Abstract
To improve the capabilities of microorganisms relevant for biodegradation, we developed a new genetic approach and applied it to the bph operon (bphEGF[orf4]A1A2A3CD[orf1]A4R) of Pseudomonas sp. strain KKS102 to enhance its biphenyl- and polychlorinated biphenyl (PCB)-degrading activity. A native promoter of the bph operon, which was under control, was replaced through homologous recombination by a series of promoters that had constitutive activity. By testing a series of promoters with various strengths, we were able to obtain strains that have enhanced degradation activity for biphenyl and PCBs. This strategy removes the rate-limiting factor associated with transcription and has the potential to improve the degradation activity of a wide variety of microorganisms involved in biodegradation.
The use of microbial metabolic potential for elimination of environmental pollutants is a promising technology. Although various factors such as pathway enzyme specificity, substrate availability, incomplete degradation pathways, and the transcription and translation of genes for bioconversion can limit efficient biodegradation, genetic engineering can be used to overcome such factors and improve degradation (3, 9, 20, 22, 30).
Among pollutants, polychlorinated biphenyls (PCBs) are the most serious pollutants, and their degradation by microorganisms has been studied extensively (9, 10). Pseudomonas sp. strain KKS102 is one of the well-characterized PCB and biphenyl degraders, and its bph gene organization, catabolic route, and regulatory mechanisms have been characterized. The bph genes are organized into an operon in the following order: bphEGF(orf4)A1A2A3CD(orf1)A4R (8, 13-15). The transcription of the bph operon is dependent on the pE promoter, which is located upstream of the bphE gene and is controlled by a negative regulator, BphS. The bphS gene is divergently orientated upstream of bphE and is separated from bphE by an insertion sequence (17). The repression mediated by BphS protein is counteracted by a meta-cleaved intermediate of biphenyl degradation (17, 18).
It is generally recognized that microorganisms can be genetically engineered to increase the rate of pollutant removal. The design of improved microorganisms includes various optimization strategies among which altering the level of transcription is a good target. In general, genes encoding catalytic activities are organized into an operon and transcription of the operon is under the control of activator(s) (6). Various efforts have been made to increase the level of transcription, e.g., creation of mutant regulators that mediate a higher level of transcription or recognize new substrates (2, 19, 21) and construction of plasmids or transposons that carry degradative genes under the control of constitutive promoters (11, 16). The transcription level should be optimized through trials with a series of promoters of different strengths if we are to exclude the rate-limiting steps associated with transcription. It can be expected that application of too strong a promoter results in a decline of overall degradation performance due to the energetic burden. In addition, once transcription reaches a threshold level, a further increase in transcription seems to be of no use, because other limitations such as translation efficiency or availability of the substrates become increasingly more important.
In this study, we developed a novel strategy to increase and optimize the level of transcription. This strategy is applicable without a detailed knowledge of the regulatory mechanisms controlling the transcription and does not involve the laborious task of handling large DNA fragments harboring entire degradation operons. This strategy is based on the integration of a series of constitutive promoters in front of the degradative operon through homologous recombination. We demonstrated the effectiveness of the strategy by enhancing the activity of the PCB and biphenyl degrader KKS102.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas sp. strain KKS102 and its derivative strains were cultivated in 1/3 Luria-Bertani (LB) medium (0.3% tryptone, 0.16% yeast extract, 0.5% NaCl) at 30°C. In the experiment addressing catabolite control, sodium succinate was added to 1/3 LB medium at a final concentration of 25 mM. Antibiotics chloramphenicol and kanamycin were used at a final concentration of 25 μg/ml each. Details regarding the bphS gene disruptant (KKSΔS) are available from reference 7.
Construction of promoters.
Promoter DNAs were prepared either synthetically (ptac, prrnB, and pEcoli promoters) or by PCR amplification (pEcore promoter). The DNA sequences of ptac and prrnB promoters were obtained from references 1 and 4, respectively. The DNA sequence of pEcoli promoter is GAATTCGGCTCGAAAATTTTTTTTCAAAACTAGTTGACAAAAAATTATGATGCATATATAATCTAGA, which has a conserved Escherichia coli −10 box and a −35 box (underlined), as well as a UP element consensus described in references 7 and 23. The pEcore promoter sequence is GAATTCGGATCCGAGTGAAGTGAGTGAAACATAGGGTTTTCACGAGTGTTTAAAAATTATGGCATGCATATAATCTAGTCGACAGATCTGGTCTAGA (17), which consists of promoter elements of pE promoter but lacks binding sites of the negative regulator, BphS. EcoRI and XbaI restriction sites for further use were arranged to flank each promoter sequence (shown in italics).
Assessment of promoter activity.
The promoters were cloned into a vector pKLZ-Z creating promoter-lacZ reporter transcriptional fusions. pKLZ-Z is a derivative of pKLZ-A (17) and differs in multicloning sites. The promoter-lacZ fusion constructs were integrated into the genome of KKS102 as described previously (17).
