Abstract
A full-length cDNA (1,434 bp) of mitogen-activated protein kinase (MAPK), a key molecule of a signal transduction cascade, was isolated from the estuarine heterotrophic dinoflagellate Pfiesteria piscicida. This cDNA (Ppmapk1) encoded a protein (PpMAPK1) of 428 amino acid residues that shared about 30 to 40% amino acid similarity with MAPKs in other organisms. Phylogenetic analysis indicated that PpMAPK1 was tightly clustered with MAPK3 in protozoans. Using reverse transcription-PCR, expression of this gene was evaluated for P. piscicida cultures grown under different conditions. While salinity shock, heat shock, starvation, and a subsequent encounter with prey did not appear to affect expression of this gene, Ppmapk1 expression level was correlated with growth rate, suggesting involvement of this gene in the regulation of cell proliferation in the organism.
Pfiesteria piscicida Steidinger et Burkholder is an ambush predator dinoflagellate implicated in major fish kills in North Carolina and Maryland estuaries (5, 6). First discovered in 1988 (6), this organism constitutes a novel genus (34) whose phylogenetic affiliation has only recently been established (24, 32). Over the last decade, P. piscicida and closely related species have been intensively studied, mainly because of a suspected capability to produce potent neurotoxins and extremely complex life cycles, issues that are still under debate (2, 11, 25, 36). This organism is of high research interest also because it is capable of kleptoplastic photosynthesis (14, 21), although its life-supporting role appears to be more limited than previously thought (21). More recently, unusually extensive RNA editing in two mitochondrial genes was found in P. piscicida as well as in other dinoflagellates (23). Another outstanding characteristic of P. piscicida is its extraordinarily rapid response to chemical cues such as fish extract or excreta (4), suggesting an efficient signal transduction mechanism. In addition, P. piscicida is able to grow in a wide range of salinity conditions (0 to 35 practical salinity units [PSU]) (35), which implies the existence of an osmoregulation mechanism (4). Rapid changes in growth and life stage transformation are also suggested by the observation of population emergence (or excystment) upon encounter of fish prey and quick disappearance (or encystment) upon removal of the prey (4). Although all these previous results imply that an efficient signal transduction network may occur in P. piscicida, regulating cellular response to the external environment and cell proliferation or transformation, no signaling molecules have been identified for this organism.
Mitogen-activated protein kinase (MAPK) is potentially a useful molecule for us in efforts to gain understanding of the molecular mechanisms by which P. piscicida interacts with the environment and changes its growth states, particularly because of the multitude of functions that have been described for this molecule in other organisms. MAPK is a key component of the evolutionarily conserved signal transduction cascades consisting of MAPK/ERK (MAPK/extracellular signal-related kinase) that is activated by a MAPK/ERK kinase (MEK) which in turn is activated by a MEK kinase (33). This generic MAPK module takes on different forms (such as MAPK3, ERK, and stress-activated protein kinase [SAPK]) to produce parallel but interwoven MAPK signaling pathways that respond to different extracellular stimuli (1). These signal pathways occur widely in eukaryotes (yeasts, plants, and animals) and are involved in regulation of a variety of cellular activities (1, 7, 30, 33). For example, MAPK is known to relay extracellular cues to transcription factors in the nucleus in yeast (e.g., Saccharomyces cerevisiae) (13), protozoan (29), and mammalian cells (39). In the nematode Caenorhabditis elegans, a MAPK homolog (MEK7) has been found to play a role in stress response (18). Recent studies demonstrated a regulatory role of MAPK in chemotaxis of amoeboid cells (27, 37). MAPK is also involved in regulation of the cell division cycle and other reproduction activities. In yeast, the organism in which MAPK is best understood, MAPK signal transduction cascades function in modulating mating pheromone response, cell cycle control, and cell integrity (10, 15, 26, 28, 38). MAPK signaling pathways also mediate cell survival, cell death, and cell cycle arrest (3, 12, 16). A MAPK (p43Ntf6) in higher plants appears to play a role in regulating cytokinesis (7, 19), and a p42 MAPK in Xenopus embryos is required for exit from the M phase of the cell cycle (9). In studying how environmental factors affect growth of P. piscicida, we isolated and characterized a MAPK gene and found that this gene was expressed in a growth rate-dependent fashion.
MATERIALS AND METHODS
Algal cultures and sample collection.
P. piscicida (CCMP1831; Provasoli-Guillard National Center of Cultures of Marine Phytoplankton, Bigelow Laboratory, West Boothbay Harbor, Maine) was grown in 15-PSU seawater (filtered with a 0.45-μm-pore-size filter and autoclaved) and supplied with the cryptophyte Rhodomonas sp. (CCMP768) as food (40). The P. piscicida cultures were maintained at a concentration of 1.0 × 104 to 1.3 × 105 P. piscicida cells/ml, with regular feeding at a prey-to-predator ratio of 2 to 5 Rhodomonas sp. cells per P. piscicida cell. A photocycle of 12 h of light and 12 h of dark was provided, with the illumination at a photon flux of 100 microeinsteins m−2 s−1. Growth was monitored by taking 1-ml samples for cell count with a Sedgewick-Rafter chamber.
