Abstract
The transglutaminase secreted by Streptoverticillium mobaraense is a useful enzyme in the food industry. A fragment of transglutaminase was secreted by Corynebacterium glutamicum when it was coupled on a plasmid to the promoter and signal peptide of a cell surface protein from C. glutamicum. We analyzed the signal peptide and the pro-domain of the transglutaminase gene and found that the signal peptide consists of 31 amino acid residues and the pro-domain consists of 45 residues. When the pro-domain of the transglutaminase was used, the pro-transglutaminase was secreted efficiently by C. glutamicum but had no enzymatic activity. However, when the plasmid carrying the S. mobaraense transglutaminase also encoded SAM-P45, a subtilisin-like serine protease derived from Streptomyces albogriseolus, the peptide bond to the C side of 41-Ser of the pro-transglutaminase was hydrolyzed, and the pro-transglutaminase was converted to an active form. Our findings suggest that C. glutamicum has potential as a host for industrial-scale protein production.
Transglutaminases (protein-glutamine γ-glutamyltransferase, EC 2.3.2.13) are a family of enzymes that catalyze an acyl transfer reaction between a γ-carboxyamide group of a glutamine residue in a peptide chain and a γ-amino group of a lysine residue, resulting in the formation of an ɛ-(γ-glutamyl) lysine cross-link (6). Transglutaminases are widely distributed, and the physiological properties of several of them have been studied. Transglutaminases derived from animals, for example, human blood coagulation factor XIII, human epidermis keratinocyte transglutaminase, guinea pig liver transglutaminase, and fish liver transglutaminase, are calcium-dependent enzymes (6, 24, 38). Calcium-independent transglutaminases have been discovered in bacteria belonging to the actinomycetes, which include, for example, Streptoverticillium cinnamoneum (4) and Streptoverticillium mobaraense. The enzyme from S. mobaraense has been especially well characterized (1, 36).
S. mobaraense transglutaminase (MTG [mature-form transglutaminase]) has been used in the food industry for the modification of proteins (9, 13, 22). It is used in binding meat or fish and gelled food products such as jelly, yogurt, and cheese. Moreover, it has great potential for use in manufacturing materials found in cosmetics, thermostable microcapsules, and carriers for immobilized enzymes. To date, it is produced by conventional fermentation, but it would be desirable to develop a more efficient system, and a number of reports have described the expression and production of MTG in host-vector systems such as Streptomyces lividans (36) and Escherichia coli (33, 39). MTG was secreted in microorganisms such as S. lividans (no more than 0.1 mg/liter) (36) and E. coli (about 5 mg/liter) (33); moreover, it was produced by an inclusion body within E. coli (39). The levels of expression in these studies were low, and it would be very difficult to produce MTG on an industrial scale via an inclusion body.
Corynebacterium glutamicum is a gram-positive, nonsporulating bacterium with a DNA content of about 56% GC (18). It is used for the industrial production of amino acids such as glutamate and lysine that have been used in human food, animal feed, and pharmaceutical products for several decades. It is nonpathogenic and produces no hazardous toxins (12, 16). Furthermore, there is much accumulated experience with the appropriate fermentation conditions. As a result, C. glutamicum should be suitable for producing a food enzyme, although little is known about industrial protein production by this organism. There are reports of secretion by C. glutamicum of heterologous proteins such as a staphyloccocal nuclease (17), protease from Dichelobacter nodosus (3), subtilisin from Bacillus (3), fibronectin-binding protein 85A from Mycobacterium tuberculosis (28), and others (25, 30). Recent reports describe two major cell surface proteins, CspA (10) and CspB (27), present in the culture medium of C. glutamicum. CspA has also been detected in Corynebacterium ammoniagenes (35). These cell surface proteins are the major proteins secreted by these strains. Bacillus brevis notably releases cell surface proteins into the culture medium (34). The promoter and signal peptide of a B. brevis cell surface protein have been used for extracellular production of heterologous gene products, and human epidermal growth factor is produced industrially with this expression system (37).
In this study, we determined the structure of the pro-MTG from S. mobaraense. According to Pasternack et al. (26), the pro-domain inhibits the activity and increases the thermostability of the enzyme. It is likely that a pro-domain is important for efficient secretion and extracellular folding of a protein.
We show that C. glutamicum secretes the pro-MTG efficiently when it is coupled to signal peptides derived from the cell surface proteins of corynebacteria. Moreover, the pro-domain is processed by a subtilisin-like protease from Streptomyces albogriseolus (31, 32), when the protease is cosecreted by C. glutamicum, and is converted into active-form MTG. We thus demonstrate that C. glutamicum can efficiently secrete two proteins derived from actinomycetes and that it has potential as a host for industrial-scale protein production.
MATERIALS AND METHODS
Bacterial strains, culture medium, and plasmids.
