Abstract
A gram-positive polychlorinated biphenyl (PCB) degrader, Rhodococcus sp. strain RHA1, metabolizes biphenyl through the 2-hydroxypenta-2,4-dienoate (HPD) and benzoate metabolic pathways. The HPD metabolic pathway genes, the HPD hydratase (bphE1), 4-hydroxy-2-oxovalerate aldolase (bphF1), and acetaldehyde dehydrogenase (acylating) (bphG) genes, were cloned from RHA1. The deduced amino acid sequences of bphGF1E1 have 30 to 58% identity with those of the HPD metabolic pathway genes of gram-negative bacteria. The order of these genes, bphG-bphF1-bphE1, differs from that of the HPD metabolic pathway genes, bphE-bphG-bphF, in gram-negative degraders of PCB, phenol, and toluene. Reverse transcription-PCR experiments indicated that the bphGF1E1 genes are inducibly cotranscribed in cells grown on biphenyl and ethylbenzene. Primer extension analysis revealed that the transcriptional initiation site exists within the bphR gene located adjacent to and upstream of bphG, which is deduced to code a transcriptional regulator. The respective enzyme activities of bphGF1E1 gene products were detected in Rhodococcus erythropolis IAM1399 carrying a bphGF1E1 plasmid. The insertional inactivation of the bphE1, bphF1, and bphG genes resulted in the loss of the corresponding enzyme activities and diminished growth on both biphenyl and ethylbenzene. Severe growth interference was observed during growth on biphenyl. The growth defects were partially restored by the introduction of plasmids containing the respective intact genes. These results indicated that the cloned bphGF1E1 genes are not only responsible for the primary metabolism of HPD during growth on both biphenyl and ethylbenzene but are also involved in preventing the accumulation of unexpected toxic metabolites, which interfere with the growth of RHA1.
Polychlorinated biphenyls (PCBs) had been used widely as industrial materials and have caused serious contamination problems worldwide. Environmental contamination by PCBs remains all over the world. The microbial degradation of PCBs is regarded as one of the most effective procedures to remove them from the environment. Many PCB-degrading bacteria have been isolated, and they commonly cometabolize PCBs through the biphenyl catabolic pathway (6) (Fig. 1). In this pathway, biphenyl is transformed to benzoate and 2-hydroxypenta-2,4-dienoate (HPD) by bphABCD gene products, and the resulting HPD is further metabolized to pyruvate and acetyl-coenzyme A (CoA) by successive reactions catalyzed by the HPD metabolic pathway enzymes, including HPD hydratase (HPDH), 4-hydroxy-2-oxovalerate aldolase (HOVA), and acetaldehyde dehydrogenase (acylating) (AADH). Detailed study of the HPD metabolic pathway has been performed in gram-negative bacteria, especially Pseudomonas sp. strain CF600 (22), Pseudomonas sp. strain KKS102 (10), and Burkholderia sp. strain LB400 (9).
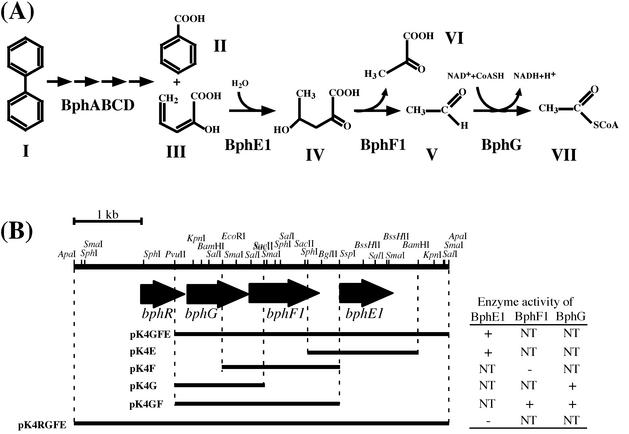
FIG. 1.
(A) Biphenyl catabolic pathway of Rhodococcus sp. strain RHA1. Compounds: I, biphenyl; II, benzoate; III, HPD; IV, 4-hydroxy-2-oxovalerate; V, acetaldehyde; VI, pyruvate; VII, acetyl-CoA. Enzymes: BphA, biphenyl dioxygenase complex; BphB, dihydrodiol dehydrogenase; BphC, 2,3-dihydroxybiphenyl dioxygenase; BphD, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase; BphE1, HPDH; BphF1, HOVA; BphG, AADH. (B) Restriction map and subclones of the 5.7-kb ApaI fragment carrying the bphRGF1E1 genes. The arrows indicate the bphR, bphG, bphF1, and bphE1 ORFs. The segment included in each subclone plasmid is indicated by a horizontal bar. The BphE1 (HPDH), BphF1 (HOVA), and BphG (AADH) activities of subclones are presented on the right. +, present; −, absent; NT, not tested.
