Abstract
Exposure of Pseudomonas aeruginosa to aminoglycosides frequently selects for recalcitrant subpopulations exhibiting an unstable, “adaptive” resistance to these antibiotics. In this study, we investigated the implication in the phenomenon of MexXY-OprM, an active efflux system known to export aminoglycosides in P. aeruginosa. Immunoblotting experiments demonstrated that the transporter MexY, but not the outer membrane pore OprM, was overproduced during the post-drug exposure adaptation period in wild-type strain PAO1. Furthermore, MexY production was dependent upon the degree of bacterial exposure to gentamicin (drug concentration). In contrast to parental strain PAO1, mutants defective in MexXY or in OprM were unable to develop adaptive resistance. Altogether, these results indicate that the resistance process requires the rapid production of MexXY and the interaction of these proteins with the constitutively produced component OprM.
Aminoglycosides remain invaluable antibiotics in the treatment of severe infections caused by Pseudomonas aeruginosa. However, their in vivo efficacy may be compromised by the development of transiently resistant subpopulations (7, 8, 15, 34). Exposure of susceptible P. aeruginosa to an aminoglycoside classically results in an early and rapid drug concentration-dependent killing followed by a phase of bacterial refractoriness characterized by a slow drug concentration-independent killing (27). This so-called adaptive resistance, which is distinct from the postantibiotic effect and which disappears when the organism is no longer in contact with the aminoglycoside, has been observed in vitro (5, 11, 19), in animal models of infection (12, 40), and in patients with cystic fibrosis (6). Because of its ephemeral and reproducible nature, adaptive resistance is not believed to result from mutational events. Ribosomal alterations and drug inactivation are not considered plausible mechanisms, either, as they would result in specific patterns of susceptibility to aminoglycosides and not cross-resistance to these antibiotics (11). Reduced intracellular accumulation of aminoglycosides, which is concomitant to adaptive resistance, was first interpreted as the consequence of lower drug uptake across the bacterial envelopes (11, 19). Supporting this assumption, pleiotropic changes in the protein profiles of the cytoplasmic membrane were detected in drug-exposed bacteria by some investigators (19). However, clear evidence for a substantial decrease in aminoglycoside transport across the inner membrane could not be obtained. The membrane potential Δψ (the driving force for drug entry) appears to be marginally diