Abstract
Zygosaccharomyces bailii ISA 1307 and the type strain of this spoilage yeast show a diploid DNA content. Together with a rather peculiar life cycle in which mitotic but no meiotic spores appear to be formed, the diploid DNA content explains the observed difficulties in obtaining auxotrophic mutants. Mitotic chromosome loss induced by benomyl and selection on canavanine media resulted in three haploid strains of Z. bailii. This new set of Z. bailii strains allows the easy isolation of recessive mutants and is suitable for further molecular genetic studies.
The genus Zygosaccharomyces is associated with a notorious resistance to stress environments (12, 22, 27). It includes different species, such as Z. bailii, Z. bisporus, and Z. rouxii, which have been frequently implicated in the spoilage of food and beverages (2). A screen for yeasts highly resistant to weak carboxylic acids involving a wide range of genera revealed that the strain ISA 1307 of Z. bailii, originally isolated from continuous production of sparkling wine (30), displayed an outstanding capacity to survive in such environments. Whereas different physiological and biochemical mechanisms have been identified to explain the resistance of Z. bailii to acidic environments (26), the molecular basis of such behavior is unknown. Molecular tools for the study of this species have begun to appear recently. These include genomic libraries and genes suitable for complementation of auxotrophic markers (17, 20). However, previous attempts to isolate auxotrophic mutants of Z. bailii failed, possibly due to a ploidy higher than haploidy (16). We therefore set out to construct a set of Z. bailii strains suitable for mutational analysis, thus widening the possibilities for further molecular genetic studies of this organism.
The strains ISA 1307 and IGC 5167 (the type strain) of the yeast Z. bailii were used. The strain W303 1A of Saccharomyces cerevisiae (MATa ade2-1 his3-11,15 ura3-1 leu2-3,112 trp1) was used as a reference for DNA quantification. Escherichia coli XL1-Blue was used as the bacterial host for plasmids (5). Yeast strains were grown in yeast extract-peptone-dextrose (21) or in synthetic medium (yeast nitrogen base [YNB]; Difco) with the necessary bases and amino acids (23). l-Canavanine (60 μg/ml; Sigma), Geneticin (G418 sulfate, 20 μg/ml; Invitrogen), 5-fluoroorotic acid (0.75 mg/ml; Sigma), or cycloheximide (0.1 and 0.4 mg/liter; Sigma) was used. E. coli strains were grown in Luria-Bertani medium at 37°C (21). Standard yeast genetic procedures were used for sporulation of Z. bailii and dissection of asci (3, 24), for DNA manipulation (20, 21), and for preparation of competent cells (10, 13). Mutations were induced by exposing yeast cells to UV light (HNS 30-W OFR; Osram) at a distance of 60 cm. For mitotic chromosome loss, cells were inoculated at 107 cells/ml in YNB containing benomyl (30 or 100 μg/ml) at 23°C. After 20 h, cells were harvested from the culture medium, washed to remove the benomyl precipitate, and inoculated at 26°C in YNB containing l-canavanine (60 μg/ml). For fluorescence microscopy analysis, cells from a mid-log growth phase were used (21). Colocalization of DsRed-NLS (red fluorescent protein tagged into the nuclei), yEGFP3-NLS (green fluorescent protein tagged into the nuclei), and DNA was performed as described elsewhere (21). Sporulated cells of Z. bailii were collected from solid media and stained with calcofluor white (18) or 4′,6′-diamidino-2-phenylidole (DAPI; Merck). For flow cytometry experiments (Partec PAS flow cytometer, equipped with an argon-ion laser emitting a 488-nm beam at 15 mW), the DNA in whole cells was stained with SYBR Green I (Molecular Probes) (9). The cell cycle histograms presented were obtained by offline analysis of list mode files with WinMDI 2.5 software.