Rhodococcus sp. strain RHA1 can efficiently transform PCB48, which consists primarily of tetrachlorobiphenyl (18). Previous studies characterized the bphAC1B and etbCbphD1 gene clusters responsible for the degradation of biphenyl to benzoate and HPD (8, 13, 14, 27). The bphE2F2 (formerly designated bphEF) genes located downstream of bphD1 were deduced to encode BphE and BphF. Introduction of bphD1 into the RHA1 mutant strain RCD1, which deleted the DNA region including etbCbphD1E2F2, however, restored the ability to grow on biphenyl (19), suggesting that the bphE2F2 genes are not essential for biphenyl degradation and that the primary HPD metabolic pathway genes may exist elsewhere. Besides biphenyl, RHA1 grows on ethylbenzene. The metabolism of ethylbenzene via propionate and HPD is estimated from the alkylbenzene in Pseudomonas (5) and Rhodococcus (25) degraders.
In the present study, we describe the unique HPD metabolic pathway genes and present evidence that they are primarily responsible for biphenyl and ethylbenzene degradation in RHA1. In addition, we have identified a putative transcriptional regulator possibly involved with the HPD metabolic pathway genes. This is to our knowledge the first report of the characterization of the HPD metabolic pathway genes from a gram-positive bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The plasmids and bacterial strains used in this study are listed in Table 1. Rhodococcus strains were grown in Luria-Bertani (LB) medium or W minimal medium (13) with biphenyl or vapor of ethylbenzene at 30°C. Escherichia coli JM109 was used as a host strain.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Rhodococcus | ||
| RHA1 | Wild type; Bph+ | 18 |
| RDE1 | Mutant derivative of RHA1; aphII gene insertion mutant of bphE1; Kmr | This study |
| RDF1 | Mutant derivative of RHA1; aphII gene insertion mutant of bphF1; Kmr | This study |
| RDG1 | Mutant derivative of RHA1; aphII gene insertion mutant of bphG; Kmr | This study |
| R. erythropolis IAM1399 | Wild type; Bph− | IAM1399 culture collection |
| Plasmids | ||
| pUC18, pUC19, pUC119 | Cloning vector; Apr | Takara Shuzo |
| pBluescript II SK(+) | Cloning vector, Apr | Stratagene |
| pBluescript II KS(+) | Cloning vector, Apr | Stratagene |
| pMS21 | pBluescript II KS(+) with 5.7-kb ApaI fragment carrying bphRGF1E1 | This study |
| pMS211 | pBluescript II SK(+) with 4.2-kb PvuII-ApaI fragment carrying bphGF1E1 | This study |
| pT7-blue | TA-cloning vector; Apr | Novagen |
| pK4 | Rhodococcus-E. coli shuttle vector, Kmr | 11 |
| pK4HKcos | pK4 containing cos region | This study |
| pK4E | pK4 with 1.7-kb SphI-BamHI fragment of RHA1 carrying bphE1 | This study |
| pK4F | pK4 with 1.8-kb EcoRI-SspI fragment of RHA1 carrying bphF1 | This study |
| pK4G | pK4 with 1.4-kb PvuII-SacII fragment of RHA1 carrying bphG | This study |
| pK4GF | pK4 with 2.5-kb PvuII-SspI fragment of RHA1 carrying bphGF1 | This study |
| pK4GFE | pK4 with 4.2-kb PvuII-ApaI fragment of RHA1 carrying bphGF1E1 | This study |
| pK4RGFE | pK4 with 5.7-kb ApaI fragment of RHA1 carrying bphRGF1E1 | This study |
| pUCKmD | pUC19 with 790-bp aphII gene fragment; Kmr | This study |
| pDE1 | pUCKmD containing a 430-bp BssHII bphE1 internal fragment | This study |
| pDF1 | pUCKmD containing a 600-bp SacII bphF1 internal fragment | This study |
| pDG1 | pUCKmD containing a 450-bp KpnI-EcoRI bphG internal fragment | This study |
| pKH402KF | pUC119 with 1.1-kb KpnI fragment of KKS102 carrying bphE | 10 |
| pBsRG6 | Cloning vector; Apr Tsr | R. van der Geize |
| pUJ1-tsr | pUC18 with insertion of tsr gene from pBsRG6; Apr Tsr | This study |
| pK4Etsr | pK4E with insertion of tsr gene from pUJI-tsr; complements the bphE1 mutant | This study |
| pK4Ftsr | pK4F with insertion of tsr gene from pUJI-tsr; complements the bphF1 mutant | This study |
| pK4Gtsr | pK4G with insertion of tsr gene from pUJI-tsr; complements the bphG mutant | This study |
Preparation of substrates.
