.49 | 2.71 | 0.54 |
| Norcamphor | 2.84 | 1.50 | 2.82 |
| Dimethyl sulfoxide | 0.42 | 0.52 | 0.54 |
Assays were performed as described in Materials and Methods. Substrates, as provided by the manufacturers, were added at 1.25 mM in the presence of 15 mM NADPH and 10 μg of enzyme at room temperature. The co-oxidation of NADPH was monitored at 340 nm. ND, no activity detected.
DISCUSSION
In the last 3 years, an increasing number of articles have reported the use of DD for the identification of regulated bacterial genes in isolated strains. The goal of this work is to assess the ability of this technique to identify specific metabolic genes in a microbial community. Previous experiments by Fleming et al. have shown that a toluene degradation gene identified by DD in a pure culture could also be detected in a reconstituted microcosm made by adding cells of a characterized species to a soil sample to a final abundance of approximately 80% of heterotrophic bacteria present (22). We set out to test the applicability of DD to the microbial community of an enrichment culture. As a test case, we chose to look for genes involved in the degradation of cyclohexanone for three reasons: first, this degradation pathway is inducible in several organisms (9, 19, 45); second, these genes have been well characterized (8, 9, 11-13, 30, 31, 34) and can be recognized by sequence similarity to gauge the success of the DD experiment; and third, the genes uncovered have potential utility for chiral biocatalysis (5, 14, 42, 51).
In this work we used high-density sampling DD (56). This approach addresses the limitations of older DD protocols, namely, the generation of false positives. It is often observed that RT-PCR bands amplified differentially from the RNA of cells grown under inducing physiological conditions do not actually reflect a difference in gene expression. These false positives are thought to arise from variability in the RT-PCR amplification process, and as such, they should arise randomly from the mRNA population. Thus, genes with unchanged levels of expression are unlikely to be sampled multiple times. In contrast, the repeated sampling of the same genes or operons is an indication that these genes are truly differentially expressed. In the DD analysis of a microbial population in which the complexity of the mRNA pool is greater than for a pure culture, the multiple sampling of a gene not actually differentially expressed is even more unlikely.
We set out to identify genes involved in the degradation of cyclohexanone in an enrichment culture and sampled six genes involved in cyclohexanone oxidation in 13 independent RT-PCRs. These genes are part of three gene clusters belonging to three different species. Two of the gene clusters encode all the genes required for the conversion of cyclohexanone into adipic acid. The organization schemes of these two clusters are very similar, with the exception of the presence of a transcriptional regulator and the surrounding genes in the Rhodococcus strain Phi2 cluster (Fig. 4).
The first cluster was sampled at the highest density with six DNA fragments identifying four genes. This gene cluster is found in Arthrobacter strain BP2, which appears to be a predominant species in the enrichment, although not accounting for more than 15% of the population. This corresponds to a sevenfold increase in the quantitative complexity of the mRNA pool (from all cells) relative to that of the Arthrobacter cyclohexanone operon. The qualitative complexity (number of different mRNA species) is much greater since 85% of the total RNA of the enrichment comes from at least 50 bacterial strains. The two other cyclohexanone monooxygenase genes present in two different Rhodococcus species (strains Phi1 and Phi2) were sampled in four and three RT-PCRs, respectively, each driven by a different primer. T-RFLP analysis showed that Rhodococcus strains Phi2 and Phi1 accounted together for less than 2% of the population. Thus, the corresponding mRNA was sampled in an RNA pool with at least a 50-fold increase in quantitative complexity. The difference in abundance between Rhodococcus strain Phi2 and Arthrobacter strain BP2 may explain the difference in the density of the sampling of their cyclohexanone degradation operons.
This work further supports the use of DD for the discovery of inducible metabolic prokaryotic genes. In one DD experiment, we identified three new Baeyer-Villiger flavin monooxygenase genes. In this work, a known metabolic pathway was used as a proof of concept case for the identification of genes in complex microbial communities. Because the multiple sampling of metabolic genes or operons is a strong lead for the identification of genes expressed under specific physiological conditions, this same approach can be applied to the discovery of other uncharacterized metabolic pathways in complex microbial populations. We believe that high-density sampling DD can be applied to other microbial populations for the discovery of enzymes that cannot be screened or selected for or for which there is insufficient sequence information for PCR amplification. The experiments described in this report provide a foundation for building on the application of DD methodology to environmental samples.
