Abstract
We have developed a PCR-oligonucleotide ligation assay to rapidly identify base substitutions in topoisomerase genes that are associated with quinolone resistance in clinical isolates of Streptococcus pneumoniae. Thirty-seven strains for which the ciprofloxacin MICs were ≥4 μg/ml and 16 strains for which the MICs were ≤2 μg/ml were assayed. Compared with sequence data, the assay correctly identified the DNA bases that encoded amino acids at the four positions most commonly associated with quinolone resistance (Ser79 and Asp83 of ParC and Ser81 and Glu85 of GyrA). Therefore, this procedure can rapidly distinguish single base substitutions associated with quinolone-resistant topoisomerases in S. pneumoniae.
Streptococcus pneumoniae is a major pathogen causing acute bacterial exacerbations of chronic bronchitis, acute otitis media, meningitis, and community-acquired pneumonia. Newer quinolones such as levofloxacin, moxifloxacin, and gatifloxacin have enhanced activity against pneumococci compared with older quinolones and are used to treat infections caused by this organism. Studies show that the mechanism of action of quinolones is inhibition of the essential bacterial type II topoisomerases, DNA gyrase and topoisomerase IV (3, 10, 13). Amino acid substitutions in the quinolone resistance determining regions (QRDRs) of these enzymes are associated with resistance to quinolones in S. pneumoniae (2, 4, 7, 10). It has also been suggested that active efflux contributes to resistance against quinolones (1, 14).
Methods to identify mutations in topoisomerases are needed since the increased use of quinolones in the clinic may increase the prevalence of resistance. We developed a rapid assay to detect point mutations in the genes that code for two of the topoisomerase subunits, GyrA and ParC. This assay utilized PCR to amplify the DNA that codes for the QRDRs of the two subunits. The oligonucleotide ligation assay (OLA) was then used to identify specific base changes in the amplified product by colorimetric detection. In PCR-OLA, a 5′-biotinylated capture probe, in which the 3′ end is the base of interest, is annealed to the target sequence. A reporter probe labeled at the 3′ end with digoxigenin is annealed to the target sequence at the next base. Ligation of the two probes occurs when there is correct base pairing between the 3′ end of the capture probe and the target, resulting in a single oligonucleotide having a biotin group at the 5′ end and digoxigenin at the 3′ end, which is then captured on a solid surface. Ligation is reported by the colorimetric alkaline phosphatase assay after reaction of an antidigoxigenin antibody conjugated to alkaline phosphatase. When a base mismatch between the 3′ end of the capture probe and the target sequence exists, ligation does not occur; therefore, only the digoxigenin-free capture probe binds the solid surface. The PCR-OLA identified point mutations in DNA for a variety of diseases such as sickle cell anemia (9) and cystic fibrosis (9) as well as for macrolide resistance in Helicobacter pylori (12). OLA requires approximately 12 h to analyze two target sequences compared with automated DNA sequencing, which requires about 33 h to complete.
MICs were determined by the NCCLS broth microdilution method (8). Genomic DNA was extracted from S. pneumoniae as previously described (12). All oligonucleotides were constructed by Sigma-Genosys (The Woodlands, Tex.). Oligonucleotides used as PCR primers flanked the sequences coding for the QRDRs and were derived from the known sequences of the gyrA gene (GenBank accession no. NC_003028) and the parC gene (GenBank accession no. Z67739). The primers were 5′-TGGGTTGAAGCCGGTTCA and 5′-TGCTGGCAAGACCGTTGG, which amplified bases 105 to 452 of the structural gene for GyrA, and 5′-CCGTCGCATTCTTTACG and 5′-AGTTGCTCCATTAACCA, which amplified bases 129 to 491 of the structural gene for ParC. Oligonucleotide capture probes (Table 1) were modified with a 5′ biotin group by incorporation of biotin-labeled phosphoramidite. Oligonucleotide reporter probes (Table 1) were modified by phosphorylation of the 5′ end and digoxigenin labeling of the 3′ end at Sigma-Genosys by proprietary methods.
TABLE 1.
Ligation probes for OLA (5′ to 3′)
| Protein | Position | Target amino acidb | Codon | Ligation probea
|
|
|---|---|---|---|---|---|
| Capture | Reporter | ||||
| ParC | 79 | Ser | TCT | TTCCACCCACACGGGGATTC | TTCTATCTATGATGCCATGGTTC |
| Tyr | TAT | TTCCACCCACACGGGGATTA | TTCTATCTATGATGCCATGGTTC | ||
| Phe | TTT | TTCCACCCACACGGGGATTT | TTCTATCTATGATGCCATGGTTC | ||
| Ala | GCT | TTCCACCCACACGGGGATGC | TTCTATCTATGATGCCATGGTTC | ||
| 83 | Asp | GAT | CGGGGATTCTTCTATCTATG | ATGCCATGGTTCGTATGTCT | |
| Tyr | TAT | CGGGGATTCTTCTATCTATT | ATGCCATGGTTCGTATGTCT | ||
| Asn | AAT | CGGGGATTCTTCTATCTATA | ATGCCATGGTTCGTATGTCT | ||
| His | CAT | CGGGGATTCTTCTATCTATC | ATGCCATGGTTCGTATGTCT | ||
| Gly | GGT | CGGGGATTCTTCTATCTATG | GTGCCATGGTTCGTATGTCT | ||
| Val | GTT | CGGGGATTCTTCTATCTATG | TTGCCATGGTTCGTATGTCT | ||
| GyrA | 84 | Ser | TCC | TATCACCCACACGGGGATTC | CTCTATTTATGAAGCCATGGTCC |
| Phe | TTC | TATCACCCACACGGGGATTT | CTCTATTTATGAAGCCATGGTCC | ||
| Ala | GCC | TATCACCCACACGGGGATGC | TCTATTTATGAAGCCATGGTCC | ||
| Tyr | TAC | TATCACCCACACGGGGATTA | TCTATTTATGAAGCCATGGTCC | ||
| Cys | TGC | TATCACCCACACGGGGATTG | TCTATTTATGAAGCCATGGTCC | ||
| 88 | Glu | GAA | ACACGGGGATTACTCTATTTATG | AAGCCATGGTCCGTATGGCT | |
| Lys | AAA | ACACGGGGATTACTCTATTTATA | AAGCCATGGTCCGTATGGCT | ||
| Gly | GGA | ACACGGGGATTACTCTATTTATG | GAGCCATGGTCCGTATGGCT | ||
Capture probes are labeled with biotin on the 5′ end. Reporter probes are phosphorylated on the 5′ end and labeled with digoxigenin on the 3′ end.
For each position, the wild-type amino acid is given first.
PCR amplification was performed under the following conditions with SuperMix (Gibco-BRL, Gaithersburg, Md.). Thermocycling conditions for parC were 40 cycles of 95°C for 3 min, 94°C for 15 s, 52°C for 30 s, and 72°C for 45 s. Cycling conditions for gyrA were 40 cycles of 95°C for 3 min, 94°C for 15 s, 45°C for 30 s, and 72°C for 30 s. Amplified products were analyzed by electrophoresis in ethidium bromide-stained 2% agarose gels for confirmation that the products, 347 bp of gyrA and 362 bp of parC, were amplified. Ligation reactions (in 20-μl mixtures) and detection of ligated products were performed as described previously (12) except that reactions were conducted in a model 9600 DNA thermocycler (Perkin-Elmer, Foster City, Calif.) for 10 cycles of 94°C for 30 s and 52°C for 5 min. Amplified products were cycle sequenced with the Big Dye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) on an automated sequencer (model 373A; Applied Biosystems).
A total of 53 S. pneumoniae strains from recent Abbott-sponsored clinical trials were analyzed by PCR-OLA and DNA sequence analysis (Table 2). The ciprofloxacin MICs for 16 of the 53 were ≤2 μg/ml, and those for the remaining 37 were ≥4 μg/ml. Thirty of the 37 S. pneumoniae strains for which the ciprofloxacin MICs were ≥4 μg/ml had at least one amino acid substitution either in ParC at Ser79 or Asp83 or in GyrA at Ser81 or Glu85. A strain for which the ciprofloxacin MIC was 32 μg/ml had a single amino acid substitution, Ser81 to Phe (Ser81-Pro), in GyrA. All 16 strains for which the MICs were ≤2 μg/ml were wild type for both parC and gyrA. Four strains for which the ciprofloxacin MICs were 4 μg/ml, two strains for which the ciprofloxacin MICs were 8 μg/ml, and one strain for which the ciprofloxacin MIC was 16 μg/ml were wild type for both parC and gyrA.
TABLE 2.