To collect samples for gene cloning, feeding was discontinued for the P. piscicida cultures for 36 h before sampling. After confirmation that very few cells of the food alga Rhodomonas sp. were present, samples were harvested using centrifugation at 3,000 × g for 20 min at 4°C and the cell pellet was resuspended in 1 ml of Trizol (Gibco BRL, Grand Island, N.Y.) and stored at −80°C until RNA extraction. In experiments to evaluate gene expression under different growth conditions, samples were collected in the same way but at differing time points (see below).
RNA extraction, cloning, and sequencing.
Frozen samples were thawed at room temperature and centrifuged at 10,000 × g for 1 min. Supernatants were transferred to a new tube, while cell pellets with about 50 μl of supernatant were frozen on dry ice for 2 min and then homogenized using a micropestle (catalog no. 84900; Fisher Scientific) until thawed. After brief centrifugation, the sample was frozen on dry ice again and homogenized. This procedure was repeated three times, and most cells were found broken when examined under the microscope. RNA was then isolated essentially as described previously (23). Throughout the study, RNA samples from all experiments were dissolved in autoclaved diethyl pyrocarbonate-treated H2O to reach a final concentration equivalent to 2 × 107 to 1 × 108 cells per ml. Following the method of Lin et al. (23), first-strand cDNA was synthesized using an amount of RNA equivalent to the same number of cells (about 105 cells) for all samples. A partial MAPK homolog was obtained by PCR amplification using a set of degenerate primers designed from the conserved regions (GR/SGAYG and EALAPHYF) of the gene in other organisms. Based on the sequence of the MAPK fragment that was cloned, specific primers were designed to clone the 5′ and 3′ ends of this gene by the RNA ligase-mediated rapid amplification of 5′ and 3′ ends technique (GeneRacer kit; Invitrogen). This technique allows only those mRNA molecules that have the methyl-Gppp cap on the 5′ end and a (poly)A tail on the 3′ end to be PCR amplified. Gene-specific primers used in this procedure were as follows: (i) for the 5′ end of the gene, 5′-CAGCTGAAATCCGTGTCGATGGAAC-3′ (PPMAPKR1) and 5′-TCACGAATCACCTTGACCGCGAC-3′ (PPMAPKR2); (ii) for the 3′ end of the gene, 5′-TACTTGCACTCCGCTGGCATTGTTC-3′ (PPMAPKF1) and 5′-CTCTTGAAGTTGTTAGATAAAGTTCC-3′ (PPMAPKF2). PCR amplification was carried out for 1 min at 95°C, followed by 35 cycles of 20 s at 94°C, 30 s at 58°C, and 40 s at 72°C. PCR products were cloned and sequenced using an ABI Prism 377XL DNA sequencer.
Phylogenetic analysis.
The nucleotide sequence of the full-length cDNA was first analyzed using the basic local alignment search tool (BLAST) program and GenBank databases for homolog search. The deduced amino acid sequence was then compared with those of related kinases from various organisms found in the search. Sequence alignments and phylogenetic analysis were performed as described previously (40). The reliability of the tree topology was evaluated by bootstrap analysis of 1,000 replicates. Because the cloned gene showed similarity to members of different subgroups of the MAPK gene family, representative members from each subgroup were included in the analysis to cluster with this dinoflagellate MAPK gene.
Feeding and starvation experiment.
When P. piscicida culture was growing in the exponential phase under a daily feeding scheme, feeding was discontinued for 36 h, and a sample was collected as a representation of the initial condition. The culture was then fed once with four to five Rhodomonas sp. cells per P. piscicida cell and harvested at 0, 4, 8, 12, 16, 20, 24, 28, 32, 36, 40, 44, 48, and 96 h after the feeding. Within 1 day, almost all of the prey algae were removed by P. piscicida, and the culture was considered starved after 36 h. All samples collected during this experiment were about 200 ml, and growth was monitored as mentioned above.
Osmotic shock and heat shock.
A P. piscicida culture was grown as described above in 15-PSU seawater, and feeding was discontinued for 28 or 36 h. For osmotic-shock experiments, a 600-ml culture was diluted with 1.2 liters of autoclaved distilled water to bring the salinity down to 5 PSU. Distilled water was added gradually (about 120 ml per min) while mixing. In parallel, another 600-ml, 15-PSU-grown culture was supplemented with 1.2 liters of 45-PSU artificial seawater to yield a salinity of 35 PSU. The 45-PSU seawater was also added at a rate of 120 ml per min. In both cases, samples of 600 ml from each culture were collected at 1, 6, and 24 h after osmotic shock for RNA extraction. The experiment was carried out twice for hypoosmotic shock and three times for hyperosmotic shock.
For the heat shock experiment, a culture was transferred from a 20°C culture room to a 30°C water bath, with a 1°C increment per min. Samples (200 ml) were collected in 1, 6, and 24 h after heat shock for RNA extraction. This experiment was separately conducted twice.