C. glutamicum ATCC 13869, C. ammoniagenes ATCC 6872, and S. mobaraense IFO13819 were used in this study. Corynebacterium spp. were grown in CM2G medium (5 g of glucose, 10 g of tryptone, 10 g of yeast extract, 5 g of NaCl, and 0.2 g of dl-methionine per liter of distilled water, adjusted to pH 7.2) at 30°C. As an MTG production medium for C. glutamicum, MMTG medium [60 g of glucose, 1 g of MgSO4, 30 g of (NH4)2SO4, 1.5 g of KH2PO4, 0.01 g of FeSO4 · 7H2O, 0.01 g of MnSO4 · 4H2O, 450 μg of thiamine hydrochloride, 450 μg of biotin, 0.15 g of dl-methionine, and 50 g of CaCO3 per liter of distilled water, adjusted to pH 7.5] was used at 30°C. Standard media and culture conditions for S. mobaraense were as described previously. E. coli JM109 was grown in Luria broth and used as an intermediate host for various plasmid constructions. C. glutamicum was transformed by electroporation as described previously (15). Antibiotics were added to final concentrations of 25 mg/liter for kanamycin (C. glutamicum and E. coli), 5 mg/liter (C. glutamicum) or 30 mg/liter (E. coli) for chloramphenicol, and 50 mg/liter for ampicillin (E. coli). The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Characteristics | Source or reference |
|---|---|---|
| pVC7 | Corynebacterium-E. coli shuttle vector; Cmr | 21 |
| pVKTG1 | pVC7 carrying fused pre-transglutaminase gene containing signal peptide of CspB | This work |
| pVKTG2 | pVC7 carrying fused pre-transglutaminase gene containing N-terminal 31 amino acid residues of CspB | This work |
| pVKTG3 | pVC7 carrying fused pre-transglutaminase gene containing N-terminal 44 amino acid residues of CspB | This work |
| pVKPTG1 | pVC7 carrying fused prepro-transglutaminase gene containing signal peptide of CspB | This work |
| pVKPTG2 | pVC7 carrying fused prepro-transglutaminase gene containing N-terminal 31 amino acid residues of CspB | This work |
| pVKPTG3 | pVC7 carrying fused prepro-transglutaminase gene containing N-terminal 44 amino acid residues of CspB | This work |
| pVKPTG0 | pVC7 carrying native prepro-transglutaminase gene expressed under the control of the cspB promoter | This work |
| pUJP45 | Plasmid carrying SAM-P45 gene | 31 |
| pVSS1 | pVC7 carrying fused prepro-SAM-P45 gene containing signal peptide of CspA of C. ammoniagenes | This work |
| pPK4 | Corynebacterium-E. coli shuttle vector; Kmr | 8 |
| pPKPTG1 | pPK4 carrying fused prepro-transglutaminase gene containing signal peptide of CspB | This work |
| pPAPTG1 | pPK4 carrying fused prepro-transglutaminase gene containing signal peptide of CspA | This work |
| pPSPTG1 | pPK4 carrying fused prepro-transglutaminase gene containing signal peptide of CspA of C. ammoniagenes | This work |
| pUC19 | E. coli cloning vector; Apr | 29 |
| pUMTG5 | pUC19 carrying transglutaminase gene containing 5′-flanking region | This work |
| pUKPTG1 | pUC19 carrying fused prepro-transglutaminase gene containing signal peptide of CspB | This work |
| pUAPTG1 | pUC19 carrying fused prepro-transglutaminase gene containing signal peptide of CspA | This work |
| pUSPTG1 | pUC19 carrying fused prepro-transglutaminase gene containing signal peptide of CspA of C. ammoniagenes | This work |
DNA manipulations.
DNA manipulations were carried out using the methods described by Sambrook et al. (29). PCR with Pyrobest DNA polymerase (Takara Shuzo, Kyoto, Japan) was performed in 50-μl reaction mixtures for 5 min at 94°C, followed by 25 cycles of 10 s at 98°C, 30 s at 55°C, and 3 min at 72°C. Nucleotide sequences were determined using a BigDye terminator cycle sequencing FS ready reaction kit and a model 377 DNA sequencer (both from Applied Biosystems).
Construction of plasmids expressing MTG or pro-MTG genes.
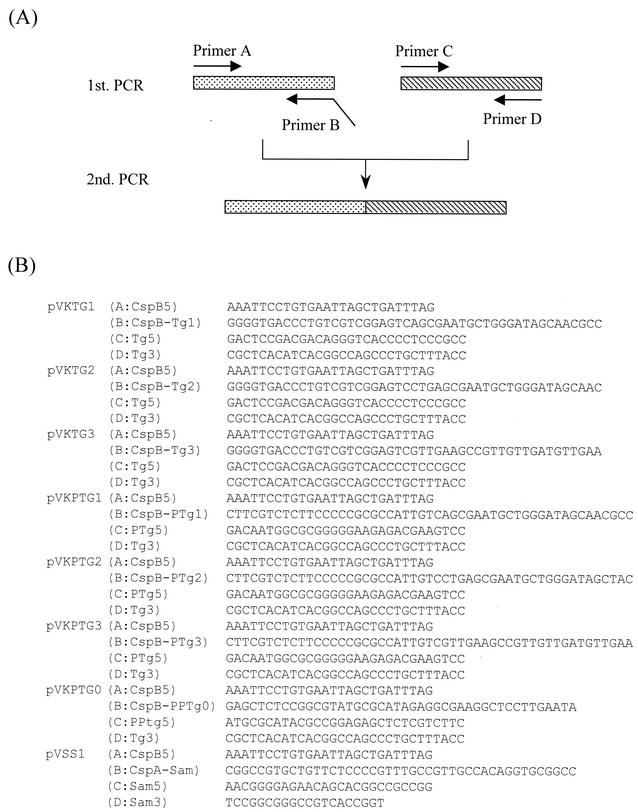
Plasmids expressing MTG genes containing the promoter and the signal sequence of C. glutamicum cspB were constructed by crossover PCR (Fig. 1). A first PCR to amplify the promoter and N-terminal regions of cspB was performed using primer A (CspB5, as the forward primer) and primer B (CspB-Tg1, CspB-Tg2, or CspB-Tg3, as the reverse primer) with chromosomal DNA of C. glutamicum as a template. Other first PCRs for amplification of the MTG gene were performed using primer C (Tg5, as the forward primer) and primer D (Tg3, as the reverse primer) with the chromosomal DNA of S. mobaraense as a template. Second PCRs were performed using primer A (CspB5, as the forward primer) and primer D (Tg3:,as the reverse primer) with the amplified DNA fragments from the first PCRs as a template (Fig. 1A). Each amplified fragment was inserted into the SmaI site of pVC7 to produce pVKTG1, pVKTG2, and pVKTG3. Then pVKTG1, pVKTG2, and pVKTG3 were digested with KpnI and XbaI, and each heterologously fused pre-MTG gene was inserted into the KpnI-XbaI site of pPK4 to produce pPKTG1, pPKTG2, and pPKTG3, respectively. All cloned fragments that had been amplified by PCR were sequenced to confirm the absence of PCR-induced errors.
FIG. 1.
Construction of plasmids expressing MTG, pro-MTG, or pro-SAM-P45 by crossover PCR. (A) Schematic representation of crossover PCR using primers A and B, or primers C and D, for the first PCR and primers A and D for the second PCR. (B) Sequences of the primers used for construction of plasmids expressing MTG, pro-MTG, or pro-SAM-P45.
