Abstract
A plasmid-borne diacetyl (acetoin) reductase (butA) from Leuconostoc pseudomesenteroides CHCC2114 was sequenced and cloned. Nucleotide sequence analysis revealed an open reading frame encoding a protein of 257 amino acids which had high identity at the amino acid level to diacetyl (acetoin) reductases reported previously. Downstream of the butA gene of L. pseudomesenteroides, but coding in the opposite orientation, a putative DNA recombinase was identified. A two-step PCR approach was used to construct FPR02, a butA mutant of the wild-type strain, CHCC2114. FPR02 had significantly reduced diacetyl (acetoin) reductase activity with NADH as coenzyme, but not with NADPH as coenzyme, suggesting the presence of another diacetyl (acetoin)-reducing activity in L. pseudomesenteroides. Plasmid-curing experiments demonstrated that the butA gene is carried on a 20-kb plasmid in L. pseudomesenteroides.
Leuconostoc species, including Leuconostoc pseudomesenteroides, are used in mixed starter cultures together with Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. lactis biovar diacetylactis for the production of many fermented dairy products, but in particular for that of cultured buttermilk, sour cream, and ripened cream butter. The volatile compound diacetyl (2,3-butanedione) is one of the main components responsible for the typical flavor of such fermented milk products. The diacetyl concentration of fresh cultured buttermilk is typically in the range of 2 to 4 mg/liter (36); however, over time, the diacetyl concentration of the product decreases. This decrease in diacetyl concentration is highly correlated with a reduction in the flavor intensity of the product. The significance of maintaining high diacetyl levels in fermented buttermilk has been demonstrated recently when a Lactococcus lactis subsp. lactis biovar diacetylactis strain with impaired α-acetolactate decarboxylase activity resulted in increased diacetyl concentration and improved product stability (10, 11, 15).
The metabolic activities and fermentation end products of Lactococcus lactis subsp. lactis, Lactococcus lactis subsp. lactis biovar diacetylactis, and Leuconostoc species are distinctly different (7, 17). The primary metabolic difference between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. lactis biovar diacetylactis is the ability of the latter organism to cometabolize glucose and citrate, which, in addition to the production of l-lactate, results in the production of substantial amounts of fermentation products, such as diacetyl, acetoin (2-hydroxy-2-butanone), 2,3-butanediol, and acetaldehyde. Leuconostoc species also have the ability to cometabolize glucose and citrate, but the excess pyruvate produced from citrate is primarily reduced to d-lactate (16). Leuconostoc species produce diacetyl and acetoin only under acidic conditions (6, 8). The cometabolism of glucose and citrate by Lactococcus lactis subsp. lactis biovar diacetylactis is well understood, and many of the enzymes and genes involved have been characterized and sequenced (9, 21, 23, 30). Studies of pyruvate and citrate metabolism in Lactococcus lactis and Leuconostoc species demonstrate that diacetyl is formed by the chemical oxidative decarboxylation of α-acetolactate with molecular oxygen and metal ions as catalysts (17, 24).
With respect to the enzymatic catabolism of diacetyl, it has been shown that diacetyl reductase activity is strain dependent in both Lactococcus lactis and Leuconostoc species (22), with the activity generally being higher for Leuconostoc species. Diacetyl is enzymatically reduced in two steps in which NAD(P)H acts as the coenzyme; initially, diacetyl is reduced to acetoin, followed by the further reduction of acetoin to 2,3-butanediol. The reduction of diacetyl to acetoin is irreversible, while the reduction of acetoin to 2,3-butanediol is reversible. These reduction reactions are mediated via the enzyme diacetyl (acetoin) reductase (acetoin:NAD+ oxidoreductase, EC 1.1.1.5); the enzyme is also sometimes referred to as acetoin reductase or 2,3-butanediol dehydrogenase. A diacetyl (acetoin) reductase from a cell extract of Leuconostoc pseudomesenteroides has been purified and characterized (25); this enzyme uses NADH as a coenzyme for the reduction of diacetyl, but NADH and NADPH function equally well as a coenzyme for the reduction of acetoin to 2,3-butanediol by the enzyme. This paper reports the molecular cloning, characterization, and inactivation of the gene butA, which encodes this diacetyl (acetoin) reductase of L. pseudomesenteroides.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Leuconostoc pseudomesenteroides CHCC2114 was obtained from the culture collection of Chr. Hansen A/S, Hørsholm, Denmark, and was routinely grown in either M17 broth supplemented with 1% lactose (32) or MRS broth (13) at 25°C. For chromosomal and plasmid purifications from L. pseudomesenteroides, dl-threonine was added to the medium to a final concentration of 40 mM. Escherichia coli DH5α and E. coli XL10-Gold Kan supercompetent E. coli were obtained from Life Technologies, Inc. (Carlsbad, Calif.), and Stratagene GmbH (Heidelberg, Germany), respectively, and were grown in Luria-Bertani (LB) medium (26) at 37°C with aeration. For L. pseudomesenteroides cloning experiments, erythromycin (ERY) at concentrations of 1 to 20 μg/ml or ampicillin (AMP) at concentrations of 50 to 150 μg/ml was added to the medium. For E. coli cloning experiments, AMP at a concentration of 25 μg/ml was added to the culture medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| L. pseudomesenteroides | ||
| CHCC2114 | ButA+ wild-type, lactose-fermenting | CHCCb |
| FPR01 | ButA+ Emr Ampr lactose-fermenting derivative of CHCC2114 | This work |
| FPR02 | ButA− lactose-fermenting derivative of FPR01 | This work |
| FPR03 | ButA− non-lactose-fermenting derivative of CHCC2114 | This work |
| FPR04 | ButA+ non-lactose-fermenting derivative of CHCC2114 | This work |
| E. coli | ||
| DH5α | Cloning host | Life Technologies, Inc. |
| XL10-Gold Kan | Expression host | Stratagene GmbH |
| Plasmids | ||
| pPCR-Script Amp | Ampr | Stratagene GmbH |
| pCR 2.1-TOPO | Ampr Kanr | Invitrogen BV |
| pDCL01 | pPCR-Script Amp derivative containing a 1,526-bp fragment carrying CHCC2114 butA | This work |
| pV2 | Emr Ampr | 19 |
| pDMP01 | pV2 derivative containing a 1,416-bp fragment EcoRI-EcoRI, corresponding to 705 bp upstream and 741 bp downstream of the protein-coding region of the CHCC2114 butA | This work |
Amp, ampicillin; Em, erythromycin; Kan, Kanamycin; r, resistance; ButA, diacetyl (acetoin) reductase.