Z. bailii is diploid but forms tetrads with mitotic spores.
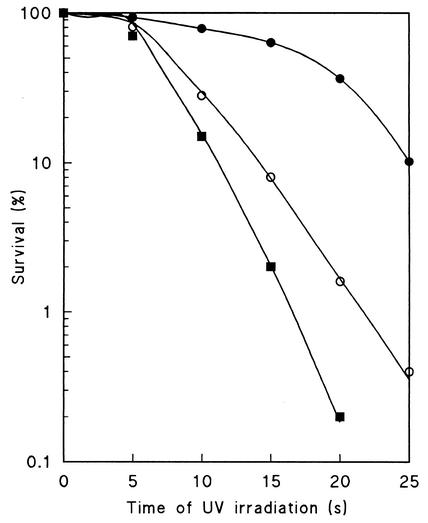
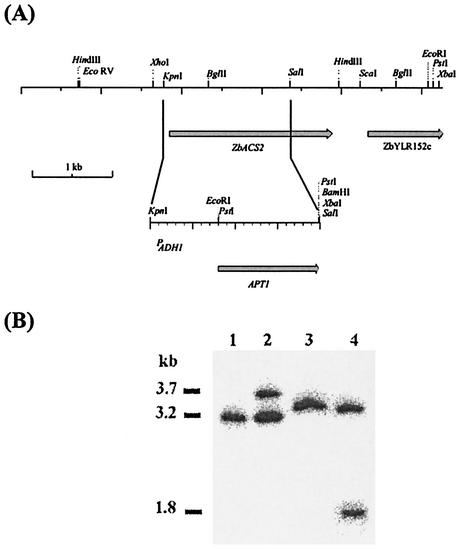
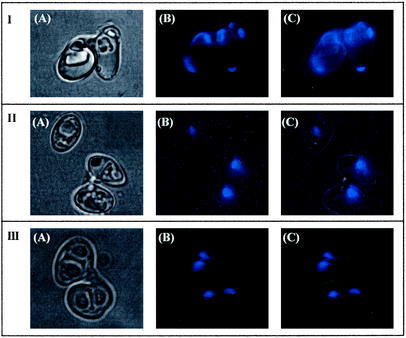
Several groups have tried to select for auxotrophic strains of Z. bailii without success (16, 17). Initial attempts to obtain Ura−, Leu−, Trp−, Ade−, or His− auxotrophic mutants of Z. bailii ISA 1307 by exposure to UV irradiation (Fig. 1) have failed. At a survival rate of 10%, around 8,000 colonies were analyzed and all were still prototrophic. Similarly, a positive selection for auxotrophic mutants (4, 29) using 5-fluoroorotic acid or 5-fluoroanthranilic acid was unsuccessful. All 5-fluoroorotic acid-resistant colonies (90 colonies per 109 cells) were still Ura+. Identical results were obtained for the type strain of Z. bailii. All together, these results pointed to a ploidy of strain ISA 1307 higher than haploidy. This idea was reinforced by the higher UV resistance of the Z. bailii strains than that of a haploid S. cerevisiae strain (Fig. 1). Most convincingly, the disruption of the ACS2 gene of Z. bailii (ZbACS2) (accession number AJ314837, encoding the acetyl coenzyme A synthetase) in Z. bailii ISA 1307 with the APT1 gene, which confers resistance to the aminoglycoside G418 (Geneticin) (11), resulted in the strain Z. bailii ISA 1307-14, which has a heterozygous ZbACS2/Zbacs2::APT1 background (Fig. 2). Furthermore, all 80 viable spores isolated from complete tetrads of this strain were G418 resistant. Moreover, all 16 spores from four complete tetrads were heterozygous for the ZbACS2/Zbacs2::APT1 alleles (F. Rodrigues, unpublished data). These observations and similar ones described by Mollapour and Piper (17) suggested that Z. bailii forms mitotic rather than meiotic spores. By using calcofluor white staining (18), we showed the presence of several bud scars in each cell involved in the conjugation process, indicating that both cells had already budded several times (Fig. 3, section I). This result points to the fact that two different cells are involved in the formation of the ascus-like structure; otherwise, if conjugation between mother and daughter cells occurs, one could expect the presence of several scars in just one of the cells. To further explain the formation of mitotic spores, the fusion of the two nuclei during this process was tested by crossing cells with different labeled nuclei (19). The DsRed-NLS and yEGFP3-NLS genes were cloned in vectors that self-replicate in Z. bailii with G418 as the dominant resistance marker. Z. bailii strains expressing each one of the fluorescent proteins were obtained, showing that the proteins were tagged into nuclei (result not shown). Unfortunately, cells expressing those fluorescent proteins lost fluorescence during the induction of sporulation, even when the strong S. cerevisiae ADH1 promoter was used. Changes in gene expression also occurred during sporulation in S. cerevisiae (7, 15); however, because no known promoter sequence of Z. bailii was up regulated under this condition, the utilization of this technology after the conjugation process was hampered. To further broaden the scope of these results, we tested whether nuclear fusion would occur during sporulation by DAPI staining of DNA (Fig. 3, section II). From 500 conjugated but not yet sporulated cells, only one showed fused nuclei. The low frequency of nuclear fusion seems to indicate that such an event does not occur during conjugation. In addition, we observed that sporulated cells of Z. bailii ISA 1307 showed only one nucleus per ascospore (Fig. 3, section III). In conclusion, the absence of nuclear fusion during sporulation in Z. bailii seems to cause mitotic spore formation.
FIG. 1.
Survival curves for Z. bailii ISA 1307 (filled circles), Z. bailii H15 (one of the isolated haploid mutants [open circles]), and S. cerevisiae W303 1A (filled squares) after irradiation with UV light.
FIG. 2.
Disruption of ZbACS2 (accession number AJ314837) by APT1. (A) Schematic representation of the APT1 expression cassette for disruption experiments. (B) Southern blot of the Z. bailii ISA 1307 wild-type strain (lanes 1 and 3) and mutant 14, which is resistant to Geneticin (Z. bailii ISA 1307-14) (lanes 2 and 4). The genomic DNA isolated was digested with the restriction enzyme HindIII (lanes 1 and 2) or PstI-XhoI (lanes 3 and 4).
FIG. 3.
Microscopy of stained Z. bailii cells. (A) Bright field; (B) blue fluorescence; (C) overlap of bright field and blue fluorescence. (Section I) Conjugated cells stained with calcofluor white; (section II) conjugated but not sporulated cells stained with DAPI; (section III) sporulated cells stained with DAPI. The fluorescence of calcofluor white and DAPI is blue.
Haploid mutants of Z. bailii.
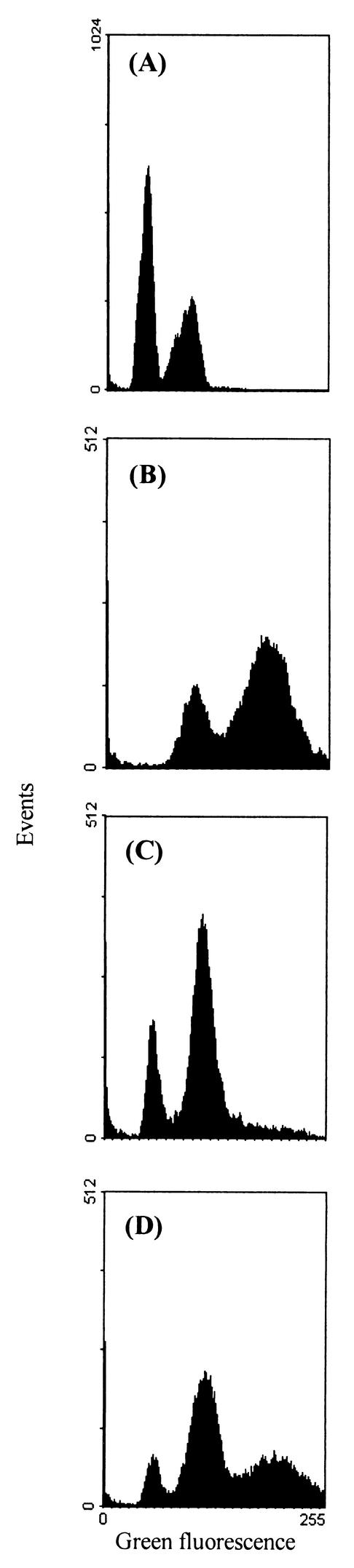
Since sporulation of Z. bailii ISA 1307 did not lead to haploid cells, haploidization was induced by growth on synthetic media containing benomyl. This drug induces mitotic chromosome loss at high frequencies (31). The CAN1 gene, encoding an arginine permease (1, 8, 25), was employed as a recessive resistance marker for the selection of haploid mutants. Canavanine, a toxic arginine analog to which can1 mutants are resistant, was used to select for spontaneous mutants. After 4 days of incubation at 30°C, colonies were found at a frequency of approximately one mutant per 1.4 × 108 cells plated. Fifty putative haploid mutants were recovered and further characterized with respect to DNA content, ability to sporulate, and production of recessive mutations. Cell DNA content was estimated by flow cytometry