Growth rate experiments.
From several cultures grown under different conditions (starvation, feeding, osmotic shock, and heat shock), samples were collected at various time points when growth rates were different. Growth rates of the cultures at the time of sample collection were calculated as μ = (lnNt − lnN0)/t, where Nt and N0 are cell concentrations at times t and 0, respectively. The samples were processed using reverse transcription-PCR (RT-PCR) to determine the expression level of Ppmapk1 in the amount of RNA equivalent to 2 × 104 cells, which was normalized to the level of cob mRNA (see below). Statistical analysis was then performed to determine the correlation between growth rate and the normalized Ppmapk1 expression level.
RT-PCR.
RT-PCR was performed to evaluate the expression of Ppmapk1. RT-PCR was carried out as follows. Total RNA equivalent to the cell number of each sample was reverse transcribed into first-strand cDNA as mentioned above. Next, 1 μl of original (for Ppmapk1) or 50-fold-diluted (for mitochondrial cytochrome b; cob) first-strand cDNA solution (equivalent to 2 × 104 or 400 cells, respectively) was used as the template and EX Taq polymerase (Takara Shuzo) was used for PCR. cob (40) was also RT-PCR amplified to normalize expression levels of Ppmapk1. At the time of this study, no well-characterized dinoflagellate housekeeping gene was available for an RT-PCR control, and our preliminary tests showed that cob differed relatively little in cells under the different physiological and growth conditions used in this study (see Fig. 3). PCR was performed at 94°C for 20 s, annealing was performed at 58°C for 30 s, and extension was performed at 72°C for 40 s for 40 cycles (for Ppmapk1) or 25 cycles (for cob). The PCR amplification of both genes had been verified by a time-step PCR protocol to be in the exponential-increase phase (40). DNA band intensity was quantified using a gel documentation system (UVP) with the standard calibration mode. The MAPK primers used were PPMAPKF1 (forward) and PPMAPKR1 (reverse) (see above), specifying a 508-bp product. The primers for cob were PPCOB1F and PPCOB1R, as described previously (23), which specified a 628-bp product. Expression of Ppmapk1 was evaluated as the band intensity of this gene normalized to that of cob (mapk/cob).
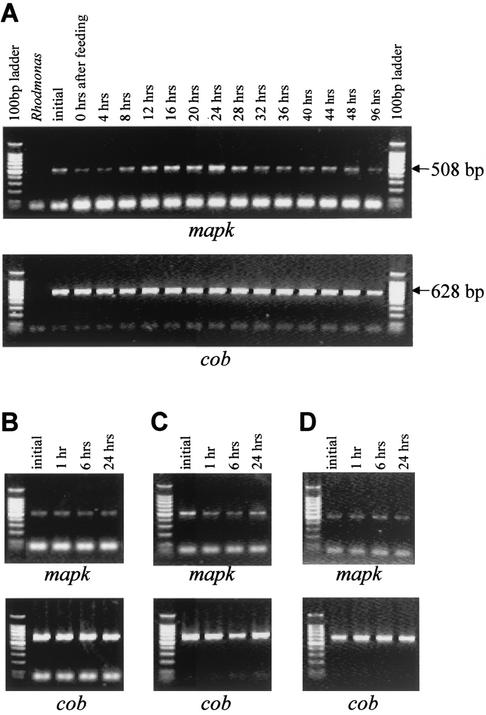
FIG. 3.
Expression of Ppmapk1 under differing feeding, salinity, and temperature conditions. Ppmapk1 expression level was determined using RT-PCR and was compared with that of cytochrome b examined simultaneously with Ppmapk1. Results from the two to three independent experiments within each treatment were similar, and only one from each treatment is shown here. (A) Feeding and starvation experiment. Samples were collected before the experiment (initial collection at about 36 h after previous feeding), at the beginning of the experiment (immediately after a feeding following a 36-h nonfeeding period), and at multiple time points after the beginning of the experiment (4 to 96 h after feeding). Rhodomonas, prey algal RNA used as a negative control. (B and C) Osmotic shock. P. piscicida cultured in 15-PSU seawater was osmotically shocked by adding autoclaved distilled water or 45-PSU artificial seawater to achieve 5 or 35 PSU, respectively, and sampled at 1, 6, and 24 h after osmotic shock. Initial samples were collected as a control about 36 and 28 h after feeding for hypoosmotic (B) and hyperosmotic (C) treatments, respectively. (D) Heat shock. Culture grown at 20°C was transferred to 30°C and samples were taken at 1, 6, and 24 h after heat shock. The initial sample was collected about 36 h after feeding. For all the experiments, RNA equivalent to the cell number was used in RT-PCR for each sample (2 × 104 cells for Ppmapk1 and 400 cells for cob).
RESULTS
Ppmapk1 gene sequence.