Chr. Hansen Culture Collection, Chr. Hansen A/S, Hørsholm, Denmark.
Enzyme purification and N-terminal amino acid sequencing.
The diacetyl (acetoin) reductase was purified from a cell extract of L. pseudomesenteroides CHCC2114 as described previously (25). The purified diacetyl (acetoin) reductase was prepared for N-terminal amino acid sequencing by using a ProSpin sample preparation cartridge (Applied Biosystems, Nærum, Denmark) in accordance with the manufacturer's instructions. The N-terminal amino acid sequence was determined at the National Food Biotechnology Centre, University College, Cork, Ireland, by Edman degradation on an automated pulsed liquid-phase protein-peptide sequencer (model 477A; Applied Biosystems, Inc., Foster City, Calif.). Liberated amino acids were detected as their phenylthiohydantoin derivatives by using a 120A analyzer (Applied Biosystems, Inc.).
Isolation, sequencing and cloning of the butA gene.
The N-terminal amino acid sequence of the diacetyl (acetoin) reductase of L. pseudomesenteroides CHCC2114 (namely, TKKVAMVTGGAQGIGEAIVRRLSADGFAVAVADL) and the consensus substrate binding region of previously sequenced diacetyl (acetoin) reductases from Bos taurus (GenBank accession no. U71200), Klebsiella terrigena (GenBank accession no. L04507), and Brevibacterium saccharolyticum (GenBank accession no. AB009078) were used to design a number of different oligonucleotide primers. Total genomic DNA (chromosomal and plasmid) from CHCC2114 was prepared as described previously (18). With the primer pair BO1 and BO2 (see Table 2) and the total genomic DNA preparation from CHCC2114 as template, a 476-bp fragment was amplified by PCR. This 476-bp fragment was sequenced and used for further primer design. Subsequently, a gene walking method (14) was used to sequence the entire butA gene, including regions upstream and downstream of the protein coding region.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Positionb | Source or reference |
|---|---|---|---|
| L. pseudomesenteroides | |||
| BO1 | 5′-AAG TTG CAA TGG TTA CCN G-3′ | 708→ | This work |
| BO2 | 5′-ACA ACG TAC AGC GAA CTT-3′ | 1183← | This work |
| CDR1 | 5′-GCA AGC GCG CAT GTG GAT C-3′ | 20→ | This work |
| CDR2 | 5′-CTT GTT GCA ATA TCA TGA TTG GC-3′ | 2182← | This work |
| CDR3 | 5′-GAC TAT AGT TGT TTC GAA TAA TAC-3′ | 657→ | This work |
| CDR10 | 5′-ATA TAT GAA TTC GCA AGC GCG CAT GTG GAT C-3′ | 20→ | This work |
| CDR11 | 5′-ATA TAT GAA TTC CTT GTT GCA ATA TCA TGA TTG GC-3′ | 2182← | This work |
| CDR15 | 5′-ATT TTT AGT TAA TTA TAC TAA TTC CTC CAA TAG TAT TAT TCG AAA C-3′ | 697← | This work |
| CDR16 | 5′-TTG GAG GAA TTA GTA TAA TTA ACT AAA AAT AAT TAA CTT TTA CCC-3′ | 1469→ | This work |
| Bos taurus | |||
| RT-DR1 | 5′-GGG TTG AAG TTG TCC ATG CGA-3′ | → | 29 |
| RT-DR2 | 5′-CAA ATC AGC AAC TGC AAC TGC-3′ | ← | 29 |
Underlined nucleotides signify EcoRI restriction sites. For CDR 15, nucleotides in bold correspond to a 15-bp sequence with homology at the 5′ end of a 741-bp fragment downstream of the coding region of butA; for CDR 16, nucleotides in bold correspond to a 15-bp sequence with homology at the 3′ end of a 705-bp fragment upstream of the coding region of butA.
For L. pseudomesenteroides primers, numbering is based on GenBank accession no. AF438200. Arrows indicate the orientation.
PCR amplifications were carried out by using either a T3 thermocycler from Biometra GmbH (Göttingen, Germany) or a Progene thermocycler from Techne (Cambridge) Ltd. (Cambridge, United Kingdom). Prior to the running of the PCR, the DNA was denatured at 94°C for 10 min. A total of 30 cycles were run, with each cycle consisting of DNA denaturation at 94°C for 1 min, followed by annealing at 50°C for 1 min and elongation at 72°C for 1 min. Pwo DNA polymerase was obtained from Roche Molecular Biochemicals (Hvidovre, Denmark). PCR products were excised from the gel and purified by using a QIAquick gel extraction kit from Qiagen GmbH (Hilden, Germany) in accordance with the manufacturer's instructions. Oligonucleotides were synthesized by TAG Copenhagen A/S (Copenhagen, Denmark) and MWG-Biotech AG (Ebersberg, Germany). Unless otherwise stated, routine DNA manipulations were performed as described previously (26).
Sequencing was performed at least twice on both strands. Samples for sequencing were prepared by using Big Dye V11 (Applied Biosystems, Inc.) and sequenced by using an ABI PRISM 310 genetic analyzer (Applied Biosystems, Inc.). The DNA and protein sequences were analyzed by using the Vector NTI program, version 6.0 (InforMax, Inc., Bethesda, Md.). Protein homology searches were performed by using the BLAST network service (1).
Expression of ButA in E. coli.