Plasmid for promoter integration.
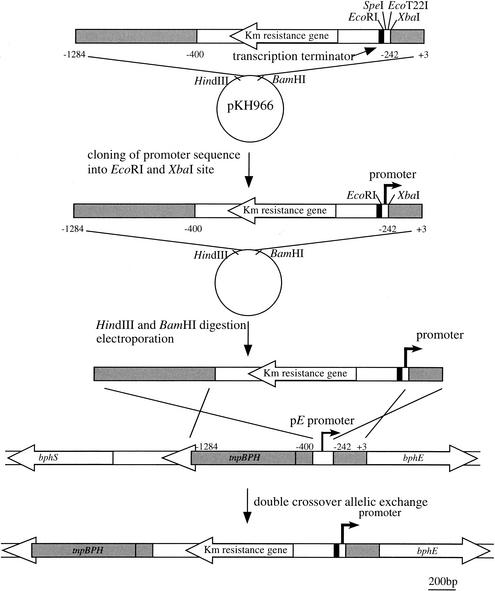
pKH966 (see Fig. 2) contains two DNA segments for double-crossover allelic DNA exchange (upstream and downstream of the pE promoter, positions −1284 to −400 and −242 to +3; +1 represents the translation initiation site of bphE). These regions were PCR amplified with a set of primers tagged with appropriate restriction sites. A DNA cassette harboring the kanamycin resistance gene and a multicloning site derived from pKLZ-Z is encompassed by these two regions.
FIG. 2.
Strategy for systematic integration of promoters. A master plasmid, pKH966, has two DNA regions for homologous recombination (shaded regions), a kanamycin resistance gene (white arrow), a transcriptional terminator (black arrow), and a cloning site. Each promoter sequence was inserted into EcoRI and XbaI sites of pKH966. A HindIII-BamHI DNA fragment of the resulting plasmid was prepared and introduced into KKS102 by electroporation. The cells are plated onto medium containing kanamycin. Double-crossover allelic exchange is expected to replace the native pE promoter with a constitutive promoter.
Integration of the promoter sequence at upstream region of bphE.
The promoter DNA sequence was ligated in the multicloning site of pKH966, and the resulting plasmid was linearized by digesting within the vector DNA sequence. The DNA was introduced into KKS102 by electrotransformation. The conditions for electrotransformation were as described previously (18). The kanamycin-resistant clones were obtained, and PCR amplification with an appropriate set of primers was performed to confirm the double-crossover homologous recombination.
Assay of BphD activity.
The assay of BphD (2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid hydrolase) activity was performed as described previously (17). In brief, cell extract was prepared and a HOPDA (2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid) solution was added. The decrease in absorbance at 434 nm was recorded. We defined 1 U of BphD activity as the activity converting 1 nmol of HOPDA per min. A molar extinction coefficient of 19,800 (26) for HOPDA was used to calculate the BphD activity. The activity was normalized by protein amount in the cell extract. The protein amount in the cell extract was measured by a protein assay kit (Bio-Rad) with bovine serum albumin as a standard.
Assay of LacZ activity.
Cells were collected by centrifugation and resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol). After disrupting cells by sonication, samples were centrifuged and the LacZ activity in the supernatant (cell extract) was measured. To cell extract, the same volume of o-nitrophenyl-β-d-galactopyranoside solution (4 mg/ml in Z buffer) was added. After incubation at 37°C, 1/4 volume of Na2CO3 (1 M) was added to terminate the reaction. The amount of o-nitrophenol produced was measured spectrophotometrically at an optical density at 420 nm (OD420). We defined 1 unit of LacZ activity as that which generates 1 nmol of o-nitrophenol per min. The activity was normalized by the amount of protein in the cell extract. The amount of protein in the cell extract was measured by a protein assay kit (Bio-Rad) with bovine serum albumin as a standard.
Northern blot analysis.
The total RNA was isolated by using RNA purification kit ISOGEN (NIPPON GENE, Tokyo, Japan). The total RNA was electrophoresed through agarose gel containing formaldehyde and blotted onto nylon membrane. The blot was hybridized with digoxigenin (DIG)-labeled DNA, and a detection system for DIG-labeled probe was used according to the protocol provided (Roche Diagnostics). The 0.9-kb EcoT22I-KpnI fragment was used to generate the bphE probe.
Biphenyl degradation activity analysis.
To examine the biphenyl degrading activity, strains were grown in 5 ml of 1/3 LB medium. The cells were grown to the stationary phase (OD600 of 1.2). To 390 μl of the culture, 10 μl of a biphenyl solution (10 mg/ml in ethanol) was added. The samples were incubated at room temperature with vigorous shaking. At each time point, samples were extracted by 1 ml of ethyl acetate containing naphthalene (5 ppm) as an internal standard for gas chromatography-mass spectrometer (GC-MS) analysis. After centrifugation, the ethyl acetate layer was analyzed by GC-MS.
PCB degradation activity analysis.
Bacteria were grown in 100 ml of 1/3 LB medium supplemented with biphenyl to an OD660 of 0.75. Kanechlor 300 was added at a final concentration of 10 mg/liter. As a negative control, heat-inactivated KKS102 cells were used. After 24 h of incubation, the 5-ml aliquot was extracted by ethyl acetate. PCBs recovered in the ethyl acetate layer were analyzed by GC-MS, as described previously (27). The percent degradation was calculated in comparison to heat-inactivated cells. In the calculations, 2,3,6,3′,4′-pentachlorobiphenyl, which is one of the most stable isomers, was selected as an internal standard.
RESULTS
Assessment of promoter strength.
Various approaches have been developed to genetically modify microorganisms to confer the desired biodegradation performance. In this study, our strategy to improve the biodegradation activity of microorganisms was to implant a promoter at the upstream region of a target operon. The bph operon involved in biphenyl and PCB degradation in Pseudomonas sp. strain KKS102 was selected as a target operon. The transcription of the operon is dependent on the pE promoter, which is located upstream of the bphE gene and regulated by a negative regulator, BphS (17).
Since we did not know what degree of transcription strength would give the maximum degradation efficiency, four different promoter DNA sequences were prepared either synthetically or by PCR amplification (Table 1). The pEcoli promoter was designed according to the consensus DNA sequence of promoter elements (−10 box, −35 box, and UP element). The ptac promoter is a promoter often used to overexpress genes in E. coli (1). prrnB is a strong promoter in E. coli for ribosomal gene transcription (4). pEcore is a core region of pE promoter and lacks BphS binding sites.
TABLE 1.
Strains used in this study
| Strain | Promoter | Relevant characteristics of the promoter or strain | Promoter-lacZ fusion strain | Source or reference |
|---|---|---|---|---|
| KH952 | pEcoli | Consensus E. coli promoter | KLZ952 | This study |
| KH967 | prrnB | Promoter for rRNA transcription | KLZ401 | 4 |
| KH968 | ptac | Promoter often used for overexpression in E. coli | KLZ301 | 1 |
| KH981 | pEcore | Core part of pE promoter | KLZ201 | This study |
| KKS102 | pE | pE promoter with operator sites for the negative regulator | KLZ12 | 17 |
| KKSΔS | pE | bphS gene-inactivated derivative of KKS102 | 17 |
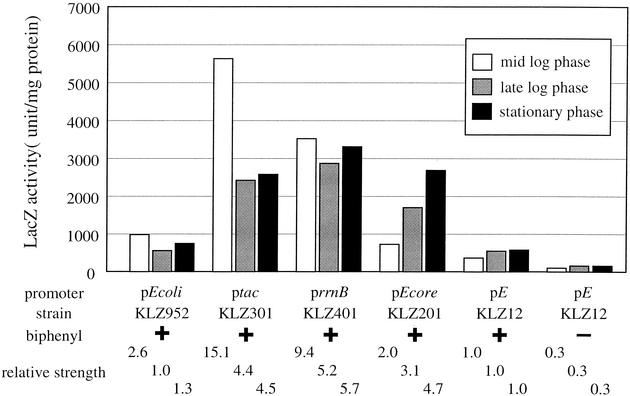
To evaluate promoter activity in KKS102, each promoter was fused with lacZ gene, and the fusion construct was integrated, as a single copy, into the genome of KKS102. To compare the activity of the constitutive promoters with that of the native promoter of bph genes, the activity of pE promoter was measured as a control. The promoter-lacZ reporter strains were grown in liquid culture, and LacZ activity was measured at mid-log phase (OD660 = 0.5), late log phase (OD660 = 1.0), and stationary phase (24 h after OD660 reached 0.5) (Fig. 1). In the pE-lacZ fusion strain, LacZ activity was 3.3-fold higher in the presence of biphenyl than in its absence. A negative regulator, BphS, mediates this inducible expression of the pE promoter (17). As shown in Fig. 1, LacZ activity of all four promoters was higher than that of the pE promoter in every growth phase tested. The ptac and prrnB promoters showed activities 4.4- to 15.1-fold higher than that of the pE promoter. The pEcore promoter showed activity 2.0- to 4.7-fold higher, and pEcoli promoter showed activity 1.0- to 2.6-fold higher than that of the pE promoter. The promoters tested differed from the native pE promoter in that they showed the same level of LacZ activity even in the absence of biphenyl (data not shown).
FIG. 1.
Assessment of the promoters used in this study by LacZ activity. Each strain harboring the promoter-lacZ fusion reporter construct in its chromosome was grown in liquid culture in the presence or absence of biphenyl. Cells were harvested at mid-log phase, late log phase, and stationary phase. LacZ activity in cell extract was assayed. The activity was normalized by the amount of protein in the reaction mixture. The results shown represent two independent experiments.