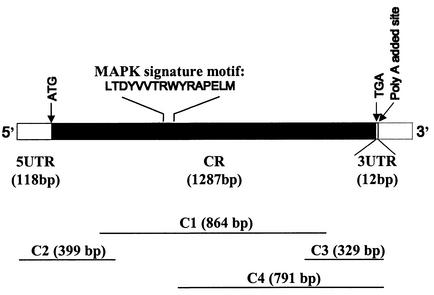
PCR using the degenerate primer set yielded a gene fragment of about 860 bp. Rapid amplification of both the 5′ and the 3′ ends led to the isolation of the full-length cDNA of Ppmapk1 (1,434 bp; GenBank accession no. AF227275). A BLAST search of the deduced protein of this gene (PpMAPK1) showed that the highest similarity was to MAPK of other organisms. Analysis of 17 clones revealed one identical Ppmapk1 sequence. Use of the specific primers designed on the basis of this sequence (PPMAPKF1 and PPMAPKR1) yielded a PCR product for cDNA from P. piscicida cultures but not for cDNA from Rhodomonas sp., although the amounts of RNA used in the RT-PCR were equivalent to the same numbers of cells for the two organisms, thus verifying the P. piscicida origin of this cloned gene. As shown in Fig. 1, the gene sequence consists of a coding region specifying 428 aa residues, a 118-bp untranslated region (UTR) at the 5′ end, and a short nucleotide tract (12 bp) of UTR at the 3′ end followed by the poly(A) tail.
FIG. 1.
Structure of the full-length MAPK cDNA of P. piscicida. The coding region (CR) consists of 1,287 bp, coding for 429 amino acid residues, flanked by a 118-bp UTR at the 5′ end (5UTR) and a short nucleotide tract (12 bp) at the 3′ end (3UTR) preceding the poly(A) tail (dotted-line open box at the 3′ end).
Phylogenetic analysis.
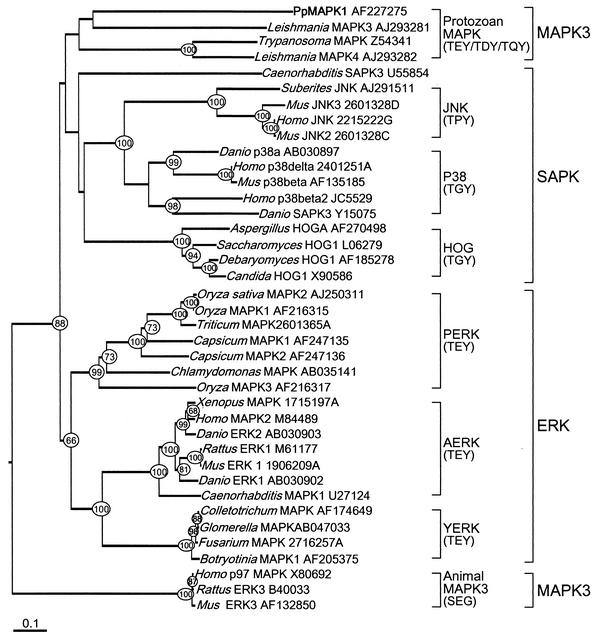
A multi-alignment of deduced amino acid sequences demonstrated the presence in PpMAPK1 of a domain unique to MAPKs, LTDYVVTRWYRAPELM, in which the TDY matched the highly conserved TXY motif for phosphorylation. The amino acid similarity of PpMAPK1 to counterparts in other organisms was about 30 to 40%. The alignment also indicated that the C and N termini of MAPK were highly variable among different organisms and that PpMAPK1 had the longer C terminus. Phylogenetic analysis using representative amino acid sequences from the three major MAPK subgroups indicated that the sequences were clustered primarily by the type of MAPK and secondarily by the group of organisms (Fig. 2). The sequences used in this analysis formed four major clades: animal MAPK3, ERK, and SAPK and protozoan MAPK3. Within clades ERK and SAPK, several subgroups were clearly identified. Closely related organisms were clustered together within the subgroups or clades. On this tree, PpMAPK1 was tightly clustered with protozoan MAPK3.
FIG. 2.
Rooted phylogenetic tree of MAPK family constructed by the neighbor-joining method (31) based on amino acid sequences. The representative members from all three major MAPK subgroups were used, and the tree was rooted by two cdc2-related protein kinases (GenBank accession no. 1815167F and AB005541). Circled numbers at nodes are bootstrap confidence values based on 1,000 replicates. Only values higher than 65% are shown. The tree was corrected by the Kimura method (17), and the scale bar denotes the number of amino acid substitutions per site. SAPK, SAPK subgroup; ERK, ERK subgroup; MAPK3, MAPK3 subgroup; JNK (c-Jun N-terminal protein kinase), animal SAPK1 cluster; p38, animal SAPK2 cluster; HOG, yeast SAPK cluster; PERK, plant ERK cluster; AERK, animal ERK cluster; YERK, yeast ERK cluster. The conserved dual phosphorylation motif TXY (threonine-variable amino acid-tyrosine) of each subfamily is shown in parentheses. Note that this motif is SEG in the animal MAPK3.