HPD was prepared enzymatically from dl-allylglycine by the method used for synthesis of 2-oxopent-4-enoate (4). Hydroxy-2-oxovalerate was prepared from 0.5 mM HPD in 1 ml of 10 mM Tris-HCl buffer (pH 7.0) using the crude HPDH produced by an E. coli transformant of pKH402KF containing bphE of Pseudomonas sp. strain KKS102 (0.2 mg of protein) (10). NAD+, NADH, acetyl-CoA, and lactate dehydrogenase were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan).
DNA manipulations and analysis.
All of the DNA techniques used, including isolation of total DNA, gene cloning, sequencing, Southern hybridization, electroporation, pulsed-field gel electrophoresis, and computer analysis, have been described previously (13, 14, 24, 27). The following primer sequences were used to amplify the bphG gene sequence in strain RHA1: forward primer, 5′-(G/A)(C/A)(C/G)AA(C/T)AT(C/T)GA(C/T)GA(G/A)TTCACC-3′, and reverse primer, 5′-GTCATGATGTC(G/C/T)A(C/T)(G/A)TTGCCGG-3′.
Analytical method.
RDE1 cells grown in 10 ml of LB medium were inoculated into 10 ml of W minimal medium containing either biphenyl or vapor of ethylbenzene and were incubated with shaking for 48 h at 30°C. At selected times, 1-ml aliquots were withdrawn, and the cells were removed by centrifugation. The supernatant was filtered through a membrane filter (pore size, 0.45 μm; Advantec, Tokyo, Japan), and the filtrate was analyzed by high-performance liquid chromatography using an Alliance 2690 system (Waters, Randolph, Mass.) and a TSKgel ODS-80TM column (inside diameter, 6 mm; length, 150 mm; Tosoh, Tokyo, Japan) at room temperature. The mobile phase was a mixture of water (50.0%), acetonitrile (49.5%), and phosphate (0.5%), and the total flow rate was 1.0 ml/min. HPD and metabolites were detected with a UV spectrophotometric detector at 265 nm.
Gene disruption.
The 430-bp BssHII, 600-bp SacII, and 450-bp KpnI-EcoRI fragments containing the internal segments of bphE1, bphF1, and bphG, respectively, were inserted into pUCKmD, which is composed of pUC19 and the aphII gene. The resulting plasmids, pDE1, pDF1, and pDG1, were independently introduced into RHA1 cells by electroporation. Transformants were selected on diluted LB agar plates containing 50 mg of kanamycin/liter and were subjected to Southern hybridization analysis in order to examine the insertion of pDE1, pDF1, and pDG1 into the chromosomal bphE1, bphF1, and bphG genes, respectively, by single crossover. In a disruption mutant, a pUCKmD segment is sandwiched between a pair of inactivated genes by terminal deletions.
RT-PCR.
RHA1 total RNA was prepared from biphenyl-, ethylbenzene-, or LB-grown cells as described by Ausubel et al. (2). Reverse transcription (RT)-PCR was carried out using a BcaBest RNA PCR kit version 1.1 (Takara Shuzo, Kyoto, Japan) according to the instructions of the manufacturer. The following primers were designed using GeneWorks software (IntelliGenetics, Mountain View, Calif.) to amplify the region spanning the boundaries of bphGF1E1, which have easily detectable sizes of ∼500 bp: bphGF1 forward primer, 5′-TCATGACAGCAGCGGCAGC-3′; bphGF1 reverse primer, 5′-TGTTGCAGCGAGACATCGG-3′; bphF1E1 forward primer, 5′-TGGTATTCCAGCTTCCTCTTGC-3′; and bphF1E1 reverse primer, 5′-CTTCCTGCCTTGCCTGATCG-3′.
Primer extension analysis.
Total RNA was isolated from biphenyl-grown cells as described above. The primer extension experiment was carried out as described previously (11). The nucleotide sequence was determined by the dideoxy termination method with an ALFexpress DNA sequencer (Pharmacia Biotech, Milwaukee, Wis.). The antisense Cy5 fluorescently labeled oligonucleotide sequence used was 5′-AATAGGTCGGTACCGATGTTGC-3′.
Enzyme assay.