Ciprofloxacin MICs and OLA results for 53 strains of S. pneumoniae
| No. of strains | Ciprofloxacin MIC (μg/ml) | Amino acid substitutionb at indicated position in:
|
Resulta | |||
|---|---|---|---|---|---|---|
| ParC
|
GyrA
|
|||||
| 79 | 83 | 81 | 85 | |||
| 5 | 0.5 | + | + | + | + | Yes |
| 5 | 1 | + | + | + | + | Yes |
| 6 | 2 | + | + | + | + | Yes |
| 4 | 4 | + | + | + | + | Yes |
| 2 | 4 | Phe | + | + | + | Yes |
| 1 | 4 | + | Asn | + | + | Yes |
| 2 | 8 | + | + | + | + | Yes |
| 1 | 8 | Phe | + | + | + | Yes |
| 1 | 8 | Phe | + | Phe | + | Yes |
| 1 | 8 | + | Asn | + | + | Yes |
| 1 | 16 | + | + | + | + | Yes |
| 1 | 16 | Phe | + | Phe | + | Yes |
| 1 | 16 | Phe | + | + | Lys | Yes |
| 1 | 16 | Tyr | + | Ala | + | Yes |
| 1 | 16 | + | Tyr | Phe | + | Yes |
| 3 | 16 | Tyr | + | Phe | + | Yes |
| 1 | 16 | Tyr | + | Tyr | + | Yes |
| 1 | 16 | Tyr | + | + | + | Yes |
| 1 | 32 | + | + | Phe | + | Yes |
| 1 | 32 | Tyr | + | Phe | + | Yes |
| 1 | 32 | Tyr | + | + | Lys | Yes |
| 3 | 32 | Phe | + | Phe | + | Yes |
| 1 | 32 | Tyr | + | + | Lys | Yes |
| 1 | 32 | + | Tyr | Phe | + | Yes |
| 1 | 32 | Tyr | + | Tyr | + | Yes |
| 2 | 64 | Phe | + | Phe | + | Yes |
| 1 | 64 | Phe | + | Tyr | + | Yes |
| 1 | 64 | Tyr | + | Phe | + | Yes |
| 1 | 64 | Tyr | Val | Phe | + | Yes |
| 1 | 64 | + | Asn | Tyr | + | Yes |
Yes, PCR-OLA in agreement with sequence results.
+, wild-type amino acid.
Compared with sequence data from the same regions, PCR-OLA correctly identified DNA sequences that code for the amino acids at four QRDR positions most frequently associated with quinolone susceptibility or resistance in 53 strains (Table 2), indicating that the OLA is a valid method for detecting mutations associated with quinolone resistance. For the seven strains without QRDR mutations for which the ciprofloxacin MICs were 4 to 16 μg/ml the gatifloxacin and moxifloxacin MICs were less than 1 μg/ml. Therefore, efflux could be responsible for the higher MICs. Alternatively, mutations outside the amplified region could be present.
Other amino acid substitutions can also contribute to decreased susceptibility to quinolones. For instance, Ser80-Pro (5), Lys137-Asn, and Ala115-Pro (2) in ParC, Asp435-Asn (5) in ParE, and Glu474-Lys (11) in GyrB have been suggested. We did not have clinical isolates containing these mutations in this study. Therefore, we constructed oligonucleotides (approximately 40-mers) from the GenBank sequence that represented a wild-type sequence and a mutant sequence at these sites to serve as surrogate templates, and we synthesized the corresponding capture probes and reporter probes. The PCR-OLA identified the correct DNA sequence when used with the oligonucleotide templates that represented rarer mutations associated with quinolone resistance as well as more common mutations (data not shown). Thus, synthetic templates can be used to validate any mutation of interest prior to screening clinical isolates.
The OLA readily identified the bases at key QRDR sites in 51 of 53 strains. It gave a high signal-to-noise ratio for each targeted sequence (Fig. 1 and Table 3). Although PCR products were obtained from the remaining two isolates, they produced very weak signals at Ser81 and Glu85 for wild-type GyrA by PCR-OLA. Polymorphisms associated with the target sequence near the amino acid of interest likely prevented proper annealing and ligation of the probes. Based on the actual sequences of these strains, new capture and reporter probes that gave a higher positive signal in the OLA were constructed (data not shown). If limited polymorphisms at critical sites such as those seen with these two strains are prevalent in clinical isolates, pools containing equal molar amounts of modified probes specific for each polymorphic allele could be used to increase the likelihood of a strong positive signal. Alternatively, if polymorphisms resulting in failed or weak OLA reactions are rare, direct DNA sequencing could be used instead; in this study, sequencing would have been done on only 2 of 106 (2%) target sequences.
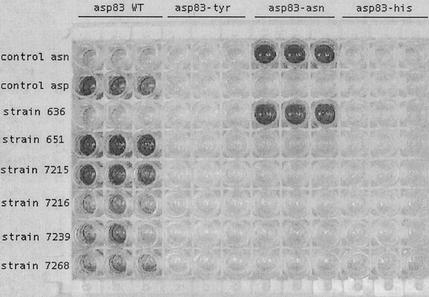
FIG. 1.