Expression of Ppmapk1.
As described above, use of primers PPMAPKF1 and PPMAPKR1 in RT-PCR yielded positive results only from P. piscicida RNA and not from the prey (Rhodomonas sp.) (Fig. 3 and 4), indicating the specificity of the primer set. RT-PCR results showed that Ppmapk1 appeared to be expressed constitutively at low levels in general. In the feeding and starvation experiment, only a faint RT-PCR band of Ppmapk1 was detected for the initial sample about 36 h after the previous feeding (0 to 4 h; Fig. 3A). Similarly low levels were constantly observed for cultures that were starved for differing periods of time (e.g., after 32 h; Fig. 3A), and the expression level decreased noticeably shortly after feeding (0 to 4 h; Fig. 3A). However, 12 to 24 h after feeding, the expression level of Ppmapk1 increased and reached its peak before it declined toward the background level again (Fig. 3A). The faint RT-PCR bands observed were not due to limited abundance of total RNA in those samples, because cob amplified under the same condition for fewer (25) cycles and with 1/50 of the cDNA amount used for Ppmapk1 yielded a strong and similarly abundant band for each reaction. The similar abundance of cob mRNA detected also verified that this gene was expressed at a more or less constant level under the conditions used in this study and that it was suitable for normalization of the Ppmapk1 expression level.
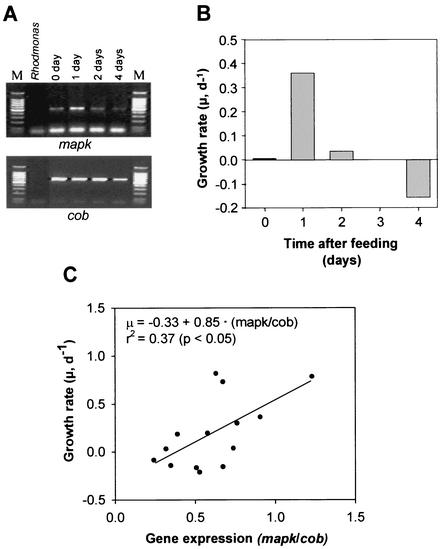
FIG. 4.
Relationship between Ppmapk1 expression level and growth rate of P. piscicida. (A) Gel electrophoresis of the RT-PCR products. The lane labeled Rhodomonas represents the prey algal RNA used as a negative control. The third lane represents samples taken immediately after feeding, and the fourth through sixth lanes represent samples taken on the indicated days after feeding. Lanes M, 100-bp DNA ladders. (B) Growth rates of the P. piscicida culture when the samples were collected for RT-PCR analysis. (C) Correlation between Ppmapk1 expression level (relative to cob) and growth rate. Data were obtained from multiple experiments. The correlation was found to be significant (P < 0.05 [t test; r = 0.61, df = 12]).
When a P. piscicida culture was osmotically shocked by the addition of distilled and deionized H2O or concentrated artificial seawater (45 PSU), no significant changes in Ppmapk1 expression in comparison to the background level were observed (Fig. 3B and C). Similarly, a low and invariable level of Ppmapk1 mRNA was seen for cultures under heat shock treatment (transfer of the culture from 20 to 30°C) (Fig. 3D).
We further examined the relationship between Ppmapk1 expression and growth rate. In the feeding and starvation experiment described above, elevation of Ppmapk1 expression in the 24 h after feeding coincided with an abrupt increase in the cell concentration of the culture. Another experiment showed that Ppmapk1 expression was at the background level when the P. piscicida culture was not fed for 36 h (day 0) and then increased remarkably 1 day after feeding (day 1) before it decreased to the background level again in subsequent days (Fig. 4A). In correspondence, the daily growth rate of this culture was near zero on day 0, 0.36 day−1 on day 1, and down to near or lower than zero subsequently (Fig. 4B). Regression of the data from several independent experiments revealed a correlation between Ppmapk1 expression level and growth rate of P. piscicida (Fig. 4C). A t test of the correlation coefficient (r = 0.61, df = 12) indicated that the correlation was significant (P < 0.05).
DISCUSSION
Identification of signal transduction pathways is critical for understanding the mechanisms by which cellular activities of unicellular organisms are regulated by environmental factors. The study reported here was the first attempt to isolate and characterize a component of a universal signal transduction pathway for dinoflagellates.
Conservation of MAPK and a unique structure in P. piscicida.
The results of this study demonstrate that P. piscicida possesses a MAPK gene, suggesting the presence of MAPK-based signal transduction pathways in this organism. Given that the MAPK gene family has been identified in a wide range of eukaryotes, the presence of MAPK in dinoflagellates was not unsuspected. The short 3′-end UTR found in P. piscicida appears to be distinct, however. How the short 3′ UTR affects Ppmapk1 mRNA is unclear, but it is tempting to speculate that it renders the mRNA less stable than a longer 3′ UTR would and results in a shorter lifetime of the mRNA and hence the relatively low mRNA abundance of this gene.