Construction of a bph constitutive strain by homologous recombination.
The strategy used to implant the promoter is shown in Fig. 2. The fundamental scheme of the strategy lies in the implantation of the promoter through double allelic homologous recombination by using the kanamycin resistance gene as a selection marker. To implant several promoters easily, we constructed a key plasmid, pKH966. The plasmid contains two DNA regions derived from the bphE upstream region (upstream and downstream of the pE promoter, −1284 to −400 and −242 to +3; +1 represents the translation initiation site of bphE) and the kanamycin resistance gene and a set of restriction sites to clone promoter DNA. pKH966 was digested by endonucleases, and each promoter DNA was cloned into the cleaved sites. The resulting plasmid was linearized by endonuclease digestion within vector DNA. KKS102 cells were transformed with the digested DNA by electroporation. Colonies grown on kanamycin-containing media were selected, and a homologous recombination event at the bphE upstream region was confirmed by PCR (data not shown).
Assessment of the strains by BphD activity.
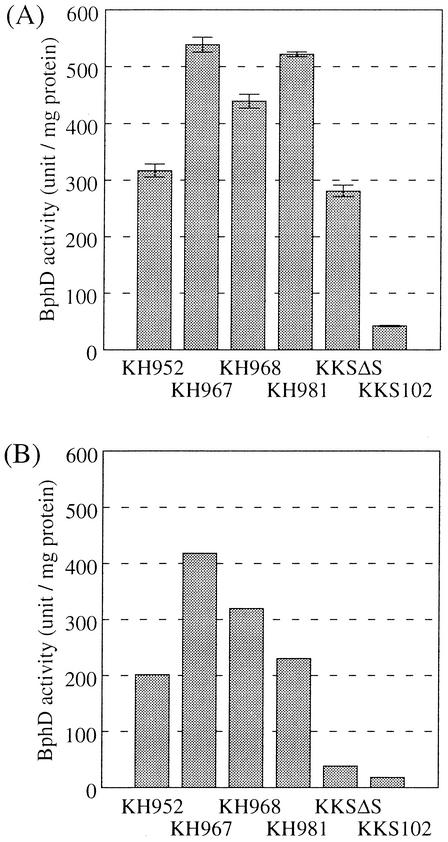
The bphD gene encoding BphD is located relatively downstream of the bph operon; thus, BphD activity could represent the expression level of the entire bph operon (17). We measured the BphD activity of the recombinant strains to assess the effect of the implanted promoters. As a control, we included strain KKSΔS, which is a KKS derivative that constitutively expresses bph genes due to inactivation of the bphS gene (17). The strains were grown in liquid culture (1/3 LB medium) until the turbidity of the culture at 600 nm reached a value of 1.2 (stationary phase). We selected the stationary phase because it is believed that the stationary phase is the prevailing status of microorganisms under natural environmental conditions (24, 29). Cells were harvested, and BphD activity in cell extract was measured as described in Materials and Methods. The result is shown in Fig. 3A. Compared to the control strain KKSΔS, KH967 and KH981 showed 1.9-fold-higher activity, KH968 showed 1.6-fold-activity, and KH952 showed comparable but higher activity.
FIG. 3.
Assessment of created strains by BphD activity. (A) Strains were grown in liquid culture in 1/3 LB medium. Cells were harvested at the stationary phase, and BphD activity in the cell extract was assayed. The activity was normalized by the amount of protein in the reaction mixture. Each bar represents the average of three parallel experiments, and vertical lines represent standard deviations. (B) Strains were grown in 1/3 LB medium supplemented with 25 mM sodium succinate for 24 h. BphD activity in the cell extract was assayed. The results shown represent two independent experiments, and the values are averages of duplicate measurements.
We also measured BphD activity of the strains grown in minimum medium (W medium) supplemented with 25 mM glucose and a trace amount of Casamino Acids (0.02%). We observed a higher activity than that of strains grown in 1/3 LB medium, but the overall activity pattern was conserved (data not shown).
Catabolite-repressive effect of succinate on promoter activity.
Recently, we found that expression of bph operon is down regulated by certain carbon sources. We tested the effect of one of the repressive carbon sources, succinate, on BphD activity. The strains were grown in 1/3 LB medium containing succinate (25 mM) for 24 h (stationary phase), and BphD activity was measured (Fig. 3B). The BphD activity of KKSΔS was more severely down regulated by the presence of succinate than were the activities of the other strains implanted with artificial promoters.
Assessment of the strains by Northern blot analysis.