Plasmids expressing pro-MTG genes were constructed by crossover PCR as described above (Fig. 1). Each amplified fragment was inserted into the SmaI site of pVC7 to produce pVKPTG1, pVKPTG2, pVKPTG3, and pVKPTG0; these were then digested with KpnI and XbaI; and each prepro-MTG gene was inserted into the KpnI-XbaI site of pPK4 to produce pPKPTG1, pPKPTG2, pPKPTG3, and pPKPTG0, respectively.
Construction of plasmids expressing the pro-MTG gene and containing the C. glutamicum CspA signal sequence.
pVKPTG1 was digested with KpnI and XbaI, and the prepro-MTG was inserted into the KpnI-XbaI site of pUC19 to produce pUKPTG1. The 373-bp MunI-NruI fragment (AATTGTCGCTTACAGTTTTTCTCAACGACAGGCTGCTAAGCTGCTAGTTCGGTGGCCTAGTGAGTGGCGTTTACTTGGATAAAAGTAATCCCATGTCGTGATCAGCCATTTTGGGTTGTTTCCATAGCAATCCAAAGGTTTCGTCTTTCGATACCTATTCAAGGAGCCTTCGCCTCTATGCGCGACACCGCATTTCGTTCCATCAAGGCTAAAGCTCAGGCTAAGCGCCGTTCCCTCTGGATTGCAGCAGGCGCTGTCCCAACCGCAATTGCGTTGACTATGTCCCTGGCACCTATGGCTTCGGCTGACAATGGCGCGGGGGAAGAGACGAAGTCCTACGCCGAAACCTACCGCCTCACGGCGGATGACGTCG), which contains the 5′-flanking region of cspB of C. glutamicum and the region coding for the 43-amino-acid signal peptide, together with the coding region of part of the pro-structure of MTG, was constructed from oligonucleotides. To convert the CspB signal peptide into the CspA peptide, the 334-bp MunI-NruI fragment of pUKPTG1 was replaced by the synthetic 373-bp MunI-NruI fragment, to generate pUAPTG1. This was digested with KpnI and XbaI, and the prepro-MTG was inserted into the KpnI-XbaI site of pPK4 to obtain pPAPTG1.
Construction of plasmids expressing the pro-MTG gene with the CspA signal sequence of C. ammoniagenes.
The 319-bp MunI-NruI fragment (AATTGTCGCTTACAGTTTTTCTCAACGACAGGCTGCTAAGCTGCTAGTTCGGTGGCCTAGTGAGTGGCGTTTACTTGGATAAAAGTAATCCCATGTCGTGATCAGCCATTTTGGGTTGTTTCCATAGCAATCCAAAGGTTTCGTCTTTCGATACCTATTCAAGGAGCCTTCGCCTCTATGAAACGCATGAAATCGCTGGCTGCGGCGCTCACCGTCGCTGGGGCCATGCTGGCCGCACCTGTGGCAACGGCAGACAATGGCGCGGGGGAAGAGACGAAGTCCTACGCCGAAACCTACCGCCTCACGGCGGATGACGTCG), which contains the 5′-flanking region of cspB of C. glutamicum and the region encoding the 25-amino-acid signal peptide of CspA of C. ammoniagenes, together with the coding region of part of the pro-structure of MTG, was constructed from oligonucleotides. To convert the CspB signal peptide into CspA of C. ammoniagenes, the 334-bp MunI-NruI fragment of pUKPTG1 was replaced by the synthetic 319-bp MunI-NruI fragment, to give pUSPTG1. This was digested with KpnI and XbaI, and the prepro-MTG was inserted into the KpnI-XbaI site of pPK4 to produce pPSPTG1.
Construction of a plasmid expressing the pro-SAM-P45 gene.
A plasmid expressing the pro-SAM-P45 gene containing the promoter of cspB of C. glutamicum and the signal sequence of cspA of C. ammoniagenes was constructed by crossover PCR (Fig. 1). A first PCR for amplification of the promoter of cspB of C. glutamicum and the signal sequence of CspA of C. ammoniagenes was performed using primer A (CspB5, as the forward primer) and primer B (CspA-Sam, as the reverse primer) with pPSPTG1 DNA as a template. Another first PCR for amplification of the pro-SAM-P45 gene was performed using primer C (Sam5, as the forward primer) and primer D (Sam3, as the reverse primer) with pUJP45 DNA as a template. A second PCR was performed using primer A (CspB5, as the forward primer) and primer D (Sam3, as the reverse primer) with the DNA amplified by the first PCR as a template (Fig. 1A). The amplified fragment was inserted into the SmaI site of pVC7 to obtain pVSS1.
Protein analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 4 to 20% gradient polyacrylamide gel was carried out as described by Laemmli (14), and gels were stained with SYPRO Orange (Bio-Rad). Proteins were electroblotted onto polyvinylidene difluoride membranes (Bio-Rad), and Western blot analysis was performed with the amplified alkaline phosphatase immune-blot assay kit (Bio-Rad). Accumulation of pro-MTG and MTG was measured by high-pressure liquid chromatography (HPLC) on a column in a 24 to 40% linear gradient of CH3CN containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min with purified MTG as a standard (39). N-terminal sequences were determined as described previously (11), by using a gas-phase protein sequencer (model PSQ) equipped with an on-line amino acid analyzer (model RF-550) (both from Shimadzu, Kyoto, Japan). Purification of MTG on a cation-exchange column (Mono S column; Amersham Pharmacia Biotech) has been described previously (39).
Enzyme assays.
MTG was assayed by the calorimetric hydroxamate procedure as described by Folk and Cole (7), and SAM-P45 was assayed as described by Suzuki et al., with N-succinyl-l-Gly-l-Pro-l-Lys-p-nitroanilide (Sigma-Aldrich) as a substrate (31).
Nucleotide sequence accession number.
The nucleotide sequence of the transglutaminase gene from S. mobaraense is deposited in the GenBank/EMBL/DDBJ database under accession no. AF531437.
RESULTS
Secretion of heterologously fused pre-MTG.