Phylogenetic analysis including various types of MAPK sequences from all major organisms indicates that PpMAPK1 is tightly clustered with its protozoan relatives, which agrees with the phylogenetic relationship derived on the basis of ribosomal rRNA genes (32). Our analysis reveals a discordance, however, in identifications of MAPK3 for animals and protozoa (Fig. 2). MAPK3 in animals is characterized by the SEG motif, whereas the protozoan homologs contain TEY, TDY, or TQY instead. The two MAPK3 clades are clearly distinct and should warrant reconsideration of their molecular identities by functional examination.
Ppmapk1 expression during starvation and feeding.
The exact function and mechanism by which Ppmapk1 works in P. piscicida cells warrant further studies using molecular and biochemical approaches. Given that Ppmapk1 is normally transcribed constitutively at low levels, this gene may constantly play an active role in mediating environmental conditions for the intracellular sites or in regulating cellular activities. Since P. piscicida has been found to respond to food supply quickly and appears to be a voracious predator (4; S. Lin, unpublished data), a MAPK signal transduction pathway may possibly be activated by starvation or feeding events. In the nematode C. elegans, a MAPK pathway was found to be associated with feeding (1). However, starvation in P. piscicida did not seem to alter expression of Ppmapk1, although the cytological effects, such as reduction in cell size (e.g., from 15.8 ± 3.8 μm to 8.7 ± 1.8 μm after 2 days of starvation [n = 200, P < 0.05]), were remarkable. Similarly, expression of this gene was not significantly changed immediately after feeding. Simultaneous measurement of cell counts showed that upon supply of algal prey, P. piscicida grazed very actively and removed the majority of the prey within 6 h (results not shown). These results indicate that starvation, encounter with food (i.e., addition of food to starved cultures), and active grazing shortly after prey addition do not have a direct influence on the expression of Ppmapk1.
Ppmapk1 expression in osmotic and heat shock.
Salinity and temperature are two important environmental factors that can potentially affect activities of P. piscicida cells, and such effects may be mediated through a MAPK pathway (20). In yeast, a MAPK (HOG1) functions as a signaling molecule for hypertonic osmotic shock. Given that yeast is a freshwater organism, induced expression of HOG1 upon exposure to high salinity (13, 14) is as expected. However, we observed no induction of Ppmapk1 expression when P. piscicida was under either hypertonic or hypotonic osmotic shock. Interestingly, this result seems to agree with the observation that P. piscicida is a euryhaline organism, growing well in conditions of salinity ranging from 5 to 35 PSU (4, 35). In our study, no unusual cell morphology or swimming pattern was seen for the culture under hypotonic osmotic shock, although an abnormal swimming pattern of cells was evident in the first 12 h under hypertonic salinity shock. No significant effects on long-term growth rates were observed in either of these cases. The possibility cannot be excluded, however, that either Ppmapk1 was involved at a posttranscriptional level or a Ppmapk1-independent signal transduction pathway was involved in the osmotic-shock responses in this species.
The heat shock treatment (from 20 to 30°C) of the P. piscicida culture did not seem to affect Ppmapk1 expression. Consistent with the absence of a Ppmapk1 signal, cell concentration of the heat-shocked culture did not decrease in comparison to that of the control. Instead, growth rate slight increased and cells became smaller (results not shown), as would be expected as a response to a temperature rise within the physiological range of the organism. This suggests that P. piscicida is tolerant of high temperature, which would be in agreement with the fact that P. piscicida has been found mostly in warmer locations and warmer seasons. Although it has been found in other areas as well (5), this organism appears to be more common in North Carolina and Maryland and tends to occur more in the summer season when water temperature is above 25°C (4, 5). Similar to the case of osmotic shocks, the possible alternative explanation of the lack of Ppmapk1 signaling in heat shock would be that an independent signal transduction unknown to us was activated in the heat shock response or Ppmapk1 was involved at a posttranscriptional level.
Association with cell proliferation and its potential as a growth rate estimator.
MAPK has been shown to be involved in the regulation of the cell division cycle. The regulatory functions range from regulation of cell cycle progression (13, 26, 38), cell cycle arrest (10, 12), and exit from M phase (9) to regulation of cytokinesis (7, 19). Our results showed that the expression level of Ppmapk1 was elevated dramatically when the growth rate of a P. piscicida population increased (Fig. 4), suggesting that PpMAPK1 might be involved in regulation of cell proliferation. The apparent association of PpMAPK1 with cell proliferation is supported by the positive correlation between Ppmapk1 expression level and growth rate found in this study. Although the data are somewhat scattered, the reason for which is not clear, the correlation is statistically significant. The correlation can be attributed to direct involvement of PpMAPK1 in cell division regulation or it may be a relationship derived from the association of PpMAPK1 with an unrecognized signal transduction pathway that is more active in actively proliferating cells. Nevertheless, the empirical correlation observed in this study can potentially be useful as a molecular marker for estimating the in situ growth rate of P. piscicida. Such a marker would be a useful alternative to the traditional cell cycle approach, in which high-frequency diel sampling is required for estimation of growth rate of microalgae through DNA analysis (8) or cell cycle protein analysis (22). However, more-extensive examination of the correlation between Ppmapk1 expression level and growth rate under different growth conditions is needed. In addition, although the primer set used in this study appeared to be specific for P. piscicida (negative results for Rhodomonas shown above; negative results for P. shumwayae and some other dinoflagellates not shown), specificity of the primer set needs to be examined further. Upon establishment of a robust correlation between growth rate and Ppmapk1 expression and a species-specific primer, application of this gene to estimations of P. piscicida growth rate in situ would become feasible.