The primer pair CDR3 and CDR2 was used in a PCR to amplify a 1,526-bp fragment corresponding to the ribosome binding site and the entire coding region of the butA gene of CHCC2114. A second primer pair, CDR1 and CDR2, was used to amplify a 2,163-bp fragment corresponding to the putative promoter −35 and −10 boxes, the ribosome binding site, and the entire coding region of the butA gene. Pwo DNA polymerase (Roche Molecular Biochemicals) was used for the PCR amplification in order to generate blunt-ended fragments. These blunt-ended PCR fragments were purified and ligated into pPCR-Script Amp; E. coli strain XL10-Gold Kan (Stratagene GmbH) was used for electroporation and cloning in accordance with the manufacturer's instructions. Transformants (white colonies) were selected by plating on LB medium containing 50 μg of AMP/ml and 0.8 mg of X-Gal (5-bromo 4-chloro-3-indolyl-β-d-galactopyranoside)/ml. Plasmids were purified from selected transformants by using a QIAprep spin miniprep kit (Qiagen GmbH) in accordance with the manufacturer's instructions. The insertion size and orientation were confirmed by cutting with SacI. A time course expression analysis of the gene was performed by inoculation of 8.75 ml of LB medium containing 50 μg of AMP/ml with 1.25 ml of an overnight culture of the E. coli transformants. After 1 h of incubation at 37°C (early exponential growth phase), expression was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to the medium at a final concentration of 2 mM; a control without IPTG was included in the experiment. Samples (1 ml) were withdrawn at hourly intervals and harvested by centrifugation (15,000 × g for 10 min at 4°C), and the cell pellet was stored at −20°C until diacetyl (acetoin) reductase activity measurement. Cell extracts of the E. coli clones were prepared by resuspension of the cell pellets in 1 ml of 50 mM sodium phosphate buffer (pH 6.0) and sonicated by using a Branson Sonifier 250 (Branson Ultrasonics, Danbury, Conn.). Cultures were sonicated for 4 min with the duty cycle set at 40% and the output control set at a microtip limit of 6. In order to prevent overheating, the samples were cooled on ice after every 1 min of treatment. Following sonication, the samples were centrifuged (15,000 × g for 10 min at 4°C), and the diacetyl (acetoin) reductase activity and protein concentration of the resulting supernatant were measured.
Gene inactivation of butA.
A two-step PCR approach was used to inactivate the butA gene by deletion of the nucleic acid sequence between positions 698 and 1471 (GenBank numbering; GenBank accession no. AF438200). The primer pairs CDR10-CDR15 and CDR11-CDR16 were used in a 30-cycle PCR to amplify a 705-bp fragment upstream and a 741-bp fragment downstream of the coding region of the butA gene; a total genomic DNA preparation from CHCC 2114 was used as template. During amplification, primer CDR10 was used to introduce an EcoRI restriction site at the 5′ end of the 705-bp fragment, while primer CDR15 was used to introduce at the 3′ end of the 705-bp fragment a 15-bp tail with homology to the 5′ end of the 741-bp fragment. Likewise, primer CDR11 was used to introduce an EcoRI restriction site at the 3′ end of the 741-bp fragment, while primer CDR16 was used to introduce at the 5′ end of the 741-bp fragment a 15-bp tail with homology to 3′ end of the 705-bp fragment. The 705- and 741-bp fragments were mixed together, and a 5-cycle PCR was run in the absence of primers in order to synthesize the full-length construct of 1,416 bp. Subsequently, the primer pair CDR10 and CDR11 was added to the reaction mixture and a 30-cycle PCR was performed in order to amplify the 1,416-bp fragment. This 1,416-bp fragment was ligated into pV2 by using T4 DNA ligase in accordance with the manufacturer's instructions (Roche Molecular Biochemicals); the fragment was then transformed into E. coli DH5α as described previously (26). Transformants (white colonies) were selected by plating on LB medium containing 50 μg of AMP/ml and 0.8 mg of X-Gal/ml. Plasmids were purified from selected transformants by using a QIAprep spin miniprep kit (Qiagen GmbH) in accordance with the manufacturer's instructions. The insertion size and orientation were confirmed by cutting with EcoRI and SacI. The constructed plasmid (5.8 kb) was designated pDMP01.
pDMP01 was electroporated into CHCC2114 in a manner based on a modification of a method described previously (12). CHCC2114 was grown at 25°C in MRS broth containing 40 mM dl-threonine, and cells were harvested at mid-exponential growth phase (optical density at 600 nm [OD600], 0.6) by centrifugation at 5,000 × g for 15 min at 4°C, washed, and resuspended to an OD600 of 20. To 40 μl of resuspended cells, 0.5 μg of pDMP01 was added, and electroporation was carried out in a Gene Pulser/MicroPulser cuvette (Bio-Rad Laboratories, Hercules, Calif.) with a 0.2-cm electrode gap, at 2.5 kV and 200 Ω. After electroporation, the cell suspension was placed on ice for 10 min, diluted with 1 ml of MRS, incubated for 1 h at 28°C, and further incubated for 2 h at 25°C. The cells were then plated on MRS containing 1 μg of ERY/ml and incubated anaerobically for 3 days at 25°C. ERY-resistant (Emr) colonies were restreaked on MRS plates containing 20 μg of ERY/ml. Finally, Emr colonies were cultured in MRS broth containing 20 μg of ERY/ml. Total genomic DNA was isolated from the Emr clones, cut with EcoRI, and subsequently Southern blotted to confirm the integration of pDMP01 into the genome of CHCC2114. One such clone in which pDMP01 had successfully integrated was designated FPR01.
Spontaneous crossing out was performed by serial propagation of FPR01 in MRS broth without the addition of ERY. In the final propagation (after 10 transfers), 1.5 ml of the culture (grown to an OD600 of 0.4 in MRS broth without the addition of ERY) was used to inoculate 30 ml of MRS broth containing 20 μg of ERY/ml. This culture was incubated for 4 h at 25°C, after which AMP was added to a final concentration of 150 μg/ml, and the culture was incubated overnight at 25°C. As AMP kills only growing cells (i.e., Emr cells) and not nondividing cells (i.e., spontaneous Ems cells), the addition of AMP can be considered an enrichment step for the subsequent isolation of spontaneous Ems cells. Cells were harvest by centrifugation (4,000 × g for 5 min at 4°C) and washed twice with fresh MRS broth. The washed cells were then plated on MRS agar and incubated anaerobically for 48 h at 25°C. Clones were randomly selected and restreaked on MRS agar with (10 μg/ml) and without ERY. Ems clones were further characterized by PCR in order to identify whether they were potential butA mutants. One such butA mutant, designated FPR02, was selected and further characterized in detail by PCR, Southern blotting, nucleotide sequencing, and diacetyl (acetoin) reductase activity measurement. Southern blots were carried out by using a GeneScreen Plus membrane (Perkin-Elmer Life Sciences, Inc., Boston, Mass.). DNA probes were labeled fluorescently by using an ECL direct nucleic acid labeling and detection system (Amersham Biosciences, Umeå, Sweden) in accordance with the manufacturer's instructions.