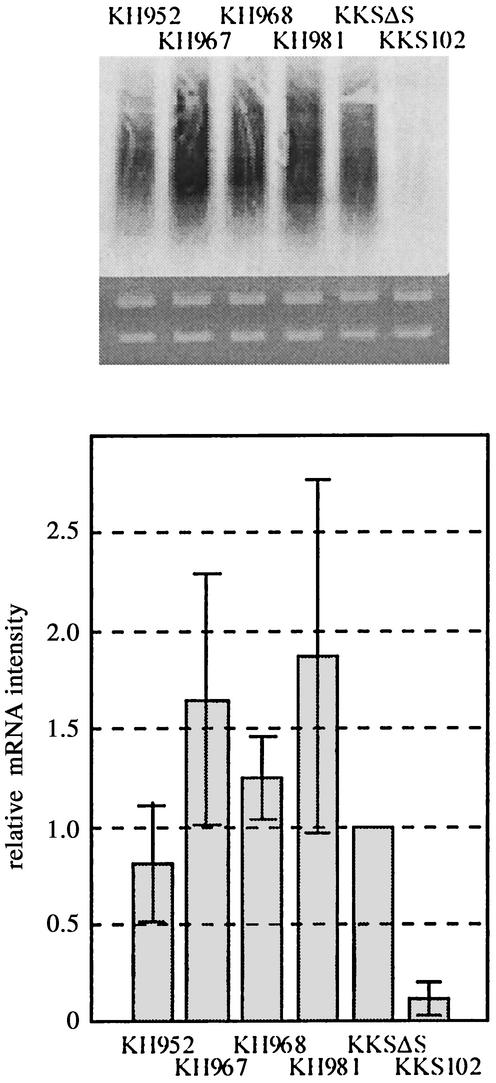
To assess the functionality of each promoter to drive transcription of bph operon, and to correlate the level of BphD activity with the level of bph transcription, the level of bphE transcription was analyzed by Northern blot analysis. Total RNA was prepared from each strain at the stationary phase, electrophoresed through agarose gel, blotted onto a membrane, and hybridized with bphE probe (Fig. 4A). The experiment was repeated three times, and the signal intensity relative to that of KKSDS was calculated (Fig. 4B). The overall hybridization signal was correlated to BphD activity.
FIG. 4.
Assessment of created strains by Northern blot analysis. (A) Each strain was grown in 1/3 LB medium to the stationary phase, and total RNA was prepared. Three micrograms of total RNA was electrophoresed through agarose gel containing formaldehyde and blotted onto a nylon membrane. The blot was hybridized with DIG-labeled bphE probe. For signal detection, CDP star was used as a substrate for chemical luminescence. The results shown represent three independent experiments. (B) The signal intensity of the blot was quantified by using Image Gauge ver. 3.1 software (Fuji Photo Film, Tokyo, Japan). The signal intensity relative to that of KKSΔS was calculated. Each bar represents the average of three independent experiments, and vertical lines represent standard deviations
Assessment of the strains by biphenyl degradation activity.
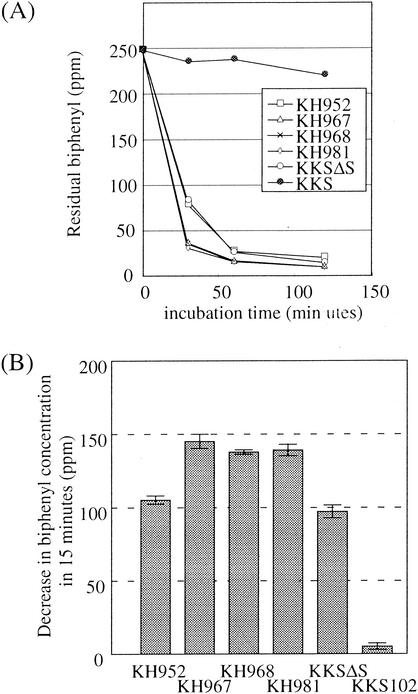
We measured the biphenyl degradation activity of the engineered strains. Each strain was grown in 5 ml of 1/3 LB medium until the turbidity at 600 nm reached a value of 1.2 (stationary phase). The culture was divided into several test tubes, and to each tube biphenyl was added at a final concentration of 250 mg/liter. After 30, 60, and 120 min of vigorous shaking at room temperature, residual biphenyl was quantified by GC-MS analysis (Fig. 5A). All the engineered strains degraded most of the biphenyl within 60 min. In contrast, the KKS102 cells degraded biphenyl only slightly in the experimental period. The higher biphenyl degradation activity of the engineered strains grown even in the absence of biphenyl emphasizes the advantage of the strategy.
FIG. 5.
Assessment of created strains by biphenyl degradation activity. Strains were cultivated in 1/3 LB medium to the stationary phase. The portions of each culture were transferred to test tubes. To the tubes, biphenyl was added at a final concentration of 250 mg/liter. The tubes were incubated at room temperature with vigorous shaking. At several time points, ethyl acetate was added to the tube and residual biphenyl was extracted. The amount of biphenyl in the ethyl acetate was analyzed by GC-MS. The cell-free 1/3 LB medium was used as a control. (A) Cells and biphenyl were reacted for 30, 60, and 120 min. The values are the averages of two parallel samples. (B) Cells and biphenyl were incubated for 15 min. Each bar represents the average of four parallel experiments, and vertical lines represent standard deviations.