Acknowledgments
We thank Timothy N. Feinstein for assistance with growing the cultures.
This research was supported by NOAA-ECOHAB grant NA86OP0491.
Footnotes
This study is dedicated to Dr. Edward J. Carpenter, the former advisor and mentor of S. Lin, for his 60th birthday. ECOHAB publication number 58.
REFERENCES
- 1.Berman, K. S., M. Hutchison, L. Avery, and M. H. Cobb. 2001. Kin-18, a C. elegans protein kinase involved in feeding. Gene 279:137-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry, J. P., K. S. Reece, K. S. Rein, D. G. Baden, L. W. Haas, W. L. Ribeiro, J. D. Shields, and R. W. Snyder. 2002. Are Pfiesteria species toxicogenic? Evidence against production of ichthyotoxins by Pfiesteria shumwayae. Proc. Natl. Acad. Sci. USA 99:10970-10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonni, A., A. Brunet, A. E. West, S. R. Datta, M. A. Takasu, and M. E. Greenberg. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286:1358-1362. [DOI] [PubMed] [Google Scholar]
- 4.Burkholder, J. M., and H. B. Glasgow, Jr. 1997. Pfiesteria piscicida and other Pfiesteria-like dinoflagellates: behavior, impacts, and environmental controls. Limnol. Oceanogr. 42:1052-1055. [Google Scholar]
- 5.Burkholder, J. M., H. B. Glasgow, and N. Deamer-Melia. 2001. Overview and present status of the toxic Pfiesteria piscicida (Dinophyceae). Phycologia 40:186-214. [Google Scholar]
- 6.Burkholder, J. M., E. J. Noga, C. H. Hobbs, and H. B. Glasgow, Jr. 1992. New “phantom” dinoflagellate is the causative agent of major estuarine fish kills. Nature 358:407-410. [DOI] [PubMed] [Google Scholar]
- 7.Calderini, O., L. Bogre, O. Vicente, P. Binarova, E. Heberle-Bors, and C. Wilson. 1998. A cell cycle regulated MAPK kinase with a possible role in cytokinesis in tobacco cells. J. Cell Sci. 111:3091-3100. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter, E. J., and J. Chang. 1988. Species specific phytoplankton growth rates via die1 DNA synthesis cycles. I. Concept of the method. Mar. Ecol. Prog. Ser. 43:105-111. [Google Scholar]
- 9.Chau, A. S.-S., and E. K. Shibuya. 1999. Inactivation of p42 mitogen-activated protein kinase is required for exit from M-phase after cyclin destruction. J. Biol. Chem. 274:32085-32090. [DOI] [PubMed] [Google Scholar]
- 10.Gachet, Y., S. Tournier, J. B. A. Millar, and J. S. Hyams. 2001. A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature 412:352-355. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, A. S., B. J. Dyer, D. Seaborn, and H. G. Marshall. 2002. Comparative toxicity of Pfiesteria spp., prolonging toxicity of P. piscicida in culture and evaluation of toxin(s) stability. Harmful Algae 1:85-94. [Google Scholar]
- 12.Gross, S. D., M. S. Schwab, A. L. Lewellyn, and J. L. Maller. 1999. Induction of metaphase arrest in cleaving Xenopus embryos by the protein kinase p90Rsk. Science 286:1365-1367. [DOI] [PubMed] [Google Scholar]
- 13.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAPK kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinstein, T. N., R. Traslavina, M.-Y. Sun, and S. Lin. 2002. Effects of light on photosynthesis, grazing, and population dynamics of the heterotrophic dinoflagellate Pfiesteria piscicida (Dinophyceae). J. Phycol. 38:659-669. [Google Scholar]
- 15.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khaled, A. R., A. N. Moor, A. Li, K. Kim, D. K. Ferris, K. Muegge, R. J. Fisher, L. Fliegel, and S. K. Durum. 2001. Trophic factor withdrawal: p38 mitogen-activated protein kinase activates NHE1, which induces intracellular alkalinization. Mol. Cell. Biol. 21:7545-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 18.Koga, M., R. Zwaal, K. Guan, L. Avery, and Y. Ohshima. 2000. A Caenorhabditis elegans MAP kinase kinase, MEK-1, is involved in stress responses. EMBO J. 19:5148-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krysan, P. K., P. J. Jester, J. R. Gottwald, and M. R. Sussman. 2002. An arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell 14:1109-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kultz, D., and M. Burg. 1998. Evolution of osmotic stress signaling via MAP kinase cascades. J. Exp. Biol. 201:3015-3021. [DOI] [PubMed] [Google Scholar]