The hydratase activity for HPD of the cell extract was assayed by monitoring the decrease in absorbance at 265 nm (A265) with a Beckman (Fullerton, Calif.) DU-7500 spectrophotometer according to the method described earlier (7). One unit of activity was defined as the amount of enzyme required to transform 1 μmol of HPD per min at 25°C. The molar extinction coefficient of HPD was taken to be 19,200 M−1 cm−1 (16). Assays for HOVA and AADH activities were performed by the method of Shingler et al. (22). HOVA activity was measured by monitoring the oxidation of NADH (A340) in the presence of excess lactate dehydrogenase. One unit of activity was defined as the amount of enzyme required to catalyze the oxidation of 1 μmol of NADH per min. AADH activity was measured by monitoring the CoA-stimulated reduction of NAD+ (A340). One unit of activity was defined as the amount of enzyme required to reduce 1 μmol of NAD+ per min.
Crude-extract preparation.
RHA1 and its derivative cells were harvested from 200-ml cultures of LB medium containing 50 mg of kanamycin/liter and were washed and resuspended in 200 ml of W minimal medium containing 0.2% biphenyl. After incubation with shaking for 20 h at 30°C, the cells were washed and suspended in 20 mM Tris-HCl buffer (pH 7.0) and disrupted by a French pressure cell (SLM-Aminco Instruments Inc., Urbana, Ill.). After centrifugation (5,000 × g; 20 min), the supernatants were used as crude extracts.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the DDBJ, EMBL, and GenBank databases under accession no. AB085906.
RESULTS
Isolation of HPD metabolic pathway genes in RHA1.
To clone the AADH gene in RHA1, PCR was performed with the primer sequences, which are highly conserved among the AADH genes of the gram-negative aromatic compound degraders Pseudomonas sp. strains CF600 (X60835) and KKS102 (D16407); Pseudomonas putida F1 (U09250), mt-2 (AF043925), NCIB9816 (U13232), and IPO1 (D63377); Pseudomonas pseudoalcaligenes KF707 (D85853); and Burkholderia sp. strain LB400 (X76500). A 350-bp PCR product was obtained from RHA1 DNA. It showed amino acid sequence identity with the AADH genes of NCIB9816 (60%) and CF600 (58%). A clone containing the AADH gene was selected from an RHA1 cosmid library by colony hybridization using the 350-bp PCR fragment as a probe. A 5.7-kb ApaI fragment of the clone that hybridized to the probe was subcloned into a plasmid, pBluescript II KS(+), to give the plasmid pMS21. The nucleotide sequence of the 5.7-kb ApaI fragment revealed four open reading frames (ORFs), ORF1 (684 bp), ORF2 (897 bp), ORF3 (1,077 bp), and ORF4 (804 bp). Shine-Dalgarno sequences were found at appropriate spacings upstream from the start codons of all the ORFs (21). The deduced amino acid sequences of ORF2, ORF3, and ORF4 showed identity with the sequences of AADH, HOVA, and HPDH from gram-negative aromatic compound degraders, respectively (Table 2), and the ORFs were designated bphG, bphF1, and bphE1, respectively (Fig. 1). The overlap of the stop codon of bphG and the start codon of bphF1 implies a translation coupling between them (12, 23). The deduced amino acid sequence of ORF1 had 27% identity with that of a new class of bacterial negative regulators, padR of the phenolic acid degrader Pediococcus pentosaceus (3) (Table 2). Thus, ORF1 was determined to be a transcriptional regulator of bphGF1E1 in RHA1 and was designated bphR (Fig. 1). In addition, HPDH activity was not conferred by pMS21 containing bphRGF1E1 but was conferred by pMS211 lacking bphR, as illustrated in Fig. 1.
TABLE 2.
Characteristics of RHA1 bph gene products
| ORF | Sizea | Similar protein (% identity) | Accession no. |
|---|---|---|---|
| 1 | 228 | P. pentosaceus negative transcriptional regulator PadR (27%) | AJ276891 |
| 2 | 299 | M. tuberculosis H37Rv hypothetical protein (61%) | Z82098 |
| Pseudomonas sp. strain CF600 AADH DmpF (53%) | X60835 | ||
| 3 | 359 | M. tuberculosis H37Rv hypothetical protein (50%) | Z82098 |
| Pseudomonas sp. strain CF600 4-hydroxy-2-oxovalerate aldolase DmpG (45%) | X60835 | ||
| 4 | 268 | M. tuberculosis H37Rv hypothetical protein (60%) | Z82098 |
| Pseudomonas sp. strain CF600 2-hydroxypenta-2,4-dienoate hydratase DmpE (37%) | X60835 |
Number of amino acid residues.