with the DNA fluorescent probe SYBR Green I (6, 9). Flow cytometry of stained exponentially growing cells of S. cerevisiae W303 1A, Z. bailii ISA 1307, Z. bailii IGC 5167, and of one isolated mutant (H15) evidenced the presence of G0/G1 and G2/M peaks (Fig. 4) indicative of different DNA contents. Cell cycle analysis revealed the mean fluorescence intensities (MFI) of the G0/G1 and G2/M peaks (Table 1) with a half-peak coefficient of variation (HPCV) lower than 7%, an indication of the high precision of the measurements. The DNA index (DNA In) was defined as the ratio between the fluorescence intensity of the G0/G1 peak of the strain to be tested and that of a reference strain. Two different DNA Ins were calculated, one in relation to S. cerevisiae W303 1A (DNA In-1) and the other in relation to Z. bailii ISA 1307 (DNA In-2) (Table 1). The values of DNA In-1 for the Z. bailii type strain and ISA 1307 indicate that they have, respectively, 1.4 and 2.1 times as much DNA as S. cerevisiae W303 1A. The DNA In-2 revealed the DNA contents of strains H15, H50, and H51 to be about half that of the parental strain (Table 1). All the other canavanine-resistant mutants isolated had DNA In-2 values ranging from 0.6 to 1 and were discarded due to their purported aneuploid status. Considering the genome sizes of the type strain of Z. bailii (7.7 Mb) (28) and of S. cerevisiae (13.1 Mb) together with the MFI of the G0/G1 peak (Table 1), the type strain of Z. bailii is, most likely, diploid. Pulsed-field gel electrophoresis showed that Z. bailii ISA 1307 has at least three more chromosomes than the type strain, two of higher masses and one of lower molecular mass (data not shown), which might explain the apparent higher DNA content of this strain than that of the type strain. All together, these results also point to a diploid DNA content of Z. bailii ISA 1307 and consequently to a haploid or near-haploid state of strains H15, H50, and H51. We next tested whether these three strains could produce recessive mutants. Indeed, a high number of spontaneous cycloheximide-resistant mutants was selected for from those strains (14, 25). The strains H15, H50, and H51 were also tested for the ability to sporulate. Whereas H15 was able to sporulate, spores were not found in the two other strains. Therefore, strains H50 and H51 were either sterile or heterothallic and were of the same mating type, since no sporulation was detected after both strains were mixed. Finally, mutations were induced in strain H15 by UV radiation (Fig. 1). As expected, because of its DNA content, this mutant was more sensitive to radiation than the wild-type strain (ISA 1307), and an auxotrophic mutant for histidine was easily isolated. Thus, this new set of Z. bailii strains is suitable for further molecular genetic studies.
