Abstract
Outbreaks of listeriosis and febrile gastroenteritis have been linked to produce contamination by Listeria monocytogenes. In order to begin to understand the physiology of the organism in a produce habitat, the ability of L. monocytogenes to attach to freshly cut radish tissue was examined. All strains tested had the capacity to attach sufficiently well such that they could not be removed during washing of the radish slices. A screen was developed to identify Tn917-LTV3 mutants that were defective in attachment to radish tissue, and three were characterized. Two of the three mutations were in genes with unknown functions. Both of the unknown genes mapped to a region predicted to contain genes necessary for flagellar export; however, only one of the two insertions caused a motility defect. The third insertion was found to be in an operon encoding a phosphoenolpyruvate-sugar phosphotransferase system. All three mutants were defective in attachment when tested at 30°C; the motility mutant had the most severe phenotype. However, not all of the mutants were defective when tested at other temperatures. These results indicate that L. monocytogenes may use different attachment factors at different temperatures and that temperature should be considered an important variable in studies of the molecular mechanisms of Listeria fitness in complex environments.
Listeria monocytogenes is a gram-positive pathogen that causes a multitude of symptoms, including septicemia, abortion, liver failure, and meningitis (50). When L. monocytogenes is not living as an intracellular invader, it can be isolated from the feces of animals as well as humans and from the soil, where it can survive as a saprophyte for 10 to 12 years. From these locations, it can contaminate the food supply in many ways. It can survive in sewage and enter the soil, where it may become associated with plants and/or farm animals before entering food-processing plants, grocery stores, and home refrigerators (2, 10). In all of these niches, L. monocytogenes can survive and even thrive, leading to contamination of foods and ingestion of the pathogen. L. monocytogenes is quite hardy under a variety of environmental stresses. It has been reported that it can grow at temperatures of between −0.4 and 50°C and withstand extremes of osmotic pressure, as evidenced by growth in 10% NaCl, and it displays a pH range of 4.3 to 9 (18, 21).
While food contamination with L. monocytogenes is most often associated with smoked meats and dairy products, fresh produce has also been implicated in outbreaks and sporadic cases of listeriosis. Specific produce-related outbreaks of listeriosis and febrile gastroenteritis caused by L. monocytogenes have been associated with contaminated cabbage, corn, and lettuce and/or celery (8, 30, 51). In addition, this organism has caused the recall of vegetables, including red peppers, sprouts, and lettuce, and potato salad (4-7, 13). L. monocytogenes has been isolated from radishes, potatoes, cucumbers, and cabbage in surveys of produce from U.S. grocery stores (29). Others have isolated it from corn and grain plants, lettuce, parsley, and watercress (46, 58). Many studies have indicated that L. monocytogenes can grow or survive on fresh or processed produce, including asparagus, broccoli, cauliflower, and leafy vegetables, and it has been isolated from many prepared salads (9, 16, 22, 28, 35). The only types of produce that seem to inhibit growth or kill L. monocytogenes are tomatoes and carrots (11, 42).
Most studies of produce-related L. monocytogenes have involved surveying for the presence of the bacterium, monitoring its growth, or testing various chemicals or conditions for killing it. Few studies have addressed the fundamental questions of what allows the bacterium to associate with produce initially and how it remains attached. In order to begin to understand the physiology of L. monocytogenes when it is associated with a plant surface, we developed a freshly cut radish model system. Radishes were chosen since L. monocytogenes has been cultured from them previously, and it has been suggested that Listeria may be present more frequently on plants that grow closer to the ground or in contact with soil (29, 45). We are interested in the fundamental physiology of L. monocytogenes in the plant environment because of the potential relevance to its entrance into processing plants and biofilm formation on plants and processing equipment and ultimately on food surfaces.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The strains used in this study are listed in Table 1. L. monocytogenes strains were routinely grown in TSYE medium, which consisted of tryptic soy broth without dextrose (Difco, Becton Dickinson, Franklin Lakes, N.J.) and 0.6% yeast extract. Listeria minimal medium was made as described previously (48). Escherichia coli was routinely cultured in Luria-Bertani medium, consisting of 1% tryptone, 0.5% yeast extract, and 0.5% NaCl. Solid media contained 1.5% agar. Motility agar contained 1% tryptone, 0.5% NaCl, and 0.5% agar. For enumerating L. monocytogenes in attachment assays, modified Oxford agar (MOX agar; Difco) was used. When necessary, the antibiotics used for L. monocytogenes were 1 μg of erythromycin/ml and 25 μg of lincomycin/ml for strains containing Tn917-LTV3 insertions and 10 μg of colistin/ml and 20 μg of moxalactam/ml to reduce background bacterial growth during mutant enrichment from radish tissue. For E. coli, when necessary, 40 μg of kanamycin/ml or 100 μg of ampicillin/ml was used. Unless otherwise indicated, L. monocytogenes was routinely incubated at 30°C, and liquid cultures were incubated with agitation. E. coli was incubated at 37°C. Bacteriophage transductions with φCU-SI153/95 (φU153) were performed with methods and media described previously (31). Phosphate-buffered saline (PBS) contained 150 mM NaCl and 10 mM sodium phosphate (pH 7.2).
TABLE 1.
Strains, bacteriophage, and plasmid used in this study
| Strain, bacteriophage, or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| L. monocytogenes | ||
| 10403 | Human clinical isolate; serotype 1/2a | D. Portnoy, University of California, Berkeley |
| F2379 | Human clinical isolate; serotype 4b | D. Portnoy |
| ScottA | Human clinical isolate; serotype 4b | D. Portnoy |
| RM2387 | Survey isolate from mint; serotype 4b | This study |
| RM2388 | Survey isolate from mint; serotype 1/2b | This study |
| RM2217 | Garlic surveillance sample; serotype 1/2b | G. Inami and M. Janda, California Department of Health Services |
| RM2218 | Oyster surveillance sample; serotype 4b | G. Inami and M. Janda |
| RM2980 | 10403::Tn917-LTV3Ω001 | This study |
| RM2981 | 10403::Tn917-LTV3Ω002 | This study |
| RM2982 | 10403::Tn917-LTV3Ω003 | This study |
| RM3094 | 10403::Tn917-LTV3Ω004 | This study |
| E. coli | ||
| DH10B | Cloning strain | D. Kaiser, Stanford University |
| Qiagen EZ | Cloning strain | Qiagen |
| Bacteriophage φCU-SI153/95 (φU153) | L. monocytogenes transducing phage | D. Hodgson, University of Warwick, Warwick, United Kingdom |
| Plasmid pDrive | Cloning vector; Kanr Ampr | Qiagen |
Aqueous suspension attachment assay.