First, to test whether C. glutamicum could secrete MTG, we constructed three plasmids, pVKTG1, pVKTG2, and pVKTG3. These have a heterologously fused pre-MTG gene, with the 5′-flanking region containing the cspB promoter of C. glutamicum, a region encoding the N-terminal 30, 31, or 44 amino acid residues, respectively (containing the 30 amino acid residues of the signal peptide of CspB of C. glutamicum), and the coding region of MTG (Fig. 2). We attempted to transform C. glutamicum ATCC 13869 with pVKTG1, pVKTG2, and pVKTG3, but transformants were obtained only with pVKTG3. These were cultured in MMTG medium at 30°C for 40 h, and the supernatants were subjected to SDS-PAGE and Western blot analysis with an anti-MTG antibody as described previously (33). As shown in Fig. 3, many MTG fragments of approximately 18 to 30 kDa were present, together with a very small amount of intact MTG of the expected molecular weight.
FIG. 2.
N-terminal amino acid sequences of heterologously fused pre-MTG or pro-MTG. Signal peptides, amino acid sequences of CspB of C. glutamicum, and amino acid sequences of the MTG or the pro-MTG are indicated by underlining, double underlining, and boxes, respectively. Cleavage sites of the signal peptides are indicated by arrows. The signal peptide encoded by the fused genes in pVKTG1, pVKTG2, pVKTG3, pVKPTG1, pPKPTG1, pVKPTG2, pPKPTG2, pVKPTG3, and pPKPTG3 is derived from CspB of C. glutamicum. The signal peptides encoded by the fused genes in pVKPTG0, pPSPTG1, and pPAPTG1 are derived from native prepro-MTG of S. mobaraense, CspA of C. glutamicum, and CspA of C. ammoniagenes, respectively. All fused genes are expressed under the control of the cspB promoter of C. glutamicum.
FIG.3.
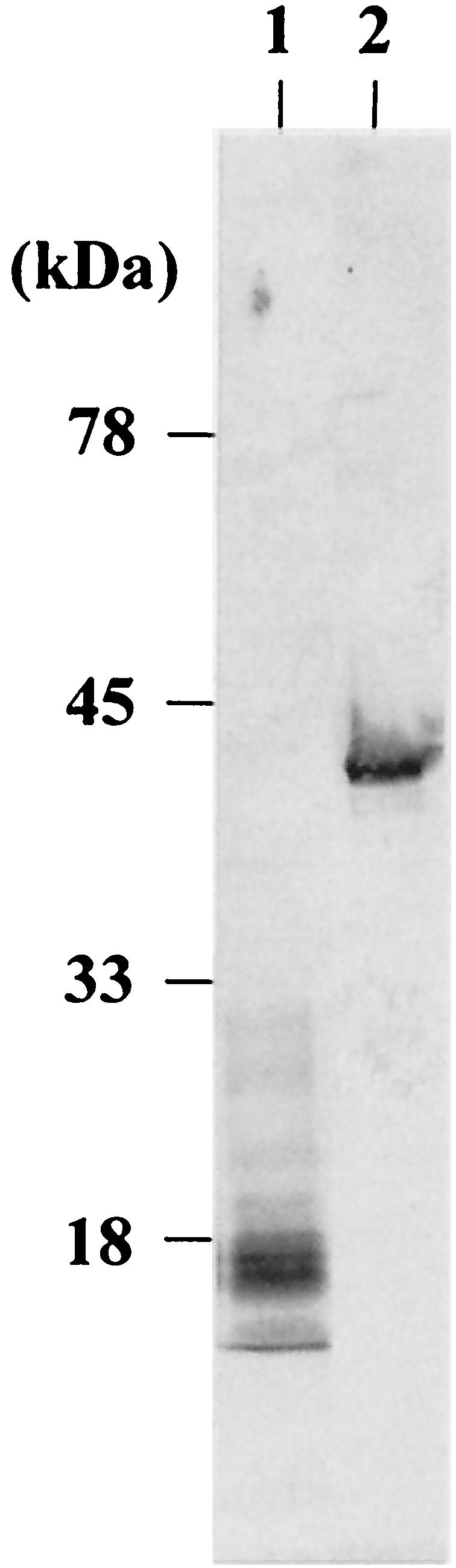
Western blot analysis using an anti-MTG antibody to detect MTG in culture supernatants. Lane 1, culture supernatant of C. glutamicum(pVKTG3); lane 2, purified MTG derived from S. mobaraense.
Sequence of the S. mobaraense prepro-MTG.
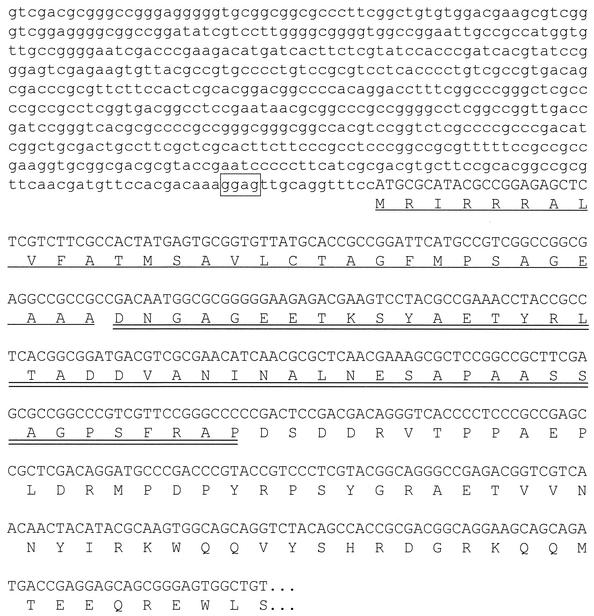
To analyze the structure of pro-MTG, we digested S. mobaraense chromosomal DNA with restriction enzymes and probed a Southern blot with a 665-bp fragment containing the N-terminal coding region of MTG. This was made by PCR using primers 5′-GACTCCGACGACAGGGTCACCCCTCCCGCC-3′ (as the forward primer) and 5′-GCGTCCGGGTCGCCGTACTTCCTCTTGTCG-3′ (as the reverse primer). We detected a SalI fragment of about 1.5 kb, containing the coding region of the prepro-MTG (data not shown). Accordingly, we cloned the 1.5-kb SalI fragment into the SalI site of pUC19 to give plasmid pUMTG5. When the nucleotide sequence of the 1.5-kb SalI fragment was determined (Fig. 4), it was found to encode the N-terminal region of MTG and its prepro-domain. The putative MTG open reading frame started with a methionine codon at nucleotide 578, and a putative Shine-Dalgarno sequence (dGGAG) was located 12 bp upstream of the translational start codon. The predicted signal peptide (31 amino acid residues) possessed the typical features observed for gram-positive bacteria. The SignalP program was able to predict the exact cleavage site (23), and the predicted pro-domain consisted of 45 amino acid residues (Fig. 4). After we had determined this sequence, Pasternack et al. published the sequence of the pro-MTG from S. mobaraense strain DSMZ (26). The two sequences are identical.