Plasmid curing of L. pseudomesenteroides CHCC2114.
Plasmid curing of L. pseudomesenteroides CHCC2114 was induced by growth at its maximum possible growth temperature; previous experiments determined that the maximum possible growth temperature of CHCC2114 was 34°C. CHCC2114 was grown at 34°C in M17 broth supplemented with 1% glucose (100 ml) to mid-exponential-phase growth (OD600, 0.45). Cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C, washed, and resuspended in the same medium to an OD600 of 0.20. A 1-ml aliquot of the cell suspension was used to inoculate 100 ml of the same medium, and the culture was incubated at 34°C until an OD600 of 0.20 was achieved. Cells were then plated on M17 agar supplemented with 1% lactose and containing 40 μg of X-Gal/ml. Following anaerobic incubation at 25°C, pinpoint white colonies were selected and plasmids were purified as described previously (2).
Measurement of diacetyl (acetoin) reductase activity.
Cell extracts of L. pseudomesenteroides were prepared by using an MM200 mixer-mill (Retsch, Haan, Germany). Cultures (50 ml) were grown in MRS broth for 48 h at 25°C, harvested by centrifugation at 5,000 × g for 15 min at 4°C, washed once with 50 mM sodium phosphate buffer (pH 6.0), and resuspended in 0.5 ml of the same buffer. In a 1.5-ml tube, 1.2 g of glass beads (diameter, 0.35 to 0.50 mm) was added to 0.5 ml of cell suspension. The tubes were shaken at 30 cycles s−1 for 6 min at 4°C in the MM200 mixer-mill and centrifuged at 12,000 × g for 10 min at 4°C. The diacetyl (acetoin) reductase activity and protein concentration were measured in the resulting supernatant. Diacetyl (acetoin) reductase activity was measured spectrophotometrically by monitoring the change in absorbance at 340 nm; an aliquot of enzyme solution (5 to 10 μl) was added to a preincubated 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.5) at 30°C. The reaction was initiated by the addition of NADH or NADPH (final concentration, 0.5 mM) and diacetyl or acetoin (final concentration, 37 mM) to the reaction mixture. The total reaction mixture volume was 1 ml, and the change in absorbance at 340 nm was monitored continuously over a period of 20 min. Controls, in which the cell extract was replaced by an equal volume of buffer, and assay blanks, in which the substrate was replaced by an equal volume of water, were also included in the experiment. One unit (1 U) of enzyme activity was defined as the amount of enzyme that converts 1 μmol of diacetyl and NAD(P)H to acetoin and NAD(P)+ per min at pH 5.5 and 30°C. Specific activity was defined as units per milligram of protein. Protein concentration was determined spectrophotometrically by using a bicinchoninic acid protein assay (Pierce, Rockford, Ill.) in accordance with the manufacturer's instructions. Bovine serum albumin (Pierce) was used as standard.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper is available at GenBank (www.ncbi.nlm.nih.gov) with the accession number AF438200.
RESULTS
Gene sequence analysis.
A total of 2,196 bp were sequenced by gene walking. An open reading frame (ORF) between positions 698 and 1,471 (GenBank numbering; accession no. AF438200) was identified as corresponding to the diacetyl (acetoin) reductase gene, butA, of L. pseudomesenteroides CHCC2114. A putative ribosome binding site, putative −35 and −10 boxes, and a TG dinucleotide located 1 bp upstream of the −10 box were identified and conform to the promoter consensus for lactococci and other gram-positive bacteria. The butA gene encodes a protein of 257 amino acids with a calculated molecular mass of 26,997 Da. The N-terminal amino acid sequence, namely, TKKVAMVTGGAQGIGEAIVRRLSADGFAVAVADL, as determined experimentally by direct amino acid sequencing of the purified protein, was identical to the predicted sequence encoded by the butA gene. Comparison of the amino acid sequence with the GenBank database showed that the product of the butA gene of L. pseudomesenteroides CHCC2114 had identities at the amino acid level of 97% to a 2,3-butanediol dehydrogenase from Bos taurus (29), 69% to an acetoin reductase from Lactococcus lactis subsp. lactis IL-1403 (5), 65% to an acetoin (diacetyl) reductase from Klebsiella pneumoniae (37), and 46% to diacetyl reductase from Lactococcus lactis subsp. lactis MG1363 (3). Comparison of the predicted amino acid sequence of the butA gene of L. pseudomesenteroides with the Prosite database demonstrated that it had the short-chain dehydrogenase-reductase family signature, namely, [LIVSPADNK]-x(12)-Y-[PSTAGNCV]-[STAGNQCIVM]-[STAGC]-K-{PC}-[SAGFYR]-[LIVMSTAGD]-x(2)-[LIVMFYW]-x(3)-[LIVMFYWGAPTHQ]-[GSACQRHM], where Y is an active-site residue (Prosite database accession no. PS00061). The short-chain dehydrogenase-reductase family is a relatively large family of enzymes, most of which are known to be NAD- or NADP-dependent oxidoreductases. Most members of this family of proteins are of about 250 to 300 amino acid residues.
A partial sequence of a second ORF, coding in the opposite orientation to the butA ORF, was also sequenced. This partial sequence, located between positions 1858 and 2196 (GenBank numbering), had amino acid identities of 93% to a protein of unknown function carried on pCI305 from Lactococcus lactis subsp. lactis UC317 (GenBank accession no. NC_002502), 91% to a DNA recombinase carried on a plasmid from Lactococcus lactis (GenBank accession no. U35629), 89% to a protein of unknown function from Listeria innocua (GenBank accession no. NC_003383), and 84% to a protein of unknown function carried on pLH1 from Lactobacillus helveticus (GenBank accession no. NC_002102).
ButA expression in E. coli.