Acknowledgments
We thank Ray Jackson for assistance in DNA sequencing, Qiong Cheng and Kristy Kostitchka for the Rhodococcus strain Phi1 cosmid library construction, and Li Liao and Mario Chen for bioinformatic assistance.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banu, S., K. Ohtani, H. Yaguchi, T. Swe, S. T. Cole, H. Hayashi, and T. Shimizu. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35:854-864. [DOI] [PubMed] [Google Scholar]
- 3.Benson, N. R., R. M. Wong, and M. McClelland. 2000. Analysis of the SOS response in Salmonella enterica serovar Typhimurium using RNA fingerprinting by arbitrarily primed PCR. J. Bacteriol. 182:3490-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaya, D., D. Vaulot, P. Amin, A. W. Takahashi, and A. R. Grossman. 2000. Isolation of regulated genes of the cyanobacterium Synechocystis sp. strain PCC 6803 by differential display. J. Bacteriol. 182:5692-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornscheuer, U. T. 2002. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 26:73-81. [DOI] [PubMed] [Google Scholar]
- 6.Bramucci, M. G., and V. Nagarajan. 2000. Industrial wastewater bioreactors: sources of novel microorganisms for biotechnology. Trends Biotechnol. 18:501-505. [DOI] [PubMed] [Google Scholar]
- 7.Brown, D. W., J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brzostowicz, P. C., M. S. Blasko, and P. E. Rouvière. 2002. Identification of two gene clusters involved in cyclohexanone oxidation in Brevibacterium epidermidis strain HCU. Appl. Microbiol. Biotechnol. 58:781-789. [DOI] [PubMed] [Google Scholar]
- 9.Brzostowicz, P. C., K. L. Gibson, S. M. Thomas, M. S. Blasko, and P. E. Rouvière. 2000. Simultaneous identification of two cyclohexanone oxidation genes from an environmental Brevibacterium isolate using mRNA differential display. J. Bacteriol. 182:4241-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bull, A. T., A. W. Bunch, and G. K. Robinson. 1999. Biocatalysts for clean industrial products and processes. Curr. Opin. Microbiol. 2:246-251. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Y. C., O. P. Peoples, and C. T. Walsh. 1988. Acinetobacter cyclohexanone monooxygenase: gene cloning and sequence determination. J. Bacteriol. 170:781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, Q., S. M. Thomas, K. Kostichka, J. R. Valentine, and V. Nagarajan. 2000. Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 by in vitro transposition. J. Bacteriol. 182:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, T., Y. Takahashi, and S. Yamamota. 1991. Manufacture of adipic acid by bio/technology. Bio Industry 8:671-678. [Google Scholar]
- 14.Colonna, S., N. Gaggero, G. Carrea, P. Pasta, V. Alphand, and R. Furstoss. 2001. Enantioselective synthesis of tert-butyl tert-butanethiosulfinate catalyzed by cyclohexanone monooxygenase. Chirality 13:40-42. [DOI] [PubMed] [Google Scholar]
- 15.Connell, N. D. 2001. Expression systems for use in actinomycetes and related organisms. Curr. Opin. Biotechnol. 12:446-449. [DOI] [PubMed] [Google Scholar]
- 16.Craig, J. E., A. Nobbs, and N. J. High. 2002. The extracytoplasmic sigma factor, final sigma E, is required for intracellular survival of nontypeable Haemophilus influenzae in J774 macrophages. Infect. Immun. 70:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donoghue, N. A., D. B. Norris, and P. W. Trudgill. 1976. The purification and properties of cyclohexanone oxygenase from Nocardia globerula CL1 and Acinetobacter NCIB 9871. Eur. J. Biochem. 63:175-192. [DOI] [PubMed] [Google Scholar]
- 19.Donoghue, N. A., and P. W. Trudgill. 1975. The metabolism of cyclohexanol by Acinetobacter NCIB 9871. Eur. J. Biochem. 60:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Entcheva, P., W. Liebl, A. Johann, T. Hartsch, and W. R. Streit. 2001. Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia. Appl. Environ. Microbiol. 67:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleming, J. T., W. H. Yao, and G. S. Sayler. 1998. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl. Environ. Microbiol. 64:3698-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraaije, M. W., N. M. Kamerbeek, W. J. van Berkel, and D. B. Janssen. 2002. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 518:43-47. [DOI] [PubMed] [Google Scholar]
- 24.Fraaije, M. W., W. J. Van Berkel, J. A. Benen, J. Visser, and A. Mattevi. 1998. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem. Sci. 23:206-207. [DOI] [PubMed] [Google Scholar]
- 25.Gravesen, A., P. Warthoe, S. Knochel, and K. Thirstrup. 2000. Restriction fragment differential display of pediocin-resistant Listeria monocytogenes 412 mutants shows consistent overexpression of a putative beta-glucoside-specific PTS system. Microbiology 146:1381-1389. [DOI] [PubMed] [Google Scholar]