FIG. 4.
Nucleotide sequence of the MTG gene from S. mobaraense, with the deduced amino acid sequence given below. The sequence is presented in the 5′-to-3′ direction. The putative Shine-Dalgarno sequence, the amino acid sequence of the signal peptide, and the amino acid sequence of the pro-domain of the MTG are boxed, underlined, and double underlined, respectively.
Secretion of the heterologously fused and the native prepro-MTG.
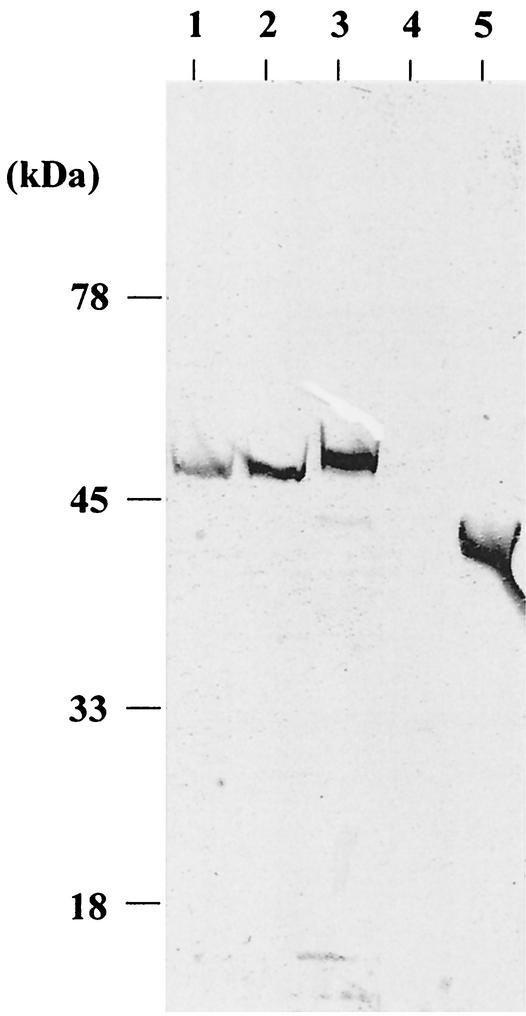
In a second experiment, we constructed four plasmids for secretion of the pro-MTG and examined secretion of the prepro-MTG in C. glutamicum. pVKPTG1, pVKPTG2, and pVKPTG3 carry the prepro-MTG gene, with the 5′-flanking region containing the promoter of cspB of C. glutamicum, the coding region of the N-terminal 30, 31, or 44 amino acid residues (containing 30 amino acid residues of the signal peptide of CspB of C. glutamicum), and the coding region of pro-MTG (Fig. 2). pVKPTG0 has the prepro-MTG gene, with the 5′-flanking region containing the promoter and the Shine-Dalgarno sequence of cspB of C. glutamicum, and the coding region of the native prepro-MTG (Fig. 2). We transformed C. glutamicum with pVKPTG1, pVKPTG2, and pVKPTG3, and transformants were obtained with each plasmid. These were cultured in MMTG medium at 30°C for 40 h, the supernatants were subjected to SDS-PAGE, and Western blot analysis was performed with an anti-MTG antibody. When the signal peptide of CspB of C. glutamicum was used, a pro-MTG with the anticipated molecular weight was detected in the culture supernatant (Fig. 5, lanes 1 to 3).
FIG. 5.
Western blot analysis of MTG in culture supernatants. Lane 1, C. glutamicum(pVKPTG1); lane 2, C. glutamicum(pVKPTG2); lane 3, C. glutamicum(pVKPTG3); lane 4, C. glutamicum(pVKPTG0); lane 5, purified MTG from S. mobaraense.
To test whether C. glutamicum could secrete MTG using the MTG signal peptide, we introduced pVKPTG0 into C. glutamicum and examined the culture supernatant by Western blotting. No pro-MTG was detected (Fig. 5, lane 4).
Production of pro-MTG using various signal peptides.
In order to increase the accumulation of pro-MTG, we tested a pPK4 vector derived from pHM1519 (20), which has a higher copy number than pVC7 derived from pAM330 (19), together with various signal peptides. Constructs pPKPTG1, pPAPTG1, and pPSPTG1 contain signal sequences derived from cspB of C. glutamicum (27), cspA of C. glutamicum (10), and cspA of C. ammoniagenes (35), respectively. The accumulations of pro-MTG in culture supernatants of transformants carrying these constructs were measured by HPLC as described in Materials and Methods. Accumulations of pro-MTG wre 152 mg/liter with plasmid pPKPTG1, 73 mg/liter with plasmid pPAPTGI, and 235 mg/liter with plasmid pPSPTG1. It is interesting that the pro-MTG level obtained with the signal peptide derived from CspA of C. ammoniagenes was higher than that obtained with the signal peptides from C. glutamicum, the host strain. As expected, the N-terminal amino acid of each secreted pro-MTG was Asp, as in the native pro-MTG (Fig. 2). This demonstrates that the signal peptides were correctly processed.
Processing of the pro-domain with a subtilisin-like protease.