- 21.Lewitus, A. J., H. B. Glasgow, Jr., and J. M. Burkholder. 1999. Kleptoplastidy in the toxic dinoflagellate Pfiesteria piscicida (Dinophyceae). J. Phycol. 35:305-312. [Google Scholar]
- 22.Lin, S., J. Chang, and E. J. Carpenter. 1995. Growth characteristics of phytoplankton determined by cell cycle proteins: PCNA immunostaining of Dunaliella tertiolecta. J. Phycol. 31:388-395. [Google Scholar]
- 23.Lin, S., H. Zhang, D. Spencer, J. Norman, and M. W. Gray. 2002. Widespread and extensive editing of mitochondrial mRNAs in dinoflagellates. J. Mol. Biol. 320:727-739. [DOI] [PubMed] [Google Scholar]
- 24.Litaker, R. W., P. A. Tester, A. Colorni, M. G. Levy, and E. J. Noga. 1999. The phylogenetic relationship of Pfiesteria piscicida, Cryptoperidiniopsoid sp. Amyloodinium ocellatum and a Pfiesteria-like dinoflagellate to other dinoflagellates and apicomplexans. J. Phycol. 35:1379-1389. [Google Scholar]
- 25.Litaker, R. W., M. W. Vandersea, S. R. Kibler, V. J. Madden, E. J. Noga, and P. A. Tester. 2002. Life cycle of the heterotrophic dinoflagellate Pfiesteria piscicida (Dinophyceae). J. Phycol. 38:442-463. [Google Scholar]
- 26.Madden, K., Y.-J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 27.Maeda, M., and R. A. Firtel. 1997. Activation of the mitogen-activated protein kinase ERK2 by the chemoattractant folic acid in Dictyostelium. J. Biol. Chem. 19:23690-23695. [DOI] [PubMed] [Google Scholar]
- 28.Metodiev, M. V., D. Matheos, M. D. Rose, and D. E. Stone. 2002. Regulation of MAPK function by direct interaction with the mating-specific Gα in yeast. Science 296:1483-1486. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima, S., S. Wang, N. Hisamoto, H. Sakai, M. Andoh, K. Matsumoto, and Y. Nozawa. 1999. Molecular cloning and expression of a stress-responsive mitogen-activated protein kinase-related kinase from Tetrahymena cells. J. Biol. Chem. 274:9976-9983. [DOI] [PubMed] [Google Scholar]
- 30.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Saldarriaga, J. F., F. J. R. Taylor, P. J. Keeling, and T. Cavalier-Smith. 2001. Dinoflagellate nuclear SSU rDNA phylogeny suggests multiple plastid losses and replacements. J. Mol. Evol. 53:204-213. [DOI] [PubMed] [Google Scholar]
- 33.Seger, R., and E. G. Krebs. 1995. The MAPK signaling cascade. FASEB J. 9:726-735. [PubMed] [Google Scholar]
- 34.Steidinger, K., J. M. Burkholder, H. B. Glasgow, Jr., C. W. Hobbs, J. K. Garrett, E. W. Truby, E. J. Noga, and S. A. Smith. 1996. Pfiesteria piscicida gen. et sp. nov. (Pfiesteriaceae fam. nov.), a new toxic dinoflagellate with a complex life cycle and behavior. J. Phycol. 32:157-164. [Google Scholar]
- 35.Sullivan, B. E., and R. A. Andersen. 2001. Salinity tolerance of 62 strains of Pfiesteria and Pfiesteria-like heterotrophic flagellates (Dinophyceae). Phycolol. Res. 49:207-214. [Google Scholar]
- 36.Vogelbein, W. K., V. J. Lovko, J. D. Shields, K. S. Reeve, P. L. Mason, L. W. Haas, and C. C. Walker. 2002. Pfiesteria shumwayae kills fish by micropredation not exotoxin secretion. Nature 418:967-970. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Y., J. Liu, and J. E. Segall. 1998. MAP kinase function in amoeboid chemotaxis. J. Cell Sci. 111:373-383. [DOI] [PubMed] [Google Scholar]
- 38.Whitehall, S., P. Stacey, K. Dawson, and N. Jones. 1999. Cell cycle-regulated transcription in fission yeast: cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol. Biol. Cell 10:3705-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, Z., P. J. Woodring, K. S. Bhakta, K. Tamura, F. Wen, J. R. Feramisco, M. Karin, J. Y. J. Wang, and P. L. Puri. 2000. P38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 20:3951-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, H., and S. Lin. 2002. Identification and quantification of Pfiesteria piscicida by using the mitochondrial cytochrome b gene. Appl. Environ. Microbiol. 68:989-994. [DOI] [PMC free article] [PubMed] [Google Scholar]