To better characterize the biphenyl degradation activity of each strain, biphenyl degradation within the initial 15 min was investigated in more detail (Fig. 5B). To note, biphenyl degradation activity was not correlated with BphD activity. The strains KH967, KH968, and KH981 showed the same level of biphenyl degradation activity. This observation shows that, as long as biphenyl is the substrate to be degraded, the level of transcription in KH968 is high enough and further increase in transcription is of no use because it is no longer a rate-limiting factor.
Assessment of the strains by PCB-degrading activity.
We measured the PCB-degrading activities of KH952 and KKS102. Strains were grown in liquid culture in the presence of biphenyl, and at mid-log phase Kanechlor 300 was added at a final concentration of 10 mg/liter. As a control, heat-inactivated KKS102 cells were used. After 24 h of incubation, PCBs were extracted by ethyl acetate and residual amount of PCBs were analyzed by GC-MS. The degradation activity of KH952 to all isomers was superior to that of KKS102 (Table 2). We noted that degradation activity was greatly enhanced toward several isomers, e.g., 2,5,4′-, 2,4,5,4′-, and 2,3,4,3′,4′-chlorobiphenyl. Of significance was the fact that KH952 degraded some isomers that were hardly degraded by KKS102, for example, 2,4,3′,4′-, 3,4,3′,4′-, and 3,4,5,2′,5′-chlorobiphenyls. However, the reasons for this unexpected expansion of substrate specificity remain to be elucidated.
TABLE 2.
PCB-degrading activity of KKS102 and KH952
| Congener | % Degradation by strain:
|
|
|---|---|---|
| KKS102 | KH952 | |
| 2,2,2,6 | 71.7 | 97.9 |
| 2,3 2,4′ | 99.6 | 99.6 |
| 3,4′ | 16.7 | 70.0 |
| 4,4′ | 85.5 | 97.5 |
| 2,6,2′ | 26.6 | 55.0 |
| 2,5,2′ | 16.4 | 69.2 |
| 2,4,2′ | 19.3 | 83.7 |
| 2,6,3′ 2,3,6 | 59.3 | 100.0 |
| 2,3,2′ 2,6,4′ | 27.3 | 75.2 |
| 2,5,3′ | 36.7 | 93.7 |
| 2,4,3′ | 100.0 | 100.0 |
| 2,5,4′ | 3.2 | 69.3 |
| 2,4,4′ | 100.0 | 100.0 |
| 3,4,2′ 2,3,3′ 2,3,4 | 64.9 | 97.7 |
| 2,3,4′ | 96.5 | 97.9 |
| 3,4,4′ | 45.4 | 94.3 |
| 2,5,2′,6′ | 0.0 | 17.4 |
| 2,4,2′,6′ | 0.0 | 13.2 |
| 2,3,6,2′ | 0.0 | 13.2 |
| 2,6,3′,5′ 2,5,2′,5′ | 0.0 | 8.9 |
| 2,4,2′,5′ | 0.0 | 15.0 |
| 2,4,2′,4′ 2,4,5,2′ 2,4,6,4′ | 0.0 | 10.4 |
| 2,3,2′,5′ | 0.0 | 0.0 |
| 2,3,2′,4′ 2,3,6,3′ | 0.0 | 49.5 |
| 2,6,3′,4′ 2,3,4,2′ 2,3,6,4′ 2,5,3′,5′ | 0.0 | 10.7 |
| 2,3,2′,3′ | 19.2 | 87.1 |
| 2,3,5,3′ 2,4,5,3′ | 39.8 | 100.0 |
| 2,3,5,4′ 2,3,3′,5′ | 4.7 | 71.9 |
| 2,4,5,4′ | 1.8 | 61.3 |
| 2,5,3′,4′ 3,4,5,2′ | 0.0 | 17.3 |
| 2,4,3′,4′ | 0.0 | 55.3 |
| 2,3,4,3′ | 67.2 | 100.0 |
| 2,3,3′,4′ 2,3,4,4′ | 55.0 | 92.5 |
| 3,4,3′,4′ | 0.0 | 26.6 |
| 2,4,5,2′,6′ 2,3,6,2′,5′ | 0.0 | 0.0 |
| 2,3,6,2′,4′ | 0.0 | 1.7 |
| 2,4,5,2′,5′ 2,3,5,2′,4′ | 0.0 | 2.4 |
| 2,4,5,2′,4′ | 0.0 | 5.6 |
| 2,4,5,2′,3′ 2,3,4,5,2′ | 0.0 | 8.9 |
| 2,3,4,2′,5′ 2,3,4,6,4′ 2,3,5,3′,5′ | 0.0 | 1.6 |
| 2,3,4,2′,4′ | 0.0 | 0.5 |
| 2,3,6,3′,4′a | 0.0 | 0.0 |
| 3,4,5,2′,5′ | 0.0 | 11.5 |
| 2,4,5,3′,4′ 2,3,4,5,3′ | 0.0 | 14.1 |
| 2,3,4,3′4′ | 6.7 | 43.8 |
2,3,6,3′,4′-pentachlorobiphenyl was used as an internal standard.