FIG. 4.
Green fluorescence histograms of S. cerevisiae W303 1A (A), Z. bailii ISA 1307 (B), Z. bailii H15 (C), and a mixture of Z. bailii ISA 1307 and Z. bailii H15 (D) after SYBR Green I staining.
TABLE 1.
Comparison of yeast genome sizes as determined by flow cytometrya
| Yeast | MFI of G0/G1 cells ± SD | HPCV (%) | MFI ratio | DNA In-1 | DNA In-2 |
|---|---|---|---|---|---|
| S. cerevisiae W303 1A | 45.6 ± 0.6 | 6.7 | 2.1 | 1.0 | 0.50 |
| Z. bailii ISA 1307 | 96.5 ± 0.7 | 7.1 | 1.8 | 2.1 | 1.00 |
| Z. bailii IGC 5167T | 64.5 ± 0.7 | 6.5 | 1.9 | 1.4 | 0.70 |
| Z. bailii H15 | 52.5 ± 2.1 | 6.3 | 2.1 | 1.1 | 0.50 |
| Z. bailii H50 | 49.5 ± 0.7 | 5.7 | 2.2 | 1.1 | 0.50 |
| Z. bailii H51 | 52.5 ± 0.7 | 6.5 | 2.1 | 1.1 | 0.50 |
Shown are MFI of cells in phase G0/G1, HPCVs, ratios between the MFI of cells in the G0/G1 and G2/M phases, and the DNA In-1 and DNA IN-2 of each yeast strain studied.
The method described herein could also be very useful for other nonconventional yeasts with a diploid DNA content, as is the case for some pathogenic yeasts. While we feel that the possibility of readily isolating recessive mutants is an important step forward, we realize that this is only a first step. The isolation of more haploid Z. bailii mutants will show us whether mating and meiosis will also become available as tools in this organism.
Acknowledgments
We thank Filipe Sansonetty for his helpful discussion on cytometry results and Anne-Marie Zeeman for her technical assistance on the karyotyping of Z. bailii strains.
Fernando Rodrigues was the recipient of a fellowship from PRAXIS XXI, and the study was supported by a research grant from the Fundação para a Ciência e Tecnologia, Lisbon, Portugal (contract PRAXIS XXI P/AGR/11135/98).
REFERENCES
- 1.Ahmad, M., and H. Bussey. 1986. Yeast arginine permease: nucleotide sequence of the CAN1 gene. Curr. Genet. 10:587-592. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, J. A. 1992. The taxonomy of the genus Saccharomyces Meyen ex Reess: a short review for non-taxonomists. Yeast 8:1-23.1496857 [Google Scholar]
- 3.Barnett, J. A., R. W. Payne, and D. Yarrow. 1990. Yeasts: characteristics and identification. University Press, Cambridge, United Kingdom.
- 4.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 5.Bullock, W. O., J. M. Fernandez, and J. M. S. Short. 1987. XL#1-Blue: a highly efficient plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 4:376-378. [Google Scholar]
- 6.Carlson, C. R., B. Grallert, R. Bernander, T. Stokke, and E. Boye. 1997. Measurement of nuclear DNA content in fission yeast by flow cytometry. Yeast 13:1329-1335. [DOI] [PubMed] [Google Scholar]
- 7.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 8.Ekwall, K., and T. Ruusala. 1991. Budding yeast CAN1 gene as a selection marker in fission yeast. Nucleic Acids Res. 19:1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortuna, M., M. J. Sousa, M. Côrte-Real, C. Leão, A. Salvador, and F. Sansonetty. 2000. Cell cycle analysis of yeasts, p. 11.13.1-11.13.9. In J. P. Robinson, Z. Darzynkiewicz, P. Dean, A. Orfao, P. Rabinovitch, H. Tanke, and L. Wheeless (ed.), Current protocols in cytometry. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 10.Gietz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5:255-269. [Google Scholar]
- 11.Hadfield, C., B. E. Jordan, R. C. Mount, G. H. Pretorius, and E. Burak. 1990. G418-resistance as a dominant marker and reporter for gene expression in Saccharomyces cerevisiae. Curr. Genet. 18:303-313. [DOI] [PubMed] [Google Scholar]
- 12.Hocking, A. D. 1996. Media for preservative resistant yeasts: a collaborative study. Int. J. Food Microbiol. 29:167-175. [DOI] [PubMed] [Google Scholar]
- 13.Inune, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 14.Kaufer, N. F., H. M. Fried, W. F. Schwindinger, M. Jasin, and J. R. Warner. 1983. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 11:3123-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y..