Strains were screened for their ability to associate with freshly cut radish tissue in an aqueous suspension. Fresh radishes from the grocery store were used within 1 day of purchase. Triangle-shaped wedges were cut with a scalpel so that each slice had one red edge. The weight of the radish slices was 0.05 to 0.2 g, and the width was approximately 1 to 2 mm. Once radish slices were prepared and weighed, they were placed individually into 10 ml of sterile water in a 15-ml conical tube. For all samples, the L. monocytogenes cells were grown at the same temperature as that at which attachment was tested. A suspension of L. monocytogenes cells in PBS at a concentration of 108 CFU/ml was prepared, and 10 μl was added to a tube containing a radish slice. In some experiments, a zero-time-point sample was taken within 5 min of the addition of cells to the radish tube.
The assay tubes were placed at an angle within a test tube rack and incubated with shaking at various temperatures. At various time points, the radish slices were removed from the tubes. When necessary, the number of L. monocytogenes cells in the assay water was determined by dilution plating on MOX agar. The radish slices were washed in test tubes (12 by 75 mm) that contained 2 ml of sterile water. Each slice was washed three times by vortexing for 1 to 2 s in three successive wash tubes before being placed in a microcentrifuge tube containing 0.3 ml of PBS. The radish slices then were crushed with a pestle connected to a cordless rotary tool (Dremel, Racine, Wis.) operated at 7,500 rpm for approximately 5 to 10 s. Immediately after crushing, 0.7 ml of PBS was added to the ground radish tissue, and the suspension was dilution plated on MOX agar. After 2 days of incubation at 30°C, the Listeria-type colonies (bluish white surrounded by a black precipitate) were counted, and the number of L. monocytogenes CFU per gram of radish tissue was calculated. Controls with no L. monocytogenes added were plated on MOX agar to ensure that the radishes were not contaminated naturally with Listeria.
Direct application assay and attachment mutant enrichment.
To select for mutants unable to attach to cut radish tissue, a pool of random Tn917-LTV3 insertion mutants of L. monocytogenes (a kind gift from D. Portnoy, University of California, Berkeley) was used (15). The pool was grown overnight at 30°C and diluted to 107 CFU/ml in PBS. Ten microliters of the cell suspension was added directly to multiple radish slices cut as described above. The slices were placed on a glass slide, which then was placed on top of two sticks laid flat in a petri plate containing a 7-cm circle of Whatman no. 1 filter paper. The filter paper was wetted with sterile water, and the entire assembly was incubated at 30°C for 1 h. The radish slices then were washed three times as described above for the attachment assay. The wash fluids were pooled and placed in a sterile flask. Seven milliliters of TSYE medium was added to the pooled wash fluids, and antibiotics were added. Erythromycin and lincomycin were added to ensure the retention of Tn917-LTV3, and colistin and moxalactam were added to reduce the growth of the background flora from the radish slices. This new culture was incubated overnight and then used to repeat the attachment, washing, and regrowth of unattached cells. In this fashion, the selection procedure was continued for 14 consecutive days. On each day, the numbers of L. monocytogenes cells in the wash fluids and on the radish slices were monitored by dilution plating on MOX agar. On days when the number of L. monocytogenes CFU in the wash fluids was the highest, 96 colonies were chosen and screened individually with the attachment assay described above.
When strains were screened for attachment by this direct application protocol, 10 μl of a suspension containing 106 CFU/ml in PBS was applied to the radish slices. After incubation for 1 h at 30°C, the radish slices were removed from the petri plate and washed and plated as described above for the radish slices in an aqueous suspension.
Cloning and sequencing of the regions surrounding Tn917-LTV3 insertions.
All of the Tn917-LTV3 insertion strains chosen for additional study were backcrossed into the parental strain by bacteriophage transduction with φU153 (31). To establish that each strain contained only one insertion, chromosomal DNA was screened by Southern blot hybridization with a probe containing a portion of the lacZ gene present in Tn917-LTV3. Cloning of the region upstream of the insertions was done as described previously (15). Briefly, the region was cloned in situ by digestion of the chromosomal DNA with either XbaI or XhoI and then religation and selection in E. coli. The region upstream of the insertions was sequenced by using the primer 5′-CAATAGAGAGATGTCACCG-3′, which is complementary to the beginning of the lacZ sequence contained in the transposon (24).
Cloning of intact regions from the chromosome.
Once the regions upstream of the transposon insertions were sequenced, they were compared with the published sequence of L. monocytogenes strain EGD-e (25, 41). PCR primers were designed on the basis of the published sequence and were used to amplify 4- to 5-kb regions surrounding the sites identified by Tn917-LTV3 insertions. The fli region was cloned by using primers 10G4rgnfor (5′-CTACGCGAAATTGATGATGTTGACTCC-3′) and 10G4rgnrev (5′-TGATTTCGTAGCCGTATGTTTCTCCTC-3′). The primers used to clone the dulcitol phosphotransferase system (PTS) region were 11A7up (5′-AGTCGCTTCATCAATTTGTTCTGCT-3′) and 11A7down (5′-ATTATTCCTGCGCTCATCGTGGTTCT-3′). The regions were amplified from strain 10403 chromosomal DNA with the Expand High-Fidelity PCR System (Roche, Indianapolis, Ind.), cloned by using a Qiagen Inc. (Valencia, Calif.) PCR Cloning Plus Kit, and sequenced to compare strain 10403 with strain EGD-e. Sequencing was done with both ABI 310 and ABI 3100 sequencers by using Big Dye Terminator version 3.0 (Applied Biosystems, Foster City, Calif.).
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was done to determine whether genes identified by transposon insertions were cotranscribed with any open reading frames (ORFs) downstream. RNA was isolated from L. monocytogenes strain 10403. Cells from 1 ml of a mid-exponential-phase culture were lysed by incubation for 10 min at room temperature in 10 mM Tris-HCl- 1 mM EDTA buffer (pH 8) containing 10 mg of lysozyme/ml and 20 U each of phage lysins Hpl 118 and Hpl 511 (37, 38). RNA was then extracted by using the Rneasy Mini Kit (Qiagen). Contaminating DNA was removed by using both an RNase-free DNase Set (Qiagen) and the DNA-free DNase kit (Ambion, Austin, Tex.). RT-PCR was performed by using a ProSTAR HF Single-Tube RT-PCR System (Stratagene, La Jolla, Calif.). For the Ω001-Ω002 region, the primers used were orf2for (5′-CCGCCAAATTGAAGTCTTACCTATGCT-3′) and orf5rev (5′-GGTATTCTTCGCTTTCTTCGGGATTGA-3′). RNA was reverse transcribed for 30 min at 42°C and immediately amplified by using a cycle program of 1 min at 95°C; 37 cycles of 30 s at 95°C, 30 s at 56°C, and 3.5 min at 68°C; and 10 min at 68°C. The expected size of the product was 2,745 bp. For the Ω003 region and its downstream neighbor, the primers used were Ω003for (5′-TCGGGAATACGGAGCAGCAGAG-3′) and Ω003rev (5′-TTAGTCCGAGTACGAAAATCAAAT-3′). The cycling program for the Ω003 region RT-PCR was 15 min at 42°C; 1 min at 95°C; 40 cycles of 30 s at 95°C, 30 s at 55°C, and 2 min at 68°C; and 10 min at 68°C. The expected size of the Ω003 region RT-PCR product was approximately 1,700 bp.