DISCUSSION
In this work we demonstrated the effectiveness of implanting a promoter to enhance degradation activity. The constructed strains expressed the bph operon at high levels not only in the absence of the pathway substrate but also in the presence of a repressive carbon source. We also demonstrated that a certain level of expression yields the highest degradation performance.
Transcription of the genes that encode a metabolic pathway is thought to be one of the bottlenecks which limit pollutant removal. In many cases, transcription of the metabolic genes is under the control of regulatory mechanisms and active transcription requires a sufficient concentration of cognate effector molecule(s). Moreover, transcription of the degradative genes is often under the control of catabolite repression that represses transcription in the presence of a favorable carbon source where the microorganism can grow rapidly (6). The transcription of degradative genes is an important step in catabolism and hence a significant event if we are to use the catabolic potential. Thus, transcription is a good target in efforts to improve the efficiency of degradation.
Strains KH967, KH968, and KH981 showed different BphD activities (Fig. 3A), while they showed the same level of biphenyl-degrading activity (Fig. 5). This indicates that transcription was no longer a rate-limiting step for biphenyl degradation in these strains. Presumably, other steps such as supplying the oxygen and/or NADH necessary for degradation, bioavailability of the substrates (28), and translation efficiency could now be rate-limiting factors. It could be postulated that the ptac promoter (KH968) was strong enough and that promoters with higher activity might not be suitable, because their use could lead to, for example, energy expenditure and instability of the genetic information. Although the reason was not clear, the presence of biphenyl at the onset of cultivation impaired the growth of strain KH967, which showed the highest BphD activity (data not shown). These observations suggest that the use of a strong promoter does not always give the desirable strain and that trials of different promoters are necessary to optimize degradation.
Implanting of a constitutive promoter was demonstrated to be effective for the creation of microorganisms with high degradation activity. This strategy is practical for a number of reasons. First, the strategy could be applicable to a wide variety of microorganisms relevant for biodegradation without needing to know how the transcription of the degradative operon is regulated. Second, the DNA element on a chromosome would be maintained more stably than on a plasmid and seems to be less likely to transfer horizontally (5), reducing the risk of dispersal of human-derived genetic information into nature. Third, implanting leads to constitutive expression. Thus, we do not have to add inducing chemicals or take catabolite control into consideration to maintain the expression level (Fig. 3B) (6). This advantage is pronounced in the degradation of PCBs that are made up of various isomers and not likely to induce bph genes effectively. Fourth, overexpression leads to the degradation of some PCB isomers that are hardly degraded by the wild-type strain; i.e., overexpression has the potential to expand the range of substrates. Although the reasons for the apparent expansion of substrate specificity are unclear, it could be speculated that rapid conversion of PCBs or PCB degradation intermediates reduces their toxic effect on cell viability. Finally, constitutive expression of the entire operon would accelerate each step of mineralization; therefore, the target compound would be mineralized more completely, preventing the accumulation of intermediates. The accumulation of PCB degradation intermediates could cause undesirable effects. The 3-chlorinated, 4-chlorinated, and 4-hydroxy HOPDAs, which were formed from PCB degradation, inhibited BphD enzyme from Burkholderia cepacia strain LB400 (25). Recently, intermediate metabolites resulting from the degradation of PCBs were shown to inhibit cell separation of Comamonas testosteroni strain TK102 (12).
This approach is applicable, provided that the host strain is subject to homologous recombination, to any genes or operons to improve biological functions in the original strain. We expect that a combination of promoter implanting and the many genetic techniques developed thus far will contribute to the effective and rapid remediation of polluted environments.
Acknowledgments
Y.O. was financially supported by research fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Y.O. is a special postdoctoral researcher at RIKEN.