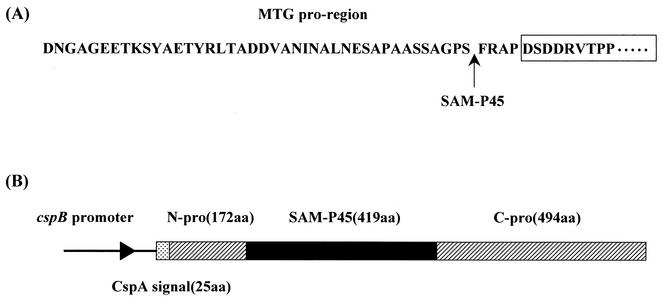
Pasternack et al. reported that the pro-domain of pro-MTG inhibits enzyme activity and increases thermostability and that Bacillus polymyxa dispase and bovine trypsin hydrolyze the peptide bond to the C side of 41-Ser and 43-Arg of the pro-domain and convert the pro-MTG to an active form (26). As noted, the pro-MTG secreted by C. glutamicum has no transglutaminase activity. Taguchi et al. have reported that SAM-P45, a subtilisin-like serine protease secreted by S. albogriseolus, hydrolyzes the pro-domain of the pro-transglutaminase from S. cinnamoneum (4) and converts it to an active form (32). Since the amino acid sequences of the pro-transglutaminase from S. cinnamoneum and S. mobaraense are 77% homologous, we tested whether the pro-MTG released by C. glutamicum could be processed by SAM-P45. The pro-MTG secreted by C. glutamicum carrying pPSPTG1 was indeed cleaved by purified SAM-P45 to the C side of 41-Ser of the pro-domain and converted to an active form. The pro-MTG was incubated with purified SAM-P45 for 2 h at a 100:1 ratio of pro-MTG to SAM-P45, and the specific activity of the purified active-form MTG, with additional Phe-Arg-Ala-Pro residues, was similar to that of the native MTG (about 23 U/mg) (39). Thus, the presence of the additional residues had no effect on the specific activity of the MTG.
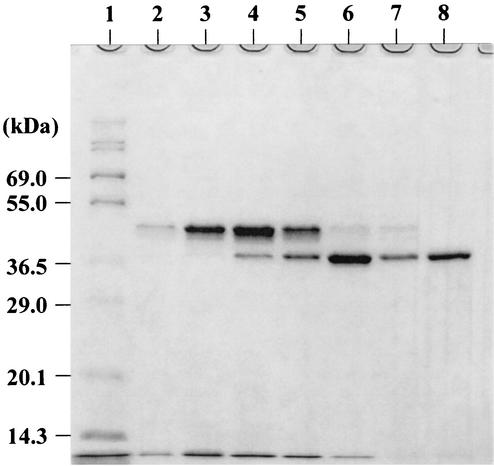
Next, we used secretion of SAM-P45 by C. glutamicum carrying pPSPTG1 to convert pro-MTG to the active form during growth. A prepro-SAM-P45 gene (Fig. 6), containing the cspB promoter of C. glutamicum and the cspA signal sequence of C. ammoniagenes, was constructed by crossover PCR and inserted into the SmaI site of pVC7 to produce pVSS1. Since pPSPTG1, the pro-MTG expression plasmid, and pVSS1, the pro-SAM-P45 expression plasmid, are compatible, we introduced pVSS1 into the C. glutamicum derivative carrying pPSPTG1. The resulting strain was cultured in MMTG medium at 30°C for 140 h. SAM-P45 activity was detected in the culture supernatant after 45 h and then gradually decreased: activity levels were 78.2 U/liter at 45 h, 70.9 U/liter at 54 h, and 58.2 U/liter at 70 h. The pro-MTG was processed by SAM-P45, and levels of active-form MTG, with added Phe-Arg-Ala-Pro residues, peaked at about 70 h (Fig. 7). The maximum yield of the active form under these conditions was 142 mg/liter. The decline after 70 h of cultivation probably occurred because MTG continues to be digested by SAM-P45, since C. glutamicum harboring only pPSPTG1 accumulates pro-MTG without any degradation after 70 h of cultivation (data not shown). The N-terminal amino acid of secreted SAM-P45 was Leu, as in native SAM-P45.
FIG. 6.
Site of cleavage of pro-MTG by SAM-P45, and schematic representation of the prepro-SAM-P45 gene expression construct. (A) The amino acid sequence of MTG is boxed, and the site of cleavage of pro-MTG by SAM-P45 is indicatedby an arrow. (B) Boxes represent the coding region of the gene. Transcription of this fusion gene is controlled by the cspB promoter. CspA signal(25aa), signal sequence of CspA derived from C. ammoniagenes; N-pro(172aa), N-terminal pro-domain of SAM-P45 derived from S. albogriseolus; SAM-P45(419aa), mature domain of SAM-P45 derived from S. albogriseolus; C-pro(494aa), C-terminal pro-domain of SAM-P45 derived from S. albogriseolus.
FIG. 7.
SDS-PAGE analysis of the active-form MTG produced by C. glutamicum carrying plasmids expressing pro-MTG and SAM-P45. Ten microliters of supernatant and an equal volume of sample buffer were applied to each slot and analyzed by SDS-PAGE. After electrophoresis, the gel was stained with SYPRO Orange as described in Materials and Methods. Lane 1, molecular weight markers; lanes 2, 3, 4, 5, 6, and 7, culture supernatants after 24, 30, 45, 54, 70, and 140 h of cultivation, respectively; lane 8, purified MTG from S. mobaraense.
DISCUSSION
Of the three plasmids that we constructed in order to test whether MTG could be secreted in C. glutamicum, only one yielded transformants in C. glutamicum. One possible explanation is that the others (pVKTG1 and pVKTG2) produce pre-MTG with transglutaminase activity within the new host and that this was lethal (2). pVKTG3 may not produce active transglutaminase because of the N-terminal 14 amino acid residues of CspB ligated to it. Another possibility is that the pre-MTGs derived from pVKTG1 and pVKTG2 cannot traverse the cytoplasmic membrane and thus remain literally “stuck” in the cytoplasmic membrane, causing death. We detected many degraded MTG fragments in the supernatant of C. glutamicum carrying pVKTG3 (Fig. 3). No proteolytic activity can be detected in C. glutamicum cultures (3, 28), and purified MTG, added at the beginning of growth of C. glutamicum, is not degraded (data not shown). There is therefore no doubt that the fragmentation of MTG occurred during protein translocation by the Sec machinery. The pro-domains of many secreted proteins are essential for correct folding as well as for secretion of the mature domain (5). At the outset of our study, no information was available about the pro-domain of MTG, so we determined whether MTG was secreted into the culture supernatant. We may suppose that mature-domain MTG on its own is only very slowly translocated, as it lacks the “folding-motive force” provided by the pro-domain. Consequently, the C-terminal regions of MTG protruding on the cytoplasmic side of the membrane may be degraded by cytoplasmic proteases, and the residual N-terminal fragments may then be released into the supernatant.