Determination of β-galactosidase activity.
Cells were prepared and assayed for β-galactosidase activity as described previously (23, 40). All cultures were harvested at the same cell densities. Units of β-galactosidase activity were defined as nanomoles of o-nitrophenol produced per minute.
Determination of generation times.
Generation times were determined by diluting early- to mid-log-phase cultures grown at 30°C in TSYE medium to a density of 108 cells/ml in PBS. Ten microliters of this suspension was added to 10 ml of sterile water containing a 0.1-g radish slice, and the Listeria-radish mixture was incubated at 30°C. The number of Listeria cells was determined by dilution plating on MOX agar. The generation times were calculated by using the formula log x = log x0 + (0.301/g)(t), where x is the final cell number, x0 is the initial cell number, t is time, and g is generation time (59).
Statistics and reproducibility.
Unless otherwise indicated, all experiments were done at least three times with three or four replicates each. All data shown are means of representative experiments with standard deviations. Student's t test was used to assess confidence intervals.
Nucleotide sequence accession numbers.
The DNA sequences reported in this study have been filed under GenBank accession numbers AF545863 and AY150228.
RESULTS
Attachment of L. monocytogenes strains to radish tissue.
Several L. monocytogenes strains were selected to assay their ability to attach to radish tissue. The strains were chosen to represent various serotypes and environmental sources (Table 1). Strains 10403, F2379, and ScottA are all human clinical isolates. Strains RM2387, RM2388, RM2217, and RM2218 were isolated during surveys of plants or oysters. Each strain was tested at least twice, and representative data (expressed as log CFU per gram of radish tissue) are shown in Table 2. Each strain was tested after 2 h of incubation with radish tissue at 30°C by the aqueous suspension attachment assay described above. The attachment assay involved washing the radish slices before crushing and plating; therefore, the data represent L. monocytogenes cells that remained attached to the radish tissue. In all instances, the starting concentration of L. monocytogenes in the water was 105 CFU/ml. This L. monocytogenes concentration was not high enough to saturate binding to the radish tissue. When the concentration of the starting inoculum was increased or decreased 10-fold, the resulting attachment data changed accordingly (data not shown).
TABLE 2.
Attachment of different strains to cut radish tissue after 2 h in aqueous suspension
| Strain | Serotype | Attachment (log CFU/g of radish tissue) |
|---|---|---|
| 10403 | 1/2a | 5.05 ± 0.15 |
| F2379 | 4b | 5.28 ± 0.10 |
| ScottA-1 | 4b | 4.84 ± 0.09 |
| ScottA-2 | 4b | 4.78 ± 0.19 |
| RM2217 | 1/2b | 5.17 ± 0.09 |
| RM2218 | 4b | 4.76 ± 0.38 |
| RM2387 | 4b | 5.12 ± 0.28 |
| RM2388 | 1/2b | 4.75 ± 0.17 |
The strains tested represented three different serotypes and attached to the plant material similarly in this assay (P > 0.05) (Table 2). Neither the serotype nor the source of the strain affected attachment. The range of the cells attached was not very large and spanned from 4.75 log CFU/g of radish for strains RM2388 (plant isolate; serotype 1/2b) and RM2218 (oyster survey isolate; serotype 4b) to 5.38 log CFU/g of radish for strain RM2387 (mint survey isolate; serotype 4b). Two different isolates of ScottA (ScottA-1 and ScottA-2; both serotype 4b) were tested and showed similar levels of attachment, 4.83 and 4.78 log CFU/g of radish. Thus, the ability to attach to radish slices appeared to be similar among the L. monocytogenes strains tested and independent of the source and serotype of the strains.
Effect of temperature on attachment to radish tissue.
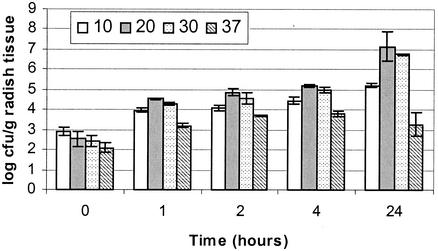
The ability of L. monocytogenes to attach to radish tissue at 10, 20, 30, and 37°C is shown in Fig. 1. The levels of attachment at both 20 and 30°C were similar over the first 4 h tested, reaching approximately 5 log CFU/g after 4 h, and they reached greater than 6 log CFU/g after 24 h. Attachment at 10°C lagged behind that at 20 and 30°C after time zero, reaching a level of 4.5 log CFU/g after 4 h. However, when tested at 24 h, the level of attachment at 10°C (∼5 log CFU/g) was equivalent to the 4-h levels at 20 and 30°C. At 37°C, the attachment levels were at least 1 log unit below those at the other three temperatures at each time point after time zero. The time-zero data were obtained within 5 min after the addition of bacteria to the radish slices. The level of attachment at 37°C after 24 h was similar to that at 4 h at the same temperature. The lower level of attachment measured at 37°C may reflect physiological changes in L. monocytogenes at 37°C relative to the other temperatures; however, it may also reflect changes in the radish tissue that affect the attachment and growth of L. monocytogenes. After 4 h at 37°C, the radish tissue was noticeably softened, and the red color on the edge was beginning to fade away. The increased degradation of the radish tissue at 24 h may have reduced the number of available attachment sites for L. monocytogenes.
FIG. 1.
Attachment of strain 10403 to radish tissue at different temperatures (degrees Centigrade). The aqueous suspension protocol was used to determine attachment levels. Cultures were originally grown at the temperatures at which they were tested. Experiments were done at least in duplicate, and representative results are shown as means and standard deviations.
Measuring attachment after 24 h of incubation is also an indirect measure of the growth of L. monocytogenes in the radish suspension. After a 1- to 2-h lag time (>6 h at 10°C), the bacteria grew quite well in the radish suspension. Therefore, more cells were available by 24 h to attach to the radish slices. The increased levels of attachment at 10, 20, and 30°C reflect the increased numbers of cells present, whereas the level of attachment at 37°C does not.