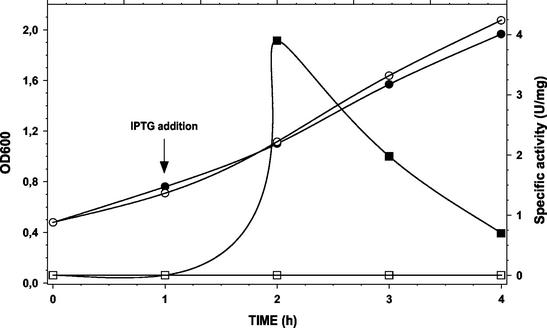
A PCR fragment of 1,526 bp, corresponding to the ribosome binding site and the entire coding region of the butA gene of L. pseudomesenteroides CHCC2114, was cloned downstream of the lac promoter of pPCR-Script Amp and transformed into E. coli. The correct orientation of the cloned fragment was confirmed by PCR. The addition of IPTG to the growth medium of exponentially growing E. coli transformants resulted in the induction of ButA activity (Fig. 1). Maximum ButA specific activity was measured 1 h after the addition of IPTG, after which the specific activity decreased. Toxicity of the expressed protein in E. coli may explain the decrease in specific activity observed 2 and 3 h after the addition of IPTG. Cloning of a larger PCR fragment of 2,163 bp, corresponding to the putative promoter −35 and −10 boxes, the ribosome binding site, and the entire coding region of the butA gene, was also attempted by using several different plasmid (pPCR-Script Amp, pCR2.1-TOPO, and pV2) and host strain (DH5α and XL10-Gold Kan) combinations, but no successful clones were obtained. The inability to clone this larger fragment containing the putative promoter sequence of the butA gene may also indicate toxicity of the expressed protein in E. coli.
FIG. 1.
Expression of L. pseudomesenteroides CHCC2114 ButA in E. coli and expression induced by the addition of IPTG (final concentration, 2 mM) to exponentially growing E. coli cells. Activity was measured in cell extracts of E. coli; 1 U of enzyme activity was defined as the amount of enzyme that converts 1 μmol of diacetyl and NAD(P)H to acetoin and NAD(P)+ per min at pH 5.5 and 30°C. •, OD600 with IPTG addition; ○, OD600 without IPTG addition; ▪, specific activity (U/mg) with IPTG addition; □, specific activity (U/mg) without IPTG addition.
Gene inactivation.
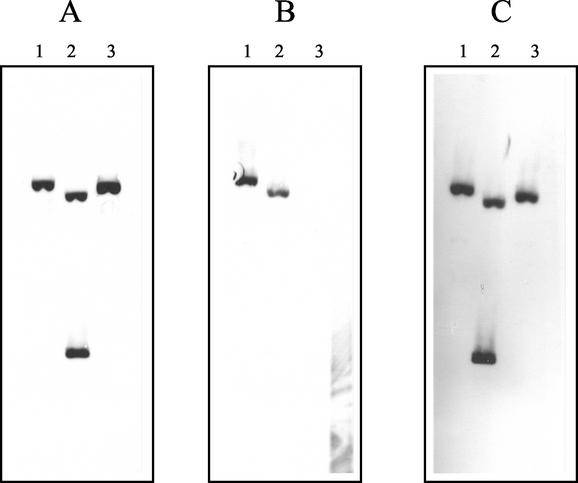
A two-step PCR approach was used to construct FPR02, a butA mutant of the wild-type strain, CHCC2114. The diacetyl reductase activities with NADH as coenzyme in cell extracts of CHCC2114 and FPR02 were 8.71 ± 0.21 and 0.73 ± 0.04 U mg−1, respectively (Table 3). These results indicate that an NADH-dependent diacetyl (acetoin) reductase gene has been deleted from the genome of FPR02. Southern blotting confirmed the deletion of the butA gene in FPR02 (Fig. 2). Probing of EcoRI-restricted total genomic DNA (chromosomal and plasmid) of FPR02 with a labeled fragment corresponding to the coding region of the butA gene failed to produce any hybridization bands. Probing of FPR02 with labeled fragments corresponding to regions upstream and downstream of the butA gene produced positive hybridization bands. Probing of the intermediate construct, FPR01, with a labeled fragment corresponding to the region upstream of the butA gene (Fig. 2) showed two distinct hybridization bands of similar intensity, indicating the homologous recombination of pDMP01 into the genome of CHCC2114. Similar results were obtained when probing of FPR01 with a labeled fragment corresponding to the region downstream of the butA gene. Probing FPR01 with a labeled fragment corresponding to the coding region of the butA gene showed a single hybridization band, also indicating the homologous recombination of pDMP01 into the genome of CHCC2114.
TABLE 3.
Diacetyl (acetoin) reductase activities of L. pseudomesenteroides strains
| Straina | Sp act (U mg−1) with the indicated substrate and coenzymeb
|
|||
|---|---|---|---|---|
| NADH
|
NADPH
|
|||
| Diacetyl | Acetoin | Diacetyl | Acetoin | |
| CHCC2114 | 8.71 ± 0.21 | 0.65 ± 0.05 | 1.39 ± 0.01 | 1.59 ± 0.00 |
| FPR02 | 0.73 ± 0.04 | 0.39 ± 0.06 | 1.95 ± 0.02 | 0.77 ± 0.03 |
| FPR03 | 0.65 ± 0.06 | 0.34 ± 0.01 | 1.27 ± 0.01 | 0.54 ± 0.01 |
| FPR04 | 7.68 ± 0.31 | 0.64 ± 0.02 | 1.59 ± 0.02 | 1.72 ± 0.06 |
CHCC2114, wild-type strain, ButA+ lactose fermenting; FPR02, DNA recombinant ButA− strain, lactose fermenting; FPR03, spontaneous ButA− strain, non-lactose fermenting; FPR04, ButA+ strain, non-lactose fermenting.
Results are presented as means ± standard deviations. One unit of enzyme activity was defined as the amount of enzyme that converts 1 μmol of diacetyl and NAD(P)H to acetoin and NAD(P)+ per min at pH 5.5 and 30°C.
FIG. 2.
Southern blot hybridization of L. pseudomesenteroides total genomic DNA (chromosomal and plasmid) restricted with EcoRI and probed with labeled PCR fragments. DNA was probed with a 486-bp fragment upstream of the butA gene (positions 20 to 505) (A), a 653-bp fragment within the coding region of the butA gene (positions 743 to 1395) (B), and a 359-bp fragment downstream of the butA gene (positions 1496 to 1854) (C). Lanes 1, CHCC2114 wild-type strain, ButA+; lanes 2, FPR01 DNA recombinant, ButA+; lanes 3, FPR02 DNA recombinant ButA− mutant.