PCR-OLA identification of quinolone resistance mutations in parC of S. pneumoniae. Shown is the 96-well colorimetric reaction for several strains of S. pneumoniae. Included were triplicate samples of strains with wild-type (WT) ParC (Asp83) and ParC with Asp83-Tyr, Asp83-Asn, and Asp83-His mutations. Top row, positive control for Asp83-Asn mutation; second row, positive control for WT Asp83; third to eighth rows, various strains of S. pneumoniae.
TABLE 3.
Ranges of optical densities for 53 isolates tested for gyrA and parC genes by PCR-OLA
| Protein | Amino acida | nb | OD492 rangec |
|---|---|---|---|
| ParC | Negative (Ser79) | 106 | 0.192-0.379 |
| Ser79 (wt) | 29 | 0.746-1.896 | |
| Phe79 | 12 | 0.676-1.650 | |
| Tyr79 | 12 | 1.016-1.971 | |
| Negative (Asp83) | 159 | 0.052-0.120 | |
| Asp83 (wt) | 47 | 0.587-1.761 | |
| Tyr83 | 2 | 1.287-1.420 | |
| Val83 | 1 | 1.350-1.412 | |
| Asn83 | 3 | 0.722-1.260 | |
| GyrA | Negative (Ser81) | 159 | 0.073-0.493 |
| Ser81 (wt) | 32 | 1.268-1.887 | |
| Phe81 | 16 | 1.225-1.972 | |
| Ala81 | 1 | 1.220-1.224 | |
| Tyr81 | 4 | 1.235-1.986 | |
| Negative (Glu85) | 53 | 0.142-0.241 | |
| Glu85 (wt) | 50 | 0.486-1.882 | |
| Lys85 | 3 | 0.715-0.902 |
Negative, all negative reactions at indicated position; wt, wild type.
n, number of target sequences.
OD492, optical density at 492 nm.
We used a single colorimetric detection system for all mutations of interest in separate reactions (Fig. 1). An alternative would be the development of a multiplex method using multicolor probes or fluorescently labeled probes with different excitation wavelengths to detect multiple mutations on a single PCR product and significantly increase the throughput of the assay. For example, multiplex OLA was used to genotype six different myostatin mutations in cattle (6). Automation can also be applied to screen multiple samples such as large collections of isolates from clinical trials or surveillance studies. In conclusion, the PCR-OLA is a rapid method that can discriminate single base pair mutations in DNA. In this study, we validated that PCR-OLA is an accurate method for identifying mutations associated with quinolone resistance in S. pneumoniae.
REFERENCES
- 1.Baranova, N., and A. Neyfakh. 1997. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1396-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bast, D. J., D. E. Low, C. L. Duncan, L. Kilburn, L. A. Mandell, R. J. Davidson, and J. C. S. de Azavedo. 2000. Fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae: contributions of type II topoisomerase mutations and efflux to levels of resistance. Antimicrob. Agents Chemother. 44:3049-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones, M. E., D. F. Sahm, N. Martin, S. Scheuring, P. Heisig, C. Thornsberry, K. Köhrer, and F.-J. Schmitz. 2000. Prevalence of gyrA, gyrB, parC, and parE mutations in clinical isolates of Streptococcus pneumoniae with decreased susceptibilities to different fluoroquinolones and originating from worldwide surveillance studies during the 1997-1998 respiratory season. Antimicrob. Agents Chemother. 44:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorgensen, J. H., L. M. Weigel, J. M. Swenson, C. G. Whitney, M. J. Ferraro, and F. C. Tenover. 2000. Activities of clinafloxacin, gatifloxacin, gemifloxacin, and trovafloxacin against recent clinical isolates of levofloxacin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karim, L., W. Coppieters, L. Grobet, A. Valentini, and M. Georges. 2000. Convenient genotyping of six myostatin mutations causing double-muscling in cattle using a multiplex oligonucleotide ligation assay. Anim. Genet. 31:396-399. [DOI] [PubMed] [Google Scholar]
- 7.Muñoz, R., and A. G. De La Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 1995. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 3rd ed. Approved standard M7-A3. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Nickerson, D. A., R. Kaiser, S. Lappin, J. Stewart, L. Hood, and U. Landegren. 1990. Automated DNA diagnostics using an ELISA-based oligonucleotide ligation assay. Proc. Natl. Acad. Sci. USA 87:8923-8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan, X., J. Ambler, S. Mehtar, and L. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan, X.-S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone, G. G., D. Shortridge, J. Versalovic, J. Beyer, R. K. Flamm, and S. K. Tanaka. 1997. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 41:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tankovic, J., B. Perichon, J. Duval, and P. Courvalin. 1996. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob. Agents Chemother. 40:2505-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeller, V., C. Janoir, M. Kitzis, L. Gutmann, and N. Moreau. 1997. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1973-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]