Selection of mutants defective in radish attachment.
We reasoned that in a pool of random mutants, we could enrich for and select those that were defective in radish attachment by using cut radish tissue as an affinity matrix to remove the attachment-competent cells from the suspension. The direct application of cells to a radish slice was chosen for the selection procedure because a greater percentage of cells attached to the radish slice in this procedure than in the aqueous suspension assay. However, even in a suspension of wild-type L. monocytogenes, not every cell attaches to the radish slice. At least 50 to 70% of the total cells applied are removed by washing. In order to produce a population enriched for attachment-defective mutants, we used a library of random pooled Tn917-LTV3 insertions of L. monocytogenes. Cells of L. monocytogenes were applied to and washed from radish slices, and then medium was added to the pooled wash fluids in order to enrich the unattached population of cells. Additional cycles of sequential attachment, washing, and regrowth were done for a period of 2 weeks. On each day, the numbers of L. monocytogenes cells both in the wash fluids and on the radish slices were monitored. On day 1, the pooled wash fluids contained 65% of the total input cells. On day 10, 88% of the input cells were removed by washing, and on day 11, 90% were removed by washing. There was a decrease in the percentage to 73% by day 14; therefore, 96 colonies were selected at random from each of the day-10 and day-11 unattached populations, for a total of 192 strains, for further screening. In addition, 96 colonies were chosen at random from a nonenriched plating of the random pooled Tn917-LTV3 insertions to determine whether the enrichment protocol had, in fact, yielded attachment mutants.
Each of the 192 strains was tested with both the direct application assay used originally to select them and the aqueous suspension assay used to screen multiple strains of L. monocytogenes. There was no significant difference in the attachment of strains tested by the direct application assay (with one exception, discussed below). Each strain bound to levels within 1 log unit of the level of the wild-type control strain.
When the 192 enriched strains were screened in the aqueous suspension assay, 5 showed reduced attachment (at least 1 log unit) to radish tissue when tested at 30°C. None of the 96 nonenriched insertion strains had a defect in attachment, indicating that the mutant enrichment protocol was successful. The five mutants were tested on motility agar, and four were nonmotile. All five mutants were analyzed by Southern blot hybridization to determine the number of transposons they contained. The four nonmotile strains each had two transposons, as indicated by two bands hybridizing to lacZ on the Southern blot (data not shown). Moreover, the bands were of the same size, suggesting that the four strains were siblings, and further analysis indicated that this was the case. All of the mutants were backcrossed into parental strain 10403 by using bacteriophage φU153 in order to ensure that the transposon was indeed causing the attachment phenotype and to separate the insertions in double-mutant strains. Several transductants obtained from the cross of the double-mutant strain into the wild type still contained two insertions, indicating that those particular transposon insertions (designated Ω001 and Ω002) probably were in close proximity to each other on the chromosome. This observation may indicate a hot spot for Tn917 transposition. The two insertions were separated, and only the strain with insertion Ω001 had a motility defect, with a 2.4-log-unit reduction in attachment at 30°C. The strain with insertion Ω002 was motile and, surprisingly, showed a reduction in attachment of 1.7 log units at 30°C. A third mutant strain, containing insertion Ω003, had no motility defect and showed a reduction in attachment of 1.8 log units at 30°C (Table 3).
TABLE 3.
Attachment of wild-type and mutant strains to radish tissue at different temperatures
| Strain | Motilitya | Attachment (log CFU/g of tissue) at the following temp (°C):
|
|||
|---|---|---|---|---|---|
| 10 | 20 | 30 | 37 | ||
| 10403 | + | 4.52 ± 0.14 | 4.87 ± 0.06 | 5.19 ± 0.33 | 3.71 ± 0.05 |
| Ω001 | − | 2.94 ± 0.19 | 3.25 ± 0.15 | 2.80 ± 0.31 | 3.87 ± 0.15 |
| Ω002 | + | 3.72 ± 0.26 | 5.01 ± 0.15 | 3.46 ± 0.12 | 4.17 ± 0.32 |
| Ω003 | + | 4.11 ± 0.09 | 4.72 ± 0.12 | 3.41 ± 0.17 | 3.93 ± 0.10 |
| Ω004b | + | 4.24 ± 0.21 | 4.56 ± 0.22 | 5.33 ± 0.21 | 3.63 ± 0.29 |
Motility was tested at 30°C. +, motile; −, nonmotile.
Transposon control strain.
The mutants were tested at temperatures other than 30°C, with variable results (Table 3). The Ω001 mutant strain attached at a lower level (1 to 2 log units) than the wild type at 10 and 20°C (P < 0.05). At 37°C, when L. monocytogenes normally does not produce flagella and thus all strains are nonmotile, attachment of the Ω001 mutant was equivalent to that of the wild type. It should be noted that wild-type strain 10403 had severely decreased motility at 37°C but retained some limited motility, as revealed on motility agar (L. Gorski, unpublished data). The Ω002 mutant also showed a decrease in attachment at both 10 and 30°C (P < 0.05), but at the other temperatures, it attached at a level similar to that of the wild type. The Ω003 mutant displayed an attachment defect at 30°C and a small but significant one at 10°C (P < 0.05) and was otherwise similar to the wild type.
Since the Ω002 insertion mutant had an attachment defect after separation of the Ω001 and Ω002 insertions, we were concerned that the act of phage transduction, used to perform the backcrosses, might have caused an attachment defect. Therefore, a Tn917-LTV3 insertion mutant with no attachment defect and not exposed to the mutant enrichment procedure was selected at random and backcrossed into strain 10403. The resulting strain (designated the Ω004 strain) had no attachment defect and was included as a transposon control strain (Table 3).
As stated above, none of the 192 mutants showed a marked attachment defect relative to the wild type in the direct application assay, with the exception of the Ω001 mutant. The values for the attachment of strain 10403 (wild type) and the Ω001, Ω002, and Ω003 mutants to radish slices at 30°C in the direct application assay were 3.9 ± 0.09, 3.3 ± 0.13, 3.8 ± 0.08, and 3.9 ± 0.06 log CFU/g of radish tissue, respectively. The initial inoculum used for this assay was different from that used for the aqueous suspension assay, so the attachment results cannot be directly compared between the two protocols. The attachment of the mutant carrying the Ω001 insertion was significantly (P < 0.05) reduced from that of the wild type, by 0.6 log unit. The Ω002 and Ω003 insertion mutants attached at levels similar to that of the wild type.
Identification of regions disrupted by Tn917-LTV3 insertions.