These results indicate that the two-step PCR gene inactivation approach resulted in a mutant, FPR02, in which only the protein-coding region of the butA gene was precisely deleted from the genome. Hybridization with pV2 showed that the antibiotic resistance marker genes Amp and Em were absent in FPR02; additional phenotypical experiments demonstrated the sensitivity of FPR02 to AMP and ERY (results not shown).
The butA gene is plasmid carried.
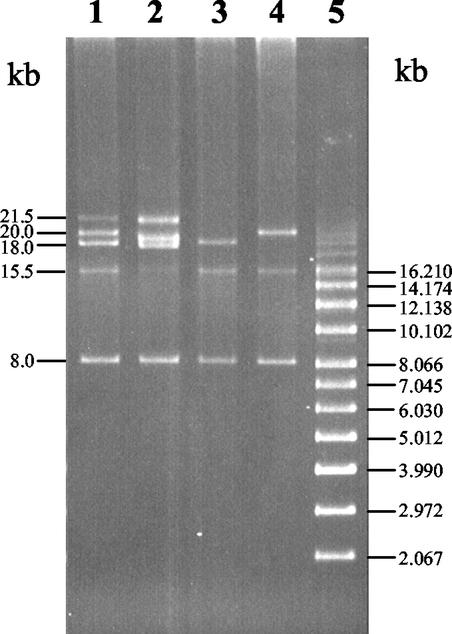
The plasmid profile of lactose-fermenting CHCC2114 showed five plasmids (Fig. 3) with estimated sizes of 21.5, 20, 18, 15.5, and 8.0 kb. Following 10 serial transfers of CHCC2114 in M17 broth in which lactose was replaced with glucose, the spontaneous non-lactose-fermenting mutants FPR03 and FPR04 were isolated. The non-lactose-fermenting mutants FPR03 and FPR04 had altered plasmid profiles compared to that of CHCC2114 (Fig. 3). The plasmid profiles of FPR03 showed the presence of the 18-, 15.5-, and 8-kb plasmids and the absence of the 21.5- and 20-kb plasmids, while the plasmid profile of FPR04 showed the presence of 20-, 15.5-, and 8-kb plasmids and the absence of 21.5- and 18-kb plasmids. The diacetyl reductase activities with NADH as the coenzyme in cell extracts of FPR03 and FPR04 were 0.65 ± 0.06 and 7.68 ± 0.31 U mg−1, respectively (Table 3). The ButA− and ButA+ phenotypes of FPR03 and FPR04, respectively, correlated with these plasmid profile differences; FPR03 contained the 18-kb plasmid but not the 20-kb plasmid, while FPR04 contained the 20-kb plasmid but not the 18-kb plasmid. These results indicate that the butA gene is carried on the 20-kb plasmid. Furthermore, additional evidence is provided by comparison of the plasmid profiles of CHCC2114 and FPR02; deletion of butA (a sequence of 774 bp) from the 20-kb plasmid is evident by the appearance of a slightly smaller plasmid of approximately 19 kb in FPR02. Sequencing (plasmid DNA) showed that the protein-encoding region of the butA gene was absent in FPR02; the upstream and downstream regions of the butA gene in FPR02 consisted of a single continuous sequence. Southern blot hybridization of plasmid DNA from CHCC2114 with a labeled probe corresponding to the protein-coding region of the butA gene confirmed that the butA gene is indeed located on the 20-kb plasmid (results not shown). Phenotypically, FPR03 and FPR04 were unable to utilize lactose; comparison of the plasmid profiles indicates that lactose utilization is associated probably with the 21.5-kb plasmid.
FIG. 3.
Plasmid profiles of L. pseudomesenteroides CHCC2114, FPR02, FPR03, and FPR04. Lane 1, CHCC2114 wild-type, ButA+, lactose-fermenting strain; lane 2, FPR02 DNA recombinant ButA− mutant, lactose-fermenting strain; lane 3, FPR03 spontaneous ButA−, non-lactose-fermenting strain; lane 4, FPR04 ButA+, non-lactose-fermenting strain; lane 5, supercoiled standard DNA plasmid molecular mass marker.
DISCUSSION
In this study, the DNA fragment carrying the butA gene of L. pseudomesenteroides CHCC2114 was subcloned, sequenced, and expressed in E. coli. Additionally, butA mutants of L. pseudomesenteroides were obtained by recombinant DNA technology and by traditional plasmid-curing methods. The nucleic acid sequencing demonstrated that the butA gene encodes a protein with a predicted molecular mass of 26,997 Da. This is in close agreement with the value of 26,910 ± 30 Da that was determined previously (25) for the purified enzyme by using-matrix assisted laser desorption-ionization-mass spectrometry. The predicted molecular mass of ButA of L. pseudomesenteroides was similar to the molecular masses of the acetoin reduc-tase of Klebsiella pneumoniae (37), the acetoin reductase of Lactococcus lactis subsp. lactis IL-1403 (5), and the diacetyl reductase of Lactococcus lactis subsp. lactis MG1363 (3), which were calculated to be 26,641, 27,081, and 27,545 Da, respectively.
The 97% identity of the butA product of L. pseudomesenteroides with the previously reported (29) sequence for a 2,3-butanediol dehydrogenase from Bos taurus (GenBank accession no. U71200) is surprising, considering the evolutionary distance between bacteria and higher eukaryotes. In our laboratory, by using reverse transcriptase PCR, bovine liver RNA as the template, and the Bos taurus primers listed in Table 2, it was not possible to verify the previously reported findings (29). The most likely explanation of the earlier reported sequence (29) is that the bovine cDNA library used in the experiments may have been contaminated with genomic DNA from L. pseudomesenteroides.