- 26.Hanna, M. N., R. J. Ferguson, Y. H. Li, and D. G. Cvitkovitch. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henne, A., R. Daniel, R. A. Schmitz, and G. Gottschalk. 1999. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl. Environ. Microbiol. 65:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henne, A., R. A. Schmitz, M. Bomeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiraishi, A., K. Masamune, and H. Kitamura. 1989. Characterization of the bacterial population structure in an anaerobic-aerobic activated sludge system on the basis of respiratory quinone profiles. Appl. Environ. Microbiol. 55:897-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwaki, H., Y. Hasegawa, M. Teraoka, T. Tokuyama, H. Bergeron, and P. C. Lau. 1999. Identification of a transcriptional activator (ChnR) and a 6-oxohexanoate dehydrogenase (ChnE) in the cyclohexanol catabolic pathway in Acinetobacter sp. strain NCIMB 9871 and localization of the genes that encode them. Appl. Environ. Microbiol. 65:5158-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamerbeek, N. M., M. J. Moonen, J. G. Van Der Ven, W. J. Van Berkel, M. W. Fraaije, and D. B. Janssen. 2001. 4-Hydroxyacetophenone monooxygenase from Pseudomonas fluorescens ACB. A novel flavoprotein catalyzing Baeyer-Villiger oxidation of aromatic compounds. Eur. J. Biochem. 268:2547-2557. [DOI] [PubMed] [Google Scholar]
- 32.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitts, C. L. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol. 2:17-25. [PubMed] [Google Scholar]
- 34.Kostichka, K., S. M. Thomas, K. J. Gibson, V. Nagarajan, and Q. Cheng. 2001. Cloning and characterization of a gene cluster for cyclododecanone oxidation in Rhodococcus ruber SC1. J. Bacteriol. 183:6478-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakey, D. L., R. K. Voladri, K. M. Edwards, C. Hager, B. Samten, R. S. Wallis, P. F. Barnes, and D. S. Kernodle. 2000. Enhanced production of recombinant Mycobacterium tuberculosis antigens in Escherichia coli by replacement of low-usage codons. Infect. Immun. 68:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang, P., L. Averboukh, and A. B. Pardee. 1993. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 21:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 38.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrs, B., S. Delagrave, and D. Murphy. 1999. Novel approaches for discovering industrial enzymes. Curr. Opin. Microbiol. 2:241-245. [DOI] [PubMed] [Google Scholar]
- 40.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 41.Martinez, E., B. Bartolome, and F. de la Cruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 42.Mihovilovic, M. D., G. Chen, S. Wang, B. Kyte, F. Rochon, M. M. Kayser, and J. D. Stewart. 2001. Asymmetric Baeyer-Villiger oxidations of 4-mono- and 4,4-disubstituted cyclohexanones by whole cells of engineered Escherichia coli. J. Org. Chem. 66:733-738. [DOI] [PubMed] [Google Scholar]
- 43.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison, M., and J. Miron. 2000. Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and pili-proteins. FEMS Microbiol. Lett. 185:109-115. [DOI] [PubMed] [Google Scholar]
- 45.Norris, D. B., and P. W. Trudgill. 1976. Multiple forms of cyclohexanone oxygenase from Nocardia globerula CL1. Eur. J. Biochem. 63:193-198. [DOI] [PubMed] [Google Scholar]
- 46.Orihuela, C. J., J. Mills, C. W. Robb, C. J. Wilson, D. A. Watson, and D. W. Niesel. 2001. Streptococcus pneumoniae PstS production is phosphate responsive and enhanced during growth in the murine peritoneal cavity. Infect. Immun. 69:7565-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulino, L. C., M. P. de Mello, and L. M. Ottoboni. 2002. Differential gene expression in response to copper in Acidithiobacillus ferrooxidans analyzed by RNA arbitrarily primed polymerase chain reaction. Electrophoresis 23:520-527. [DOI] [PubMed] [Google Scholar]
- 48.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Seow, K. T., G. Meurer, M. Gerlitz, E. Wendt-Pienkowski, C. R. Hutchinson, and J. Davies. 1997. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J. Bacteriol. 179:7360-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart, J. D. 2000. Organic transformations catalyzed by engineered yeast cells and related systems. Curr. Opin. Biotechnol. 11:363-368. [DOI] [PubMed] [Google Scholar]
- 52.Taxman, D. J., and J. P. Ting. 2001. Identification of novel Mycoplasma hyorhinis gene fragments by differential display analysis of co-cultures. J. Microbiol. Methods 44:217-223. [DOI] [PubMed] [Google Scholar]
- 53.Trudgill, P. W. 1990. Cyclohexanone 1,2-monooxygenase from Acinetobacter NCIMB 9871. Methods Enzymol. 188:70-77. [DOI] [PubMed] [Google Scholar]
- 54.Trudgill, P. W. 1990. Cyclopentanone 1,2-monooxygenase from Pseudomonas NCIMB 9872. Methods Enzymol. 188:77-81. [DOI] [PubMed] [Google Scholar]
- 55.Van Der Werf, M. J. 2000. Purification and characterization of a Baeyer-Villiger mono-oxygenase from Rhodococcus erythropolis DCL14 involved in three different monocyclic monoterpene degradation pathways. Biochem. J. 347:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walters, D. M., R. Russ, H. Knackmuss, and P. E. Rouvière. 2001. High-density sampling of a bacterial operon using mRNA differential display. Gene 273:305-315. [DOI] [PubMed] [Google Scholar]
- 57.Welsh, J., K. Chada, S. S. Dalal, R. Cheng, D. Ralph, and M. McClelland. 1992. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 20:4965-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]