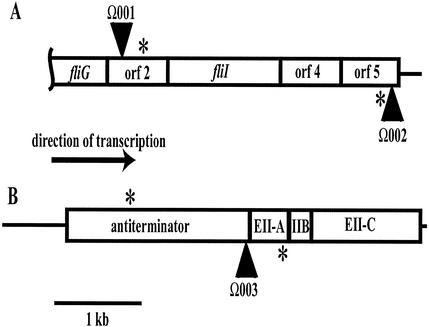
The regions upstream of the Ω001, Ω002, and Ω003 insertions were cloned and partially sequenced to identify the genes that were disrupted. These sequences were then compared with the published sequence of L. monocytogenes strain EGD-e. PCR primers were designed to amplify 4 to 5 kb of the regions flanking the insertions. These regions were then cloned from strain 10403, sequenced, and compared to the sequence of strain EGD-e. The physical maps of the regions are illustrated in Fig. 2. The Ω001 and Ω002 insertions, which were inserted in different directions, both were located in genes with no known functions but mapped to a region predicted to contain genes necessary for flagellar assembly. The motility defect in the Ω001 insertion strain could be the result of a disruption of either the target gene (ORF 2) or the fliI gene immediately downstream, if those genes are cotranscribed. The fliI gene product is necessary for the export of flagellum-related proteins (21). Scanning electron microscopy of the Ω001 insertion strain indicated the absence of flagella on its surface (data not shown). ORF 4 in Fig. 2A is predicted to encode a transglycosylase. Both the 10403 and the EGD-e sequences indicated that there is a translation terminator immediately downstream of ORF 5, and the results of RT-PCR experiments suggested that ORF 5 is indeed the end of the operon (data not shown). The Ω001 and Ω002 insertions, which were determined by phage transduction to be physically linked (see above), were in fact 3,147 bp apart from each other on the 10403 chromosome. The DNA sequence of this region is 99.3% identical between strains 10403 and EGD-e.
FIG. 2.
Physical structures of the regions disrupted by Tn917-LTV3 insertions. (A) Chromosomal region of the Ω001 and Ω002 insertions. (B) Chromosomal region of the Ω003 insertion. The gene names given are those identified in the L. monocytogenes EGD-e sequencing project. Unknown genes are labeled as ORFs. Tn917-LTV3 insertions are indicated by triangles. The placement of the triangle on the sequence indicates the orientation of the insertion. The arrow indicating the direction of transcription is relevant for both panels A and B. The asterisks indicate the positions of primers used for RT-PCR to determine cotranscription with genes downstream of the insertions. EII-A, IIB, and EII-C, enzymes IIA, IIB, and IIC, respectively.
The ORF disrupted by Ω003 is predicted to encode a transcription antiterminator of the BglG family (41). This type of antiterminator is associated with the phosphoenolpyruvate-sugar PTS and is regulated in a substrate-specific manner to allow transcription of the genes necessary to produce products to transport that substrate (27, 55). Downstream of the ORF disrupted by Ω003 (Fig. 2B) are ORFs predicted to encode the enzyme IIA, IIB, and IIC components of the PTS for the sugar galactitol (dulcitol). The antiterminator is usually the first gene in an operon encoding specific transport proteins (27). The DNA sequence of the identified PTS region is 99.4% identical between strains 10403 and EGD-e.
RT-PCR of disrupted regions.
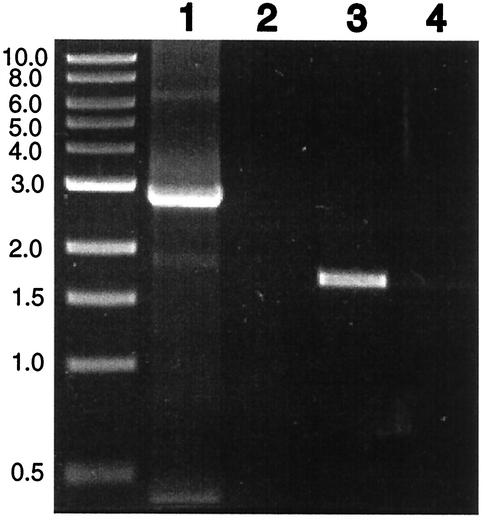
To determine whether the genes disrupted by the transposon insertions were cotranscribed with ORFs downstream and hence whether the mutations could have polar effects, RT-PCR was performed with whole-cell RNA on the two regions identified and illustrated in Fig. 2. Since Ω001 and Ω002 were inserted within 3 kb of each other in a potential operon, primers that hybridized to regions within the disrupted ORFs but between the insertion sites were designed. The results shown in Fig.3 illustrate that these genes are normally cotranscribed, since an expected 2.745-kb PCR product resulted from the mRNA. Figure 3 also shows that the gene disrupted by the Ω003 insertion is cotranscribed with its immediate downstream neighbor (expected 1.7-kb PCR product). The fact that the no-RT control lanes (Fig. 3, lanes 2 and 4) were devoid of PCR products indicates that the resulting bands in the other lanes were from mRNA and not contaminating DNA in the preparation. Therefore, both the Ω001 and the Ω003 insertion mutations could have polar effects on other genes in their respective operons. It follows then that the mutations responsible for the attachment phenotypes (and, in the case of Ω001, the motility defect) could be the result of disruptions of these genes or genes downstream from them.
FIG. 3.
RT-PCR of insertion regions. RNA was isolated from strain 10403 for RT-PCR. Primers used to determine the cotranslation of insertions Ω001 and Ω002 were orf2for and orf5rev (lanes 1 and 2), and the expected product was 2.745 kb. Primers used to determine the cotranslation of insertion Ω003 and the gene downstream of it were Ω003for and Ω003rev (lanes 3 and 4), and the expected product was 1.7 kb. The locations of the primers on the chromosome are indicated by asterisks in Fig. 2. Lanes 2 and 4 are controls that contained no RT to test the purity of the RNA preparations. Size markers are in kilobases.
Temperature-dependent expression of targeted promoters.
Because of the variable attachment phenotypes of the Ω002 and Ω003 insertion mutants at different temperatures, we wondered about the effect of temperature on gene expression. The Tn917-LTV3 insertion element contains a promoterless lacZ gene; therefore, if the transposon is inserted in the correct orientation, the activity of the affected promoter can be measured by the production of β-galactosidase. The Ω002 and Ω003 insertions are in the correct orientation for such a measurement. The amount of β-galactosidase activity produced is shown in Table 4. Although some of the values were quite low, the background values were even lower. When the assay was performed with the Ω001 insertion strain, which contains the lacZ gene in the opposite orientation, β-galactosidase activity was less than 0.1 U (data not shown). Since the two affected genes are cotranscribed, the lacZ Ω002 insertion is expressed by the same upstream promoter as the Ω001 insertion. Transcription of the Ω001-Ω002 operon was lowest at 37°C. The Ω003 insertion and its operon were expressed at all temperatures tested, but the lowest expression occurred at 30°C, the temperature at which the Ω003 mutant displayed its most severe attachment defect (Table 3).