The inactivation of the butA gene in CHCC2114 was performed by using a two-step PCR approach in which the entire protein-coding region of the butA gene was deleted. With NADH as coenzyme, the butA mutant, FPR02, and the wild-type strain, CHCC2114, had diacetyl reductase activities of 0.73 ± 0.04 and 8.71 ± 0.21 U mg−1, respectively. However, with NADPH as coenzyme, the diacetyl reductase activities for FPR02 and CHCC2114 were 1.95 ± 0.02 and 1.39 ± 0.01 U mg−1, respectively. The detection of diacetyl-reducing activity with NADPH as coenzyme indicates either the presence in L. pseudomesenteroides of a second diacetyl reductase in which NADPH acts as the coenzyme or the presence of another NADPH-dependent enzyme which has diacetyl reductase activity as a side activity. Indeed, in a previous study by the authors (25), anion exchange chromatography separated the diacetyl-reducing activities of a cell extract of CHCC2114 into two distinct activity peaks; NADH- and NADPH-dependent diacetyl-reducing activities eluted at 0.15 and 0.25 M NaCl, respectively. The presence of two diacetyl reductases in a Leuconostoc species has been described in a previous report (22) in which crude extracts of L. paramesenteroides 9-1 were found to possess diacetyl-reducing activities specific for NADH and NADPH. The presence of more than one diacetyl reductase activity has been reported also in other organisms, with two different enzymes reported in Lactococcus lactis subsp. lactis biovar diacetylactis (formerly Streptococcus diacetylactis) (9) and between two and five enzymes for various species of Bacillus (33). The diacetyl-reducing activities of crude extracts of two strains of another Leuconostoc species, Leuconostoc lactis NCW1 and S3, have been reported to be specific only for NADPH (22). The diacetyl reductases of Enterobacter aerogenes (4, 31) and Kluyveromyces marxianus (27) have been reported to be specific for NADH, while for E. coli, the enzyme is specific for NADPH (28).
Plasmid-curing experiments, plasmid profiling, and Southern blotting (results not shown) indicate that the butA gene in L. pseudomesenteroides is carried on a 20-kb plasmid. This is the first report of the butA gene being plasmid carried in lactic acid bacteria. In Lactococcus lactis subsp. lactis IL-1403, the gene is chromosomally carried (5). In Lactococcus lactis subsp. lactis IL-1403, butA (putative) lies downstream of a putative 2,3-butanediol dehydrogenase gene (butB) and upstream of a putative ABC transporter ATP binding gene (yjcA). In Lactococcus lactis subsp. lactis MG1363, butA (experimental) is also carried on the chromosome (3) but lies downstream of a putative outer membrane lipoprotein precursor gene (plpC) and upstream of another putative outer membrane lipoprotein precursor gene (plpD). Interestingly, at the amino acid level, there is only 45% identity between the butA of Lactococcus lactis subsp. lactis IL-1403 and the butA of Lactococcus lactis subsp. lactis MG1363, reflecting considerable divergence within this gene. In Leuconostoc species, the technologically important genes encoding citrate and lactose utilization are also plasmid carried (18, 20, 34, 35). Phenotypically, FPR03 and FPR04 were unable to utilize lactose; comparison of the plasmid profiles indicates that lactose utilization is associated with the 21.5-kb plasmid. Downstream of the butA gene, a second ORF, coding in the opposite orientation to the butA gene, was partially sequenced. This partial sequence had 91% identity at the amino acid level to a DNA recombinase carried on a plasmid from Lactococcus lactis (GenBank accession no. U35629). Based on this significant homology, the ORF coding in the opposite direction to the butA gene is putatively identified as a DNA recombinase.
The butA mutants reported here might be useful in improving the flavor stability of certain fermented dairy products, such as cultured buttermilk, sour cream, and ripened cream butter, where the characteristic butter flavor note is due to the presence of diacetyl. Leuconostoc species, including Leuconostoc pseudomesenteroides, are used in mixed starter cultures together with Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. lactis biovar diacetylactis for the production of these fermented dairy products. The principal roles of Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. lactis biovar diacetylactis are acidification and diacetyl formation, while the role of the Leuconostoc component is the degradation of the acetaldehyde produced by the Lactococcus strains; acetaldehyde is an undesirable flavor compound in cultured buttermilk, sour cream, and ripened cream. However, in addition to degrading acetaldehyde, Leuconostoc species also reduce diacetyl into the relatively flavorless compounds acetoin and 2,3-butanediol due to these species' diacetyl reductase activity. Therefore, reduction of diacetyl by Leuconostoc species reduces the flavor stability of the fermented dairy product. However, the application of the butA mutant strains FPR02 or FPR03 in the production of fermented dairy products could be expected to improve the flavor stability by their inability to reduce diacetyl. Even though FPR02 differs from CHCC2114 by the deletion of just 774 bp from the 20-kb plasmid and contains no foreign DNA, it is considered to be genetically modified within the European Union (EU Council Directive 90/220/EEC) due to the techniques employed in its construction (electroporation and PCR). In contrast, FPR03 is not considered to be genetically modified according to EU legislation because it was produced by plasmid curing. The performance of FPR02 and FPR03 in milk fermentations are currently being investigated in our laboratory.
Acknowledgments
We thank Per Strøman, Martin B. Pedersen (Chr. Hansen A/S., Hørsholm, Denmark), and Eric Emond (Chr. Hansen Inc., Milwaukee, Wis.) for invaluable help and advice on many of the scientific aspects of this paper. Thanks are also due to Per Jensen, Slagteriskolen, Roskilde, Denmark, for kindly providing bovine liver samples and to Aine Healy, National Food Biotechnology Centre, University College, Cork, Ireland, for the N-terminal amino acid sequencing.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aungpraphapornchai, P., H. G. Griffin, and M. J. Gasson. 1999. Cloning, DNA sequence analysis, and deletion of a gene encoding diacetyl-acetoin reductase from Lactococcus lactis. DNA Seq. 10:163-172. [DOI] [PubMed] [Google Scholar]
- 4.Blomqvist, K., M. Nikkola, P. Lehtovaara, M.-L. Suihko, U. Airaksinen, K. B. Stråby, J. K. C. Knowles, and M. E. Penttila. 1993. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J. Bacteriol. 175:1392-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genet. Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogan, T. M. 1981. Constitutive nature of the enzymes of citrate metabolism in Streptococcus lactis ssp. diacetylactis. J. Dairy Res. 48:489-495. [Google Scholar]
- 7.Cogan, T. M., and K. N. Jordan. 1994. Metabolism of Leuconostoc bacteria. J. Dairy Sci. 77:2704-2717. [Google Scholar]
- 8.Cogan, T. M., M. O'Dowd, and D. Mellerick. 1981. Effects of pH and sugar on acetoin production from citrate by Leuconostoc lactis. Appl. Environ. Microbiol. 41:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crow, V. L. 1990. Properties of 2,3-butanediol dehydrogenases from Lactococcus lactis subsp. lactis in relation to citrate fermentation. Appl. Environ. Microbiol. 56:1656-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curic, M. 1998. Improving and stabilizing the aroma in fermented dairy products. Ph.D. thesis. Technical University of Denmark, Lyngby, Denmark.