RHA1 contains three linear plasmids, pRHL1 (1,100 kb), pRHL2 (450 kb), and pRHL3 (330 kb) (14, 20). Southern hybridization analyses were performed to localize the genes among the replicons in RHA1, which were separated by pulsed-field gel electrophoresis. The bphRGF1E1 gene probe hybridized to none of the linear plasmids but to the origin of electrophoresis, suggesting a chromosomal localization of the bphRGF1E1 genes in RHA1.
Transcription of bphRGF1E1.
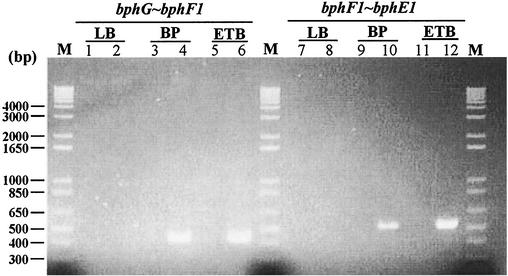
To examine the operon structure of bphGF1E1, RT-PCR experiments were performed with total RNA extracted from RHA1 cells (Fig. 2). Two primer sets, whose PCR products were expected to extend from bphG to bphF1 and from bphF1 to bphE1, were employed. The PCR products with the expected sizes for bphG-bphF1 (430 bp) and bphF1-bphE1 (534 bp) were obtained from the total RNA of cells grown on biphenyl and ethylbenzene. No PCR product was obtained from the cells grown in LB medium. These results suggest that bphGF1E1 are simultaneously transcribed as an operon and are transcriptionally induced during the degradation of biphenyl or ethylbenzene.
FIG. 2.
Agarose gel electrophoresis of RT-PCR products across the boundaries of bphGF1E1 genes. RHA1 cells were grown on biphenyl (BP) or ethylbenzene (ETB) or in LB medium. Molecular size markers are in lanes M. The sizes of the marker fragments are presented on the left. The carbon sources used are shown above the lane numbers. The boundaries indicated on the top were amplified using the primer sequences described in Materials and Methods. No detectable products were obtained in control reactions with each pair of primers from which RT had been omitted (lanes 1, 3, 5, 7, 9, and 11) or in reactions performed with LB medium-grown cells (lanes 2 and 8).
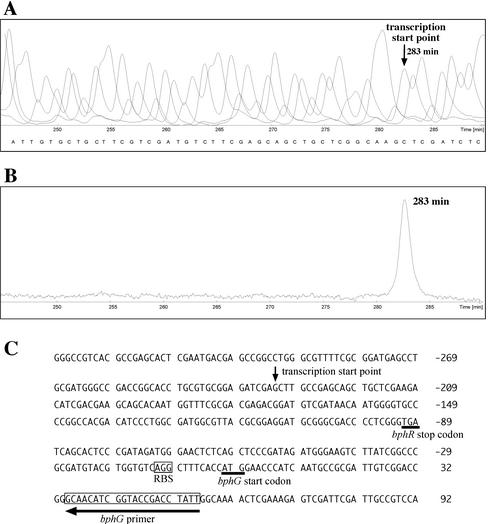
Primer extension analysis using an oligonucleotide complementary to the 5′ terminus of the bphG coding sequence was performed with RNA extracted from RHA1 cells grown on biphenyl. The 5′ end of the transcript was mapped 232 nucleotides upstream of the bphG start codon (Fig. 3). This site lies within the center of the coding sequence of bphR. Consensus procaryotic promoter sequences, including E. coli and Bacillus promoters, however, were not identified in the vicinity of the bphG transcription start point.
FIG. 3.
Identification of the 5′ end of the RHA1 bphGF1E1 transcript. (A) Nucleotide sequence obtained with cloned bphG and fluorescent bphG primer. (B) Primer extension product obtained by using RNA from biphenyl-grown RHA1 cells as a template and the bphG primer. The retention time of the product is indicated. (C) Nucleotide sequence of the upstream region of bphG. The vertical arrow indicates the transcription start point estimated from panels A and B. The horizontal arrow indicates the position of the bphG primer (boxed). The deduced ribosome-binding site (RBS) for bphG is enclosed in a box. The stop codon of bphR and the start codon of bphG are underlined.
Enzyme activities of bphGF1E1 gene products.
The 4.2-kb PvuII-ApaI fragment including bphGF1E1 was inserted downstream of the lac promoter in a vector, pBluescript II SK(+), to form pMS211. The crude extract of E. coli JM109 containing pMS211 did not show any enzyme activities encoded by bphGF1E1. However, this fragment inserted downstream of the lac promoter in a vector, pK4 conferred HPDH activity in the crude extract on Rhodococcus erythropolis IAM1399. Then, each of the RHA1 bphGF1E1 genes was inserted downstream of the lac promoter in pK4 to construct pK4G, pK4F, and pK4E, and the resulting plasmids were introduced into strain IAM1399. The introduction of pK4G and pK4E conferred activity in the crude extract of AADH (4 mU/mg of protein) and HPDH (0.48 U/mg of protein), respectively, on IAM1399. However, HOVA activity was not found in IAM1399 carrying pK4F. Then, the 2.5-kb PvuII-SspI fragment containing bphGF1 was inserted downstream of the lac promoter in pK4, and the resulting plasmid, pK4GF, was introduced into IAM1399. The transformant carrying pK4GF exhibited the activity of HOVA (42 mU/mg).