TABLE 4.
β-Galactosidase expression of Ω002 and Ω003 insertions at various temperatures
| Temp (°C) | β-Galactosidase expression (nmol of o-nitrophenol produced/min)a of the following insertion:
|
|
|---|---|---|
| Ω002 | Ω003 | |
| 10 | 2.9 ± 0.1 | 10.8 ± 0.7 |
| 20 | 4.2 ± 0.1 | 7.7 ± 0.5 |
| 30 | 3.1 ± 0.5 | 4.9 ± 0.7 |
| 37 | 1.2 ± 0.1 | 20.4 ± 11 |
Normalized for cell density.
Growth studies with the Ω003 insertion strain.
Since the region surrounding the Ω003 insertion was predicted to contain a sugar transport PTS operon, we hypothesized that the attachment defect in that mutant strain could be a growth defect, which would limit the quantity of cells recovered in the attachment assay. Therefore, the generation time of the Ω003 insertion strain in an aqueous radish suspension was measured as described above. The doubling times of the wild-type (strain 10403) and Ω003 strains at 30°C were not significantly different, with the wild type and the Ω003 mutant doubling every 57 and 61 min, respectively. Therefore, the possible transport defect in the Ω003 strain did not result in a growth defect during the attachment assay.
Since the operon affected in the Ω003 strain is proposed to be involved in the transport of dulcitol, it was important to determine whether the Ω003 strain could grow on dulcitol. However, we determined that even wild-type L. monocytogenes does not grow on dulcitol, either as a sole carbon and energy source or cometabolized in TSYE medium. Saklani-Jusforgues et al. (49) reported that an L. monocytogenes Tn917lac insertion mutant was incapable of growth on the sugar arabitol and had a disruption in the dulcitol transport operon, specifically in a gene three ORFs downstream from the gene disrupted by Ω003 (25, 41). The Ω003 strain was incapable of utilizing arabitol as a sole carbon and energy source, nor could it cometabolize arabitol as a supplement to TSYE medium. However, the Ω003 mutant did grow on minimal medium with glucose as the sole carbon and energy source. In comparison, wild-type strain 10403 grew on minimal medium containing either glucose or arabitol, suggesting that the operon mutated in the Ω003 strain is required for arabitol metabolism.
DISCUSSION
Outbreaks caused by L. monocytogenes-contaminated foods and the serious illnesses and fatalities that occur in susceptible individuals illustrate the importance of understanding the fundamental mechanisms of L. monocytogenes survival, growth, and virulence as they relate to food production. To understand how the bacterium establishes a niche and persists on food-processing equipment, the attachment of L. monocytogenes to abiotic surfaces, such as stainless steel and rubber, has been studied (32, 52, 53, 57). We have developed a model system with cut radish tissue to study the molecular mechanisms involved in L. monocytogenes attachment to plant surfaces. This is the first report of which we are aware to begin to study the details of the molecular interactions of L. monocytogenes with plant tissue. Radishes were chosen for this study for several reasons. L. monocytogenes has been isolated from radishes, and the organism may be more likely to be found on root vegetables or on portions of plants closer to the ground (29, 45). Radishes grow relatively quickly, lending themselves well to in situ experiments for future studies. Also, radishes usually are not cooked before consumption.
Seven strains of L. monocytogenes representing different serotypes and sources were tested for the ability to attach to radish tissue at 30°C (Table 2). The ability was intrinsic and reproducible in all of the strains tested. Therefore, with our limited set of strains, plant strains were not better adapted for plant attachment than nonplant strains. This result suggests that the mechanisms involved in this interaction with radish tissue are present in most strains of L. monocytogenes.
Temperature regulates many virulence and environmental genes in L. monocytogenes, sometimes with resulting cell surface changes (3, 12, 14, 34, 36, 43). These changes could affect attachment, so the ability of L. monocytogenes strain 10403 to attach to radish tissue was tested at temperatures of between 10 and 37°C. Temperature did play a role in the attachment phenotype (Fig. 1). The attachment levels at 37°C were low at all times tested. A possible explanation for this result is temperature-regulated physiological changes. Motility and flagellar biosynthesis are, for example, down-regulated at 37°C. Therefore, cells may not be able to move to potential attachment sites.
We used the direct application method for the mutant enrichment protocol, since a greater proportion of cells remained associated with the radish slices with this method than in aqueous suspension. Therefore, fewer cycles of attachment and regrowth were necessary to produce an increase in the number of cells removed by washing. Also, since we had speculated that flagella might play a role in attachment, we searched for other genes associated with attachment. We reasoned that in the aqueous suspension assay, motility-related mutants would dominate the enrichment culture. It should be noted that the enrichment for mutants was done at 30°C, thereby selecting for mutations important for attachment at 30°C. The fact that the mutants ultimately chosen for further analysis had different attachment phenotypes at various temperatures does not change the conclusion that defects were expressed at 30°C. As mentioned above, there are examples of physiological changes in L. monocytogenes grown at different temperatures, so perhaps it is not surprising that different components play roles in attachment to different surfaces at different temperatures. These results indicate the need for future studies of attachment of L. monocytogenes in food production environments and the importance of temperature as a factor. While successful, the enrichment protocol used here to select mutants did have drawbacks. Attachment-defective mutants with associated growth defects would not be enriched by this method, since they would be overgrown by growth-competent strains. Also, there is a risk of a mutant becoming dominant in the culture. Indeed, we did see the latter effect, since the strain carrying insertions Ω001 and Ω002 was represented four times in the 192 mutants picked.
The most severe attachment defect that we observed was that of the Ω001 insertion mutant, which was nonmotile as well as attachment defective. Flagella have been shown to act as mediators of attachment in several species of bacteria, including L. monocytogenes attachment to stainless steel (19, 44, 47, 57). In addition, motility has been shown to play a role in attachment in the initiation of biofilm formation (33). The Ω001 insertion is downstream of the gene predicted to encode FliG, which is a component of the flagellar motor, and upstream of the gene predicted to encode FliI (56). In Salmonella enterica serovar Typhimurium and Caulobacter crescentus, the fliI gene product has been demonstrated to be an ATPase necessary for the transport of flagellar components, and deletion of that gene leads to a nonmotile phenotype (20, 54). The cotranscription of the Ω001 ORF with fliI (Fig. 3) makes the identity of the precise cause of the defect unclear.