- 11.Curic, M., B. Stuer-Lauridsen, P. Renault, and D. Nilsson. 1999. A general method for selection of α-acetolactate decarboxylase-deficient Lactococcus lactis mutants to improve diacetyl formation. Appl. Environ. Microbiol. 65:1202-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David, S., G. Simons, and W. M. de Vos. 1989. Plasmid transformation by electroporation of Leuconostoc paramesenteroides and its use in molecular cloning. Appl. Environ. Microbiol. 55:1483-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 14.Harrison, R. W., J. C. Miller, M. J. D'Souza, and G. Kampo. 1997. Easy gene walking. BioTechniques 22:650-653. [DOI] [PubMed] [Google Scholar]
- 15.Henriksen, C. M., D. Nilsson, S. Hansen, and E. Johansen. 1999. Industrial applications of genetically modified microorganisms: gene technology at Chr. Hansen A/S. Int. Dairy J. 9:17-21. [Google Scholar]
- 16.Hugenholtz, J. 1993. Citrate metabolism in lactic acid bacteria. FEMS Microbiol. Rev. 12:165-178.
- 17.Hugenholtz, J., and M. J. C. Starrenburg. 1992. Diacetyl production by different strains of Lactococcus lactis subsp. lactis var. diacelylactis and Leuconostoc spp. Appl. Microbiol. Biotechnol. 38:17-22. [Google Scholar]
- 18.Johansen, E., and A. Kibenich. 1992. Characterization of Leuconostoc isolates from commercial mixed strain mesophilic starter cultures. J. Dairy Sci. 75:1186-1191. [Google Scholar]
- 19.Johansen, E., P. Strøman, and E. B. Hansen. 1995. Genetic modification of lactic acid bacteria used in the production of food, p. 85-88. In Unanswered safety questions when employing GMO's. Coordination Commission Risk Assessment Research, Hoorn, The Netherlands.
- 20.Lin, J., P. Schmitt, and C. Diviès. 1991. Characterization of a citrate-negative mutant of Leuconostoc mesenteroides subsp. mesenteroides: metabolic and plasmidic properties. Appl. Microbiol. Biotechnol. 34:628-631. [DOI] [PubMed] [Google Scholar]
- 21.Marugg, J. D., D. Goelling, U. Stahl, A. M. Ledeboer, M. Y. Toonen, W. M. Verhue, and C. T. Verrips. 1994. Identification and characterization of the α-acetolactate synthase gene from Lactococcus lactis subsp. lactis biovar diacetylactis. Appl. Environ. Microbiol. 60:1390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellerick, D., and T. M. Cogan. 1981. Induction of some enzymes of citrate metabolism in Leuconostoc lactis and other heterofermentative lactic acid bacteria. J. Dairy Res. 48:497-502. [Google Scholar]
- 23.Phalip, V., C. Monnet, P. Schmitt, P. Renault, J. J. Godon, and C. Divies. 1994. Purification and properties of the alpha-acetolactate decarboxylase from Lactococcus lactis subsp. lactis NCDO 2118. FEBS Lett. 351:95-99. [DOI] [PubMed] [Google Scholar]
- 24.Ramos, A., N. K. Jordan, T. Cogan, and H. Santos. 1994. 13C nuclear magnetic resonance studies of citrate and glucose cometabolism in Lactococcus lactis. Appl. Environ. Microbiol. 60:1739-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rattray, F. P., M. Walfridsson, and D. Nilsson. 2000. Purification and characterization of a diacetyl reductase from Leuconostoc pseudomesenteroides. Int. Dairy J. 10:781-789. [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schwarz, J. G., and Y. D. Hang. 1994. Purification and characterization of diacetyl reductase from Kluyveromyces marxianus. Lett. Appl. Microbiol. 18:272-276. [Google Scholar]
- 28.Silber, P., H. Chung, P. Gargiulo, and H. Schulz. 1974. Purification and properties of a diacetyl reductase from Escherichia coli. J. Bacteriol. 118:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smania, A. M., and C. E. Argarana. 1997. Molecular cloning and characterization of a cDNA encoding a bovine butanediol dehydrogenase. Gene 197:231-238. [DOI] [PubMed] [Google Scholar]
- 30.Snoep, J. L., M. J. Teixeira de Mattos, M. J. C. Starrenburg, and J. Hugenholtz. 1992. Isolation, characterization, and physiological role of pyruvate dehydrogenase complex and α-acetolactate synthase of Lactococcus lactis subsp. lactis bv. diacetylactis. J. Bacteriol. 174:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Størmer, F. C. 1975. 2,3-Butanediol biosynthetic systems in Aerobacter aerogenes. Methods Enzymol. 41:519-533. [DOI] [PubMed] [Google Scholar]
- 32.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ui, S., H. Masuda, and H. Muraki. 1983. Stereospecific and electrophoretic natures of bacterial 2,3-butanediol dehydrogenases. J. Ferment. Technol. 61:467-471. [Google Scholar]
- 34.Vaughan, E. E., S. David, A. Harrington, C. Daly, G. F. Fitzgerald, and W. M. de Vos. 1995. Characterization of plasmid-encoded citrate permease (citP) genes from Leuconostoc species reveals high sequence conservation with the Lactococcus lactis citP gene. Appl. Environ. Microbiol. 61:3172-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan, E. E., S. David, and W. M. de Vos. 1996. The lactose transporter in Leuconostoc lactis is a new member of the LacS subfamily of galactoside-pentose-hexuronide translocators. Appl. Environ. Microbiol. 62:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vedamuthu, E. R. 1994. The dairy Leuconostoc: use in dairy products. J. Dairy Sci. 77:2725-2737. [Google Scholar]
- 37.Wardwell, S. A., Y. T. Yang, H. Y. Chang, K. Y. San, F. B. Rudolph, and G. N. Bennett. 2001. Expression of the Klebsiella pneumoniae CG21 acetoin reductase gene in Clostridium acetobutylicum ATCC 824. J. Ind. Microbiol. Biotechnol. 27:220-227. [DOI] [PubMed] [Google Scholar]