Disruption of bphGF1E1 genes in RHA1.
The bphE1, bphF1, and bphG disruption mutants RDE1, RDF1, and RDG1, respectively, were constructed by the insertion of pDE1, pDF1, and pDG1 containing the respective gene segments with truncations at both termini, as described in Materials and Methods.
The crude extracts of RDE1, RDF1, and RDG1 showed diminished or trace activity of HPDH (133 mU/mg of protein), HOVA (5.90 mU/mg of protein), and AADH (<0.1 mU/mg of protein), respectively, in comparison with those of RHA1 (504, 26.0, and 4.0 mU/mg of protein, respectively). These results suggested that the cloned bphGF1E1 genes are primarily responsible for HPD metabolism.
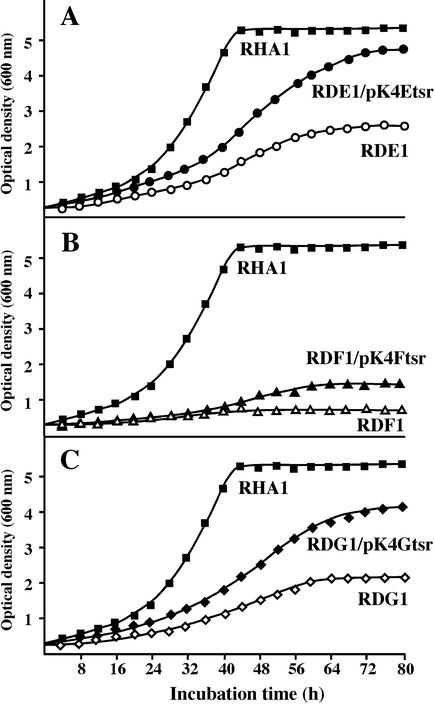
The growth on ethylbenzene of RDE1, RDF1, and RDG1 was examined, and all the mutant strains showed diminished growth (Fig. 4), which was deduced to be supported by propionate generated from ethylbenzene. The growth defects of RDE1, RDF1, and RDG1 were restored to some extent by the introduction of pK4Etsr, pK4Ftsr, and pK4Gtsr containing the respective intact genes. These results suggested that ethylbenzene is metabolized through the HPD metabolic pathway encoded by bphGF1E1 in RHA1. The growth of RDF1, however, is also diminished. The metabolite accumulated in RDF1 may be toxic enough to strictly repress the growth of RDF1. The introduction of intact genes into RDF1 and RDG1 did not restore the growth deficiency completely. These results might be due to either the poor expression of an intact gene inserted in a vector or the polar effect of a disruption on the downstream gene(s). We extracted the metabolites from ethylbenzene during incubation with RDE1, and they were separated by high-performance liquid chromatography. HPD was detected after 24 h but disappeared after 48 h, and an alternate metabolite appeared, suggesting that HPD was converted to an unknown substance in the absence of BphE1.
FIG. 4.
Growth of bphGF1E1 disruption mutants on ethylbenzene. The strains RDE1, RDF1, and RDG1, which are disruption mutants of bphE1 (A), bphF1 (B), and bphG (C), respectively, and their derivatives, RDE1/pK4Etsr, RDF1/pK4Ftsr, and RDG1/pK4Gtsr, which have plasmids containing the respective intact genes, together with RHA1 were grown in W minimal medium at 30°C on ethylbenzene.
None of the mutants grew on biphenyl in a liquid medium. On a solid medium, they grew slightly on biphenyl only at a low concentration (Table 3). Their growth defects were restored to some extent when the plasmids containing the respective intact genes were introduced. These results suggested that the bphGF1E1 genes are primarily responsible for HPD metabolism in biphenyl degradation. HPD was not detected in the metabolites produced from biphenyl during the incubation with RDE1 even after a short period (24 h), suggesting that HPD was transformed to some unknown metabolite in the absence of BphE1.
TABLE 3.