The gene affected by the Ω002 insertion shows no homology to known genes. The physical structure of the fliI operon in L. monocytogenes is different from those described for gram-positive and gram-negative bacteria, so we can make no assumptions about the function of the ORF affected by Ω002 based on context (1, 26, 54). Since other genes in the Ω001-Ω002 region are important for flagellar assembly and function, the gene disrupted by Ω002 may play a role in these mechanisms. The gene disrupted by Ω002 may be involved in the synthesis of an ancillary flagellar component necessary for attachment to surfaces. This operon is expressed at very low levels at 37°C (Table 4), a fact which may explain why neither Ω001 nor Ω002 strains had attachment defects at 37°C.
The operon affected by the Ω003 insertion was predicted to encode a sugar transport PTS. Despite the fact that the genome sequence indicated that this operon was important for dulcitol transport, L. monocytogenes could not grow on dulcitol. Rather, our data as well as those of Saklani-Jusforgues et al. (49) suggest that this operon is important for arabitol transport. The growth of this mutant in an aqueous radish suspension resulted in a generation time equivalent to that of the wild type at 30°C, so we concluded that the defect in attachment at 30°C and the slight defect at 10°C were not the result of a growth defect during the course of the assay. The Ω003 mutant attached at a level similar to that of the wild type at the other temperatures tested. It remains unclear what role a sugar transport system would play in attachment. Readily metabolized sugars, such as glucose, cellobiose, fructose, and mannose, have been shown to regulate virulence gene expression in L. monocytogenes, and it has been speculated that the organism senses the presence of some sugars not present in a mammalian host and down-regulates virulence gene expression outside the host (14, 39). It is possible that a sugar transport system affects other cellular systems via catabolite repression, although little is known about such systems in L. monocytogenes (14, 17).
The measurement of promoter activity by β-galactosidase assays (Table 4) indicated that the Ω003 operon was expressed at the lowest level at 30°C. However, interpreting β-galactosidase expression for this insertion may be problematic because the disrupted gene is predicted to encode a transcription antiterminator of the BglG type, the disruption of which may lead to misregulation of the operon (and hence the lacZ Ω003 insertion). On the other hand, since the Ω003 insertion for the antiterminator is at the extreme 3′ end of the gene (Fig. 2), the gene product may be partially functional.
Of the three mutants identified, only the Ω001 mutant showed a defect in the direct application assay (attachment value, 3.3 ± 0.13 log CFU/g of radish tissue). This result was somewhat unexpected, since all of the mutants were originally selected by that assay. It is possible that slight differences in attachment were enhanced and magnified during the multiple enrichment steps. It is also possible that different mutations selected for in the complex, competitive enrichment culture environment are not important when the strains are screened individually. Different types of attachment assays may be required to identify different mechanisms of attachment involving multiple cell surface factors.
The fact that no mutant completely defective in attachment was detected in this screening does not preclude the possibility that such a mutant exists. A mutant showing no attachment at all may have been lost in our screening for several reasons, including the possibility that potential growth defects may have contributed to such a mutant being outcompeted by other mutants in the enrichment steps. Furthermore, the results of this study indicate that temperature may play a key role in the type and function of attachment factors available to L. monocytogenes. Future studies should focus on individual insertion mutant libraries instead of random pools, and these should be screened at multiple temperatures to identify the molecular factors involved in the L. monocytogenes-plant tissue interaction.
Acknowledgments
We thank D. Wood and D. Bailey for scanning electron microscopy expertise. We thank D. Portnoy, D. Hodgson, N. Freitag, G. Inami, and M. Janda for strains and advice.
This work was funded by the U.S. Department of Agriculture (Agricultural Research Service CRIS project number 5325-42000-040-00D).
REFERENCES
- 1.Albertini, A. M., T. Caramori, W. D. Crabb, F. Scoffone, and A. Galizzi. 1991. The flaA locus of Bacillus subtilis is part of a large operon coding for flagellar structures, motility functions, and an ATPase-like polypeptide. J. Bacteriol. 173:3573-3579. [DOI] [PMC free article] [PubMed]
- 2.Al-Ghazali, M. R., and S. K. Al-Azawi. 1990. Listeria monocytogenes contamination of crops grown on soil treated with sewage sludge cake. J. Appl. Bacteriol. 69:642-647. [DOI] [PubMed] [Google Scholar]
- 3.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 20September2001. Boskovich fresh cut recalls fresh, halved and seeded red bell peppers. SafetyAlterts.com. [Online.] http://209.58.136.120./recall/f/014/f45110.htm.
- 5.Anonymous. 21September2001. Mrs. Crockett's kitchens recalls certain potato salad products. SafetyAlerts.com. [Online.] http://209.58.136.120./recall/f/00/crocketts.htm.
- 6.Anonymous. 28April1999. Product cut salad products. SafetyAlerts.com. [Online.] http://209.58.136.120./rcls/fda/99/apr/f339%2D342%2D9.htm.
- 7.Anonymous. 7September1998. Sprout recall. FSNet. [Online.] http://131.104.232.9/fsnet/1998/9-1998/fs-09-07-98-01.txt.