In the second experiment, we determined the structure of pro-MTG (Fig. 4) and were successful in achieving its secretion by using the signal peptide of CspB of C. glutamicum (Fig. 5, lanes 1 to 3). It is therefore clear that the pro-domain is indispensable for the secretion of MTG. However, no pro-MTG was detected in the culture supernatant of C. glutamicum carrying pVKPTG0 (Fig. 5, lane 4), which carries the prepro-MTG gene with the cspB promoter of C. glutamicum and the signal peptide of S. mobaraense MTG. To date, signal peptides derived from four gram-positive bacteria, Bacillus amyloliquefaciens (30), Cellulomonas fimi (25), Staphylococcus aureus (17), and Bacillus subtilis (3), have been shown to be functional in C. glutamicum; however, it is clear that the S. mobaraense MTG signal peptide is ineffective. It is interesting that the C. ammoniagenes CspA signal peptide was more effective than the CspB or CspA signal peptide of C. glutamicum in promoting secretion of pro-MTG by C. glutamicum. It may be that the structure of that construct is particularly amenable to translocation by the Sec machinery.
The pro-MTG secreted by C. glutamicum had no transglutaminase activity. However, as in the case of the pro-transglutaminase secreted by S. cinnamoneum (32), it was cleaved by SAM-P45 from S. albogriseolus at the C side of 41-Ser of the pro-domain and converted to an active form. We could detect SAM-P45 activity in the supernatant of C. glutamicum harboring a pro-SAM-P45 expression plasmid. Its N-pro-domain was correctly processed, no doubt by an autoproteolytic reaction, as there is no proteolytic activity in cultures of native C. glutamicum (3, 28).
In this work, we have succeeded in achieving efficient secretion of active-form MTG by using C. glutamicum as a host, and the amount accumulated (142 mg/liter) was greater than those obtained with other hosts (33, 36). Our results thus demonstrate that C. glutamicum can secrete heterologous exoproteins derived from actinomycetes and that it has potential as a host for the industrial production of such heterologous proteins.
Acknowledgments
We are grateful to S. Taguchi for his kind gifts of purified SAM-P45 and plasmid pUJP45. We also thank Y. Usuda and H. Kawasaki for helpful discussions.
REFERENCES
- 1.Ando, H., M. Adachi, K. Umeda, A. Matsuura, M. Nonaka, R. Uchio, H. Tanaka, and M. Motoki. 1989. Polymerization and characteristics of a novel transglutaminase derived from microorganisms. Agric. Biol. Chem. 53:2619-2623. [Google Scholar]
- 2.Bassford, P., and J. Beckwith. 1979. Escherichia coli mutants accumulating the precursor of a secreted protein in the cytoplasm. Nature 277:538-541. [DOI] [PubMed] [Google Scholar]
- 3.Billman-Jacobe, H., L. Wang, A. Kortt, D. Stewart, and A. Radford. 1995. Expression and secretion of heterologous proteases by Corynebacterium glutamicum. Appl. Environ. Microbiol. 61:1610-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duran, R., M. Junqua, J. M. Schmitter, C. Gancet, and P. Goulas. 1998. Purification, characterization, and gene cloning of transglutaminase from Streptoverticillium cinnamoneum CBS 683.68. Biochimie 80:313-319. [DOI] [PubMed] [Google Scholar]
- 5.Eder, J., and A. R. Fersht. 1995. Pro-sequence-assisted protein folding. Mol. Microbiol. 16:609-614. [DOI] [PubMed] [Google Scholar]
- 6.Folk, J. E. 1980. Transglutaminases. Annu. Rev. Biochem. 49:517-531. [DOI] [PubMed] [Google Scholar]
- 7.Folk, J. E., and P. W. Cole. 1966. Mechanism of action of guinea pig liver transglutaminase. I. Purification and properties of the enzyme: identification of a functional cysteine essential for activity. J. Biol. Chem. 241:5518-5525. [PubMed] [Google Scholar]
- 8.Hirano, S., M. Sugimoto, E. Nakano, M. Izui, A. Hayakawa, Y. Yoshihara, and T. Nakamatsu. July 2000. Methods of producing l-lysine. U.S. patent 6,090,597.
- 9.Ikura, K., R. Sasaki, and M. Motoki. 1992. Use of transglutaminase in quality-improvement and processing of food proteins. Agric. Food Chem. 2:389-407. [Google Scholar]
- 10.Joliff, G., L. Mathieu, V. Hahn, N. Bayan, F. Duchiron, M. Renaud, E. Shechter, and G. Leblon. 1992. Cloning and nucleotide sequence of the csp1 gene encoding PS1, one of the two major secreted proteins of Corynebacterium glutamicum: the deduced N-terminal region of PS1 is similar to the Mycobacterium antigen 85 complex. Mol. Microbiol. 6:2349-2362. [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi, Y., H. Kojima, T. Tanaka, Y. Takatsuka, and Y. Kamio. 1997. Characterization of a second lysine decarboxylase isolated from Escherichia coli. J. Bacteriol. 179:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krämer, R. 1994. Secretion of amino acids by bacteria: physiology and mechanism. FEMS Microbiol. Rev. 12:75-94. [Google Scholar]
- 13.Kuraishi, C., J. Sakamoto, and T. Soeda. 1996. The usefulness of transglutaminase for food processing, p. 29-38. In G. R. Takeoka, R. Teranishi, P. J. Williams, and A. Kobayashi (ed.), Bio/technology for improved foods and flavors. ACS Symposium Series 637. American Chemical Society, Washington, D.C.