REFERENCES
- 1.Amann, E., J. Brosius, and M. Ptashne. 1983. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25:167-178. [DOI] [PubMed] [Google Scholar]
- 2.Cebolla, A., C. Sousa, and V. de Lorenzo. 1997. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J. Biol. Chem. 272:3986-3992. [DOI] [PubMed] [Google Scholar]
- 3.Chen, W., F. Bruhlmann, R. D. Richins, and A. Mulchandani. 1999. Engineering of improved microbes and enzymes for bioremediation. Curr. Opin. Biotechnol. 10:137-141. [DOI] [PubMed] [Google Scholar]
- 4.Csordas-Toth, E., I. Boros, and P. Venetianer. 1979. Structure of the promoter region for the rrnB gene in Escherichia coli. Nucleic Acids Res. 7:2189-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 6.Diaz, E., and M. A. Prieto. 2000. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11:467-475. [DOI] [PubMed] [Google Scholar]
- 7.Estrem, S. T., T. Gaal, W. Ross, and R. L. Gourse. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 95:9761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda, M., Y. Yasukochi, Y. Kikuchi, Y. Nagata, K. Kimbara, H. Horiuchi, M. Takagi, and K. Yano. 1994. Identification of the bphA and bphB genes of Pseudomonas sp. strains KKS102 involved in degradation of biphenyl and polychlorinated biphenyls. Biochem. Biophys. Res. Commun. 202:850-856. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa, K. 2000. Engineering dioxygenases for efficient degradation of environmental pollutants. Curr. Opin. Biotechnol. 11:244-249. [DOI] [PubMed] [Google Scholar]
- 10.Furukawa, K. 1994. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation 5:289-300. [DOI] [PubMed] [Google Scholar]
- 11.Gallardo, M. E., A. Ferrandez, V. De Lorenzo, J. L. Garcia, and E. Diaz. 1997. Designing recombinant Pseudomonas strains to enhance biodesulfurization. J. Bacteriol. 179:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiraoka, Y., T. Yamada, K. Tone, Y. Futaesaku, and K. Kimbara. 2002. Flow cytometry analysis of changes in the DNA content of the polychlorinated biphenyl degrader Comamonas testosteroni TK102: effect of metabolites on cell-cell separation. Appl. Environ. Microbiol. 68:5104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikuchi, Y., Y. Nagata, M. Hinata, K. Kimbara, M. Fukuda, K. Yano, and M. Takagi. 1994. Identification of the bphA4 gene encoding ferredoxin reductase involved in the biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J. Bacteriol. 176:1689-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuchi, Y., Y. Yasukochi, Y. Nagata, M. Fukuda, and M. Takagi. 1994. Nucleotide sequence and functional analysis of the meta-cleavage pathway involved in biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J. Bacteriol. 176:4269-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lajoie, C. A., A. C. Layton, and G. S. Sayler. 1994. Cometabolic oxidation of polychlorinated biphenyls in soil with a surfactant-based field application vector. Appl. Environ. Microbiol. 60:2826-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohtsubo, Y., M. Delawary, K. Kimbara, M. Takagi, A. Ohta, and Y. Nagata. 2001. BphS, a key transcriptional regulator of bph genes involved in polychlorinated biphenyl/biphenyl degradation in Pseudomonas sp. KKS102. J. Biol. Chem. 276:36146-36154. [DOI] [PubMed] [Google Scholar]
- 18.Ohtsubo, Y., Y. Nagata, K. Kimbara, M. Takagi, and A. Ohta. 2000. Expression of the bph genes involved in biphenyl/PCB degradation in pseudomonas sp. KKS102 induced by the biphenyl degradation intermediate, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid. Gene 256:223-228. [DOI] [PubMed] [Google Scholar]
- 19.Pavel, H., M. Forsman, and V. Shingler. 1994. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J. Bacteriol. 176:7550-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pieper, D. H., and W. Reineke. 2000. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 11:262-270. [DOI] [PubMed] [Google Scholar]
- 21.Ramos, J. L., A. Wasserfallen, K. Rose, and K. N. Timmis. 1987. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science 235:593-596. [DOI] [PubMed] [Google Scholar]
- 22.Reineke, W. 1998. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu. Rev. Microbiol. 52:287-331. [DOI] [PubMed] [Google Scholar]
- 23.Ross, W., S. E. Aiyar, J. Salomon, and R. L. Gourse. 1998. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J. Bacteriol. 180:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seah, S. Y., G. Labbe, S. Nerdinger, M. R. Johnson, V. Snieckus, and L. D. Eltis. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 275:15701-15708. [DOI] [PubMed] [Google Scholar]
- 26.Seah, S. Y., G. Terracina, J. T. Bolin, P. Riebel, V. Snieckus, and L. D. Eltis. 1998. Purification and preliminary characterization of a serine hydrolase involved in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 273:22943-22949. [DOI] [PubMed] [Google Scholar]
- 27.Shimura, M., G. Mukerjee-Dhar, K. Kimbara, H. Nagato, H. Kiyohara, and T. Hatta. 1999. Isolation and characterization of a thermophilic Bacillus sp. JF8 capable of degrading polychlorinated biphenyls and naphthalene. FEMS Microbiol. Lett. 178:87-93. [DOI] [PubMed] [Google Scholar]
- 28.Sikkema, J., J. A. de Bont, and B. Poolman. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorne, S. H., and H. D. Williams. 1997. Adaptation to nutrient starvation in Rhizobium leguminosarum bv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J. Bacteriol. 179:6894-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timmis, K. N., and D. H. Pieper. 1999. Bacteria designed for bioremediation. Trends Biotechnol. 17:200-204. [DOI] [PubMed] [Google Scholar]