- 8.Aureli, P., G. C. Fiorucci, D. Caroli, G. Marchiaro, O. Novara, L. Leone, and S. Salmaso. 2000. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria moncytogenes. N. Engl. J. Med. 342:1236-1241. [DOI] [PubMed] [Google Scholar]
- 9.Berrang, M. E., R. E. Brackett, and L. R. Beuchat. 1989. Growth of Listeria monocytogenes on fresh vegetables stored under controlled atmosphere. J. Food Prot. 52:702-705. [DOI] [PubMed] [Google Scholar]
- 10.Beuchat, L. R. 1996. Listeria monocytogenes: incidence on vegetables. Food Control 7:223-228. [Google Scholar]
- 11.Beuchat, L. R., and R. E. Brackett. 1991. Behavior of Listeria monocytogenes inoculated into raw tomatoes and processed tomato products. Appl. Environ. Microbiol. 57:1367-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brackett, R. E. 1999. Incidence and behavior of Listeria monocytogenes in products of plant origin, p. 631-655. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 14.Brehm, K., M.-T. Ripio, J. Kreft, and J.-A. Vázquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by β-glucosides. J. Bacteriol. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camilli, A., D. A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlin, F., and C. Nguyen-The. 1994. Fate of Listeria monocytogenes on four types of minimally processed green salads. Lett. Appl. Microbiol. 18:222-226. [Google Scholar]
- 17.Christensen, D. P., A. K. Benson, and R. W. Hutkins. 1998. Cloning and expression of the Listeria monocytogenes ScottA ptsH and ptsI genes, coding for HPr and enzyme I, respectively, of the phosphotransferase system. Appl. Environ. Microbiol. 64:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, M., M. Jones, and C. Holyoak. 1990. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Microbiol. 69:63-72. [DOI] [PubMed] [Google Scholar]
- 19.DeFlaun, M. F., B. M. Marshall, E.-P. Kulle, and S. B. Levy. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl. Environ. Microbiol. 60:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, F., and R. M. Macnab. 1996. Enzymatic characterization of FliI. J. Biol. Chem. 271:31981-31988. [DOI] [PubMed] [Google Scholar]
- 21.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farber, J. M., S. L. Wang, Y. Cai, and S. Zhang. 1998. Changes in populations of Listeria monocytogenes inoculated on packaged fresh-cut vegetables. J. Food Prot. 61:192-195. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari, E., S. M. H. Howard, and J. A. Hoch. 1986. Effect of stage 0 sporulation mutations on subtilisin expression. J. Bacteriol. 166:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. deDaruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, D. O., K.-D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueño, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. dePablos, J.-C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 26.Goodfellow, I. G., C. E. Pollitt, and R. E. Scockett. 1996. Cloning of the fliI gene from Rhodobacter sphaeroides WS8 by analysis of a transposon mutant with impaired motility. FEMS Microbiol. Lett. 142:111. [DOI] [PubMed] [Google Scholar]
- 27.Görke, B., and B. Rak. 1999. Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J. 18:3370-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heisick, J. E., F. M. Harrell, E. H. Peterson, S. McLaughlin, D. E. Wagner, I. V. Wesley, and J. Bryner. 1989. Comparison of four procedures to detect Listeria spp. in foods. J. Food Prot. 52:154-157. [DOI] [PubMed] [Google Scholar]
- 29.Heisick, J. E., D. E. Wagner, M. L. Nierman, and J. T. Peeler. 1989. Listeria spp. found on fresh market produce. Appl. Environ. Microbiol. 55:1925-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, J. L., K. Shands, G. Friedland, P. Ecking, and D. W. Fraser. 1986. An outbreak of type 4b Listeria monocytogenes infection involving patients from eight Boston hospitals. Arch. Intern. Med. 146:520-524. [PubMed] [Google Scholar]
- 31.Hodgson, D. A. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312-323. [DOI] [PubMed] [Google Scholar]
- 32.Kalmokoff, M. L., J. W. Austin, X.-D. Wan, G. Sanders, S. Banerjee, and J. M. Farber. 2001. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 91:725-734. [DOI] [PubMed] [Google Scholar]
- 33.Korber, D. R., J. R. Lawrence, and D. E. Caldwell. 1994. Effect of motility on surface colonization and reproductive success of Pseudomonas fluorescens in dual-dilution continuous culture and batch culture systems. Appl. Environ. Microbiol. 60:1421-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leimeister-Wachter, M., E. Domann, and T. Chakraborty. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, C.-M., S. Y. Fernando, and C.-I. Wei. 1996. Occurrence of Listeria monocytogenes, Salmonella spp., Escherichia coli and E. coli O157:H7 in vegetable salads. Food Control 7:135-140. [Google Scholar]
- 36.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse, I. I., and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loessner, M. J., A. Schneider, and S. Scherer. 1996. Modified Listeria bacteriophage lysin genes (ply) allow efficient overexpression and one-step purification of biochemically active fusion proteins. Appl. Environ. Microbiol. 62:3057-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loessner, M. J., A. Schneider, and S. Scherer. 1995. A new procedure for efficient recovery of DNA, RNA, and proteins from Listeria cells by rapid lysis with a recombinant bacteriophage endolysin. Appl. Environ. Microbiol. 61:1150-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Moszer, I., P. Glaser, and A. Danchin. 1995. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology 141:261-268. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen-The, C., and B. M. Lund. 1991. The lethal effect of carrot on Listeria species. J. Appl. Bacteriol. 70:479-488. [DOI] [PubMed] [Google Scholar]
- 43.Nichols, D. S., K. A. Presser, J. Olley, T. Ross, and T. A. McMeekin. 2002. Variation of branched-chain fatty acids marks the normal physiological range for growth in Listeria monocytogenes. Appl. Environ. Microbiol. 68:2809-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 45.Petran, R. L., E. A. Zottola, and R. B. Gravani. 1988. Incidence of Listeria monocytogenes in market samples of fresh and frozen vegetables. J. Food Sci. 53:1238-1240. [Google Scholar]
- 46.Porto, E., and M. N. U. Eiroa. 2001. Occurrence of Listeria monocytogenes in vegetables. Dairy Food Environ. Sanit. 21:282-286. [Google Scholar]
- 47.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 48.Premaratne, R. J., W.-J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saklani-Jusforgues, H., E. Fontan, and P. L. Goossens. 2001. Characterization of a Listeria monocytogenes mutant deficient in d-arabitol fermentation. Res. Microbiol. 152:175-177. [DOI] [PubMed] [Google Scholar]
- 50.Schlech, W. F., III. 2000. Epidemiology and clinical manifestations of Listeria monocytogenes infection, p. 473-479. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 51.Schlech, W. F., III, P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 52.Smoot, L. M., and M. D. Pierson. 1998. Effect of environmental stress on the ability of Listeria monocytogenes Scott A to attach to food contact surfaces. J. Food Prot. 61:1293-1298. [DOI] [PubMed] [Google Scholar]
- 53.Smoot, L. M., and M. D. Pierson. 1998. Influence of environmental stress on the kinetics and strength of attachment of Listeria monocytogenes ScottA to buna-N rubber and stainless steel. J. Food Prot. 61:1286-1292. [DOI] [PubMed] [Google Scholar]
- 54.Stephens, C., C. Mohr, C. Boyd, J. Maddock, J. Gober, and L. Shapiro. 1997. Identification of the fliI and fliJ components of the Caulobacter flagellar type III protein secretion system. J. Bacteriol. 179:5355-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stülke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 56.Thomas, D., D. G. Morgan, and D. J. DeRosier. 2001. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J. Bacteriol. 183:6404-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vatanyoopaisarn, S., A. Nazli, C. E. R. Dodd, C. E. D. Rees, and W. M. Waites. 2000. Effect of flagella in initial attachment of Listeria monocytogenes to stainless steel. Appl. Environ. Microbiol. 66:860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weis, J., and H. P. R. Seeliger. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 30:29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White, D. 1995. The physiology and biochemistry of prokaryotes. Oxford University Press, New York, N.Y.