Growth characteristics of bphGF1E1 gene mutants
| Strain | Growth on carbon source (biphenyl)a |
|---|---|
| RHA1/pK4 | +++ |
| RDE1 | + |
| RDF1 | + |
| RDG1 | + |
| RDE1/pK4Etsr | ++ |
| RDF1/pK4Ftsr | ++ |
| RDG1/pK4Gtsr | ++ |
10 mg per plate. +, weak growth; ++, good growth; +++, very good growth.
DISCUSSION
In the present study, we cloned and characterized the HPD metabolic pathway genes in a gram-positive aromatic degrader for the first time. The gene order of RHA1, bphG-bphF1-bphE1, differs from that of the gram-negative bacteria, bphE-bphG-bphF (9, 10, 22). A BLAST search also indicated the existence in Mycobacterium tuberculosis H37Rv of a set of uncharacterized genes similar to bphGF1E1. The gene order of these Mycobacterium genes is, however, identical to that of the bphEGF genes in gram-negative bacteria. In addition, RHA1 bphGF1E1 are accompanied by a putative transcriptional regulatory gene, bphR. These results suggest the unique evolutionary history of HPD metabolic pathway genes in RHA1.
RHA1 has another set of putative HPD metabolic genes, bphE2F2, located downstream of bphD1. The following facts, however, support the notion that the bphGF1E1 genes are primarily responsible for the HPD metabolic pathway in RHA1. (i) bphF1 has much more identity to its counterparts in gram-negative PCB degraders and is accompanied by the neighboring bphG gene. (ii) The enzyme activities conferred by the bphGF1E1 genes were lost by the respective gene disruptions, which strictly diminished growth on biphenyl. (iii) The complementation of each gene disruption by an intact gene restored the growth defects of disruption mutants on biphenyl.
In addition to bphGF1E1, the benABC genes are also located on the chromosome (11). These genes are involved in the lower metabolic pathway for biphenyl degradation. On the other hand, the upper pathway genes, including bphAC1B and etbCbphD1, are located on the linear plasmids (14, 20). A pair of gene clusters containing the bphABCD genes on a linear plasmid were also reported in the PCB-degrading R. erythropolis strain TA421 (1). The requirement of the lower metabolic pathway for a variety of mono- and polyaromatic compounds might have caused the responsible genes to be located on the chromosome, which is more stable than the linear plasmids in a cell.
The physical association of HOVA and AADH and the requirement of the latter for the activity of the former have been indicated in the phenol-degrading Pseudomonas sp. strain CF600 (17). This notion was supported by the results of the expression of the bphF gene in IAM1399. The bphG mutant lacking AADH activity, however, showed HOVA activity of 23.5 mU/mg of protein, equivalent to that of RHA1 (26.0 mU/mg of protein). RHA1 may have residual metabolic activity toward acetaldehyde which is not accompanied by the reduction of NAD+, representing the activity of AADH.
The transcription start point of the bphGF1E1 operon lies at the center of the bphR coding sequence. The transcription start site within the preceding regulatory gene has been reported on the ORF0 gene in a PCB degrader, P. pseudoalcaligenes KF707, which has identity with GntR-type transcriptional regulators (26). ORF0 was suggested to be responsible for the transcriptional activation of bphD and bphX1X2X3, equivalent to bphEGF (26). A PCB degrader, Pseudomonas sp. strain KKS102, also has a GntR-type regulator gene, bphS, upstream of the bphEGF genes, which was shown to negatively regulate the expression of the bphEGF genes (15).
The severe interference with growth on biphenyl by the gene disruption indicates the formation of an unknown toxic metabolite in the absence of the bphGF1E1 genes. A metabolic activity toward HPD other than those of the bphGF1E1-encoded enzymes was suggested by the analysis of metabolites. The metabolic activity toward HPD in RDE1 during growth on biphenyl seems to be stronger than that during growth on ethylbenzene. In addition, such severe interference with RDE1 during growth on biphenyl was not observed during growth on ethylbenzene. Some enzyme(s) induced during growth on biphenyl seems to be involved in the formation of a toxic metabolite from HPD. Thus, the cloned bphGF1E1 genes seem not only to be responsible for the primary metabolism of HPD during growth on both biphenyl and ethylbenzene but also to be involved in preventing the accumulation of an unexpected toxic metabolite which interferes with the growth of RHA1.
Acknowledgments
We are grateful to K. Ueda and T. Beppu (Department of Applied Biological Sciences, Nihon University) for the kind gift of a plasmid, pUCKmD. We thank W. Kitagawa and R. van der Geize for helpful suggestions.
This study was supported by the Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN) in Japan.
REFERENCES
- 1.Arai, H., S. Kosono, K. Taguchi, M. Maeda, E. Song, F. Fuji, S.-Y. Chung, and T. Kudo. 1998. Two sets of biphenyl and PCB degradation genes on a linear plasmid in Rhodococcus erythropolis TA421. J. Ferment. Bioeng. 86:595-599. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Barthelmebs. L., B. Lecomte, C. Divies, and J.-F. Cavin. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 182:6724-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collinsworth, W. L., P. J. Chapman, and S. Dagley. 1973. Stereospecific enzymes in the degradation of aromatic compounds by Pseudomonas putida. J. Bacteriol. 113:922-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton, R. W., and K. N. Timmis. 1986. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putida RE204. J. Bacteriol. 168:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa, K. 1994. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation 5:289-300. [DOI] [PubMed] [Google Scholar]
- 7.Harayama, S., M. Rekik, K.-L. Ngai, and L. N. Ornston. 1989. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J. Bacteriol. 171:6251-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauschild, J. E., E. Masai, K. Sugiyama, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1996. Identification of an alternative 2,3-dihydroxybiphenyl 1,2-dioxygenase in Rhodococcus sp. strain RHA1 and cloning of the gene. Appl. Environ. Microbiol. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofer, B., S. Backhaus, and K. N. Timmis. 1994. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene 144:9-16. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi, Y., Y. Yasukochi, Y. Nagata, M. Fukuda, and M. Takagi. 1994. Nucleotide sequence and functional analysis of the meta-cleavage pathway involved in biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J. Bacteriol. 176:4269-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitagawa, W., K. Miyauchi, E. Masai, and M. Fukuda. 2001. Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Bacteriol. 183:6598-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau, P. C. K., H. Bergeron, D. Labbé, Y. Wang, R. Brousseau, and D. T. Gibson. 1994. Sequence and expression of the todGIH genes involved in the last three steps of toluene degradation by Pseudomonas putida F1. Gene 146:7-13. [DOI] [PubMed] [Google Scholar]
- 13.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masai, E., K. Sugiyama, N. Iwashita, S. Shimizu, J. E. Hauschild, T. Hatta, K. Kimbara, K. Yano, and M. Fukuda. 1997. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene 187:141-149. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsubo. Y., M. Delawary, K. Kimbara, M. Takagi, A. Ohta, and Y. Nagata. 2001. BphS, a key transcriptional regulator of bph genes involved in PCB/biphenyl degradation in Pseudomonas sp. KKS102. J. Biol. Chem. 276:146-154. [DOI] [PubMed] [Google Scholar]
- 16.Pollard, J. R., and T. D. H. Bugg. 1998. Purification, characterization and reaction mechanism of monofunctional 2-hydroxypentadienoic acid hydratase from Escherichia coli. Eur. J. Biochem. 251:98-106. [DOI] [PubMed] [Google Scholar]
- 17.Powlowski, J., L. Sahlman, and V. Shingler. 1993. Purification and properties of the physically associated meta-cleavage pathway enzymes 4-hydroxy-2-oxo-ketovalerate aldolase and aldehyde dehydrogenase (acylating) from Pseudomonas sp. strain CF600. J. Bacteriol. 175:377-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seto, M., K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seto, M., N. Okita, K. Sugiyama, E. Masai, and M. Fukuda. 1996. Growth inhibition of Rhodococcus sp. strain RHA1 in the course of PCB transformation. Biotechnol. Lett. 18:1193-1198. [Google Scholar]
- 20.Shimizu, S., H. Kobayashi, E. Masai, and M. Fukuda. 2001. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 67:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shine, J., and L. Dalgarno. 1975. Determination of cistron specificity in bacterial ribosomes. Nature 254:34-38. [DOI] [PubMed] [Google Scholar]
- 22.Shingler, V., J. Powlowski, and U. Marklund. 1992. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J. Bacteriol. 174:711-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soonyoung, H., S.-J. Kim, C.-K. Kim, Y. Kim, S.-J. Kim, and Y.-C. Kim. 1999. The PhnIJ genes encoding acetaldehyde dehydrogenase (acylating) and 4-hydroxy-2-oxovalerate aldolase in Pseudomonas sp. DJ77 and their evolutionary implications. Biochem. Biophys. Res. Commun. 256:469-473. [DOI] [PubMed] [Google Scholar]
- 24.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 25.Warhurst, A. M., K. F. Clarke, R. A. Hill, R. A. Holt, and C. A. Fewson. 1994. Metabolism of styrene by Rhodococcus rhodochrous NCIMB13259. Appl. Environ. Microbiol. 60:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe, T., R. Inoue, K. Kimura, and K. Furukawa. 2000. Versatile transcription of biphenyl catabolic bph operon in Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 275:31016-31023. [DOI] [PubMed] [Google Scholar]
- 27.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]