- 16.MacKenzie, D. A., B. M. Pearson, L. J. Fuller, and M. H. Keenman. 1987. Stability of chimaeric vectors based on plasmids from the yeast Zygosaccharomyces bailii. Microbios Lett. 36:55-63. [Google Scholar]
- 17.Mollapour, M., and P. Piper. 2001. Targeted gene deletion in Zygosaccharomyces bailii. Yeast 18:173-186. [DOI] [PubMed] [Google Scholar]
- 18.Pringle, J. R. 1991. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194:732-735. [DOI] [PubMed] [Google Scholar]
- 19.Rodrigues, F., M. van Hemert, H. Y. Steensma, M. Côrte-Real, and C. Leão. 2001. Red fluorescent protein (DsRed) as a reporter in Saccharomyces cerevisiae. J. Bacteriol. 183:3791-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigues, F., A.-M. Zeeman, C. Alves, M. J. Sousa, H. Y. Steensma, M. Côrte-Real, and C. Leão. 2001. Construction of a genomic library of the food spoilage yeast Zygosaccharomyces bailii and isolation of the β-isopropylmalate dehydrogenase gene (ZbLEU2). FEMS Yeast Res. 1:67-71. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1998. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Schuller, D., M. Côrte-Real, and C. Leão. 2000. A differential medium for the enumeration of the spoilage yeast Zygosaccharomyces bailii in wine. J. Food Prot. 63:1570-1575. [DOI] [PubMed] [Google Scholar]
- 23.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 24.Sherman, F., and J. Hicks. 1991. Micromanipulation and dissection of asci. Methods Enzymol. 194:21-37. [DOI] [PubMed] [Google Scholar]
- 25.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 26.Sousa, M. J., L. Miranda, M. Côrte-Real, and C. Leão. 1996. Transport of acetic acid in Zygosaccharomyces bailii: effects of ethanol and their implications on the resistance of the yeast to acidic environments. Appl. Environ. Microbiol. 62:3152-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas, D. S., and R. R. Davenport. 1985. Zygosaccharomyces bailii—a profile of characteristics and spoilage activities. Food Microbiol. (London) 2:157-169.
- 28.Torok, T., C. Royer, D. Rockhold, and A. D. King. 1992. Electrophoretic karyotyping of yeasts, and southern blotting using whole chromosomes as templates for the probe preparation. J. Gen. Appl. Microbiol. 38:313-325. [Google Scholar]
- 29.Toyn, J. H., P. L. Gunyuzlu, W. H. White, L. A. Thompson, and G. F. Hollis. 2000. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast 16:553-560. [DOI] [PubMed] [Google Scholar]
- 30.Wium, H., M. Malfeito-Ferreira, V. Loureiro, and S. Aubyn. 1990. A rapid characterization of yeast contaminants associated with sparkling wine production. Ind. Bevande. 19:504-506. [Google Scholar]
- 31.Wood, J. S. 1982. Genetic effects of methyl benzimidazole-2-yl-carbamate on Saccharomyces cerevisiae. Mol. Cell. Biol. 2:1064-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]