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Liebl, W., A. Bayerl, B. Schein, U. Stillner, and K. H. Schleifer. 1989. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol. Lett. 53:299-303. [DOI] [PubMed] [Google Scholar]
- 16.Liebl, W. 1991. The genus Corynebacterium—nonmedical, p. 1157-1171. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 17.Liebl, W., A. J. Sinskey, and K.-H. Schleifer. 1992. Expression, secretion, and processing of staphylococcal nuclease by Corynebacterium glutamicum. J. Bacteriol. 174:1854-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malumbres, M., J. A. Gil, and J. F. Martin. 1993. Codon preference in Corynebacteria. Gene 134:15-24. [DOI] [PubMed] [Google Scholar]
- 19.Miwa, K., H. Matsui, M. Terabe, S. Nakamori, K. Sano, and H. Momose. 1984. Cryptic plasmids in glutamic acid-producing bacteria. Agric. Biol. Chem. 48:2901-2903. [Google Scholar]
- 20.Miwa, K., K. Matsui, M. Terabe, K. Ito, M. Ishida, H. Takagi, S. Nakamori, and K. Sano. 1985. Construction of novel shuttle vectors and a cosmid vector for the glutamic acid-producing bacteria Brevibacterium lactofermentum and Corynebacterium glutamicum. Gene 39:281-286. [DOI] [PubMed] [Google Scholar]
- 21.Moriya, M., H. Matsui, K. Yokozeki, S. Hirano, A. Hayakawa, M. Izui, and M. Sugimoto. March1997. Amplification of gene using artificial transposon. Japan patent publication JP9070291.
- 22.Motoki, M., and N. Nio. 1983. Cross-linking between different food proteins by transglutaminase. J. Food Sci. 48:561-566. [Google Scholar]
- 23.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Hijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi, K., K. Ishikawa, K. Yokoyama, T. Ohtsuka, N. Nio, and E. Suzuki. 2001. Crystal structure of red sea bream transglutaminase. J. Biol. Chem. 276:12055-12059. [DOI] [PubMed] [Google Scholar]
- 25.Paradis, F. W., D. G. Warren, D. G. Kilburn, and R. C. Miller, Jr. 1987. The expression of Cellulomonas fimi cellulase genes in Brevibacterium lactofermentum. Gene 61:199-206. [DOI] [PubMed] [Google Scholar]
- 26.Pasternack, R., S. Dorsch, J. T. Otterbach, I. R. Robenek, S. Wolf, and H.-L. Fuchsbauer. 1998. Bacterial pro-transglutaminase from Streptoverticillium mobaraense: purification, characterization and sequence of zymogen. Eur. J. Biochem. 257:570-576. [DOI] [PubMed] [Google Scholar]
- 27.Peyret, J. L., N. Bayan, G. Joliff, T. Glik-Krzywicki, L. Mathieu, E. Shechter, and G. Leblon. 1993. Characterization of the cspB gene encoding PS2, an ordered surface-layer protein in Corynebacterium glutamicum. Mol. Microbiol. 9:97-109. [DOI] [PubMed] [Google Scholar]
- 28.Salim, K., V. Haedens, J. Content, G. Leblon, and K. Huygen. 1997. Heterologous expression of the Mycobacterium tuberculosis gene encoding antigen 85A in Corynebacterium glutamicum. Appl. Environ. Microbiol. 63:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Smith, M. D., J. L. Flickinger, D. W. Lineberger, and B. Schmidt. 1986. Protoplast transformation in coryneform bacteria and introduction of an α-amylase gene from Bacillus amyloliquefaciens into Brevibacterium lactofermentum. Appl. Environ. Microbiol. 51:634-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki, M., S. Taguchi, S. Yamada, S. Kojima, K. Miura, and H. Momose. 1997. A novel member of the subtilisin-like protease family from Streptomyces albogriseolus. J. Bacteriol. 179:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taguchi, S., K. Arakawa, K. Yokoyama, H. Takagi, and H. Momose. Overexpression and purification of microbial pro-transglutaminase from Streptoverticillium cinnamoneum and in vitro processing by Streptomyces albogriseolus proteases. J. Biosci. Bioeng., in press. [DOI] [PubMed]
- 33.Takehana, S., K. Washizu, K. Ando, S. Koikeda, K. Takeuchi, H. Matsui, M. Motoki, and H. Takagi. 1994. Chemical synthesis of the gene for microbial transglutaminase from Streptoverticillium and its expression in Escherichia coli. Biosci. Biotechnol. Biochem. 58:88-92. [DOI] [PubMed] [Google Scholar]
- 34.Udaka, S., N. Tsukagoshi, and H. Yamagata. 1989. Bacillus brevis, a host bacterium for efficient extracellular production of useful proteins. Biotechnol. Genet. Eng. Rev. 7:113-146. [DOI] [PubMed] [Google Scholar]
- 35.Usuda, Y., H. Kawasaki, and T. Utagawa. 2001. Characterization of the cell surface protein of Corynebacterium ammoniagenes. Biochim. Biophys. Acta 1522:138-141. [DOI] [PubMed] [Google Scholar]
- 36.Washizu, K., K. Ando, S. Koikeda, S. Hirose, A. Matsuura, H. Takagi, M. Motoki, and K. Takeuchi. 1994. Molecular cloning of the gene for microbial transglutaminase from Streptoverticillium and its expression in Streptomyces lividans. Biosci. Biotechnol. Biochem. 58:82-87. [DOI] [PubMed] [Google Scholar]
- 37.Yamagata, H., K. Nakahama, Y. Suzuki, A. Kailua, N. Tsukagoshi, and S. Udaka. 1989. Use of Bacillus brevis for efficient synthesis and secretion of human epidermal growth factor. Proc. Natl. Acad. Sci. USA 86:3589-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasueda, H., K. Nakanishi, Y. Kawakawa, K. Nagase, M. Motoki, and H. Matsui. 1995. Tissue-type transglutaminase from red sea bream (Paris major): sequence analysis of the coda and functional expression in Escherichia coli. Eur. J. Biochem. 232:411-419. [DOI] [PubMed] [Google Scholar]
- 39.Yokoyama, K., N. Nakamura, K. Saguaro, and K. Kubota. 2000. Overproduction of microbial transglutaminase in Escherichia coli, in vitro refolding, and characterization of the refolded form. Biosci. Biotechnol. Biochem. 64:1263-1270. [DOI] [PubMed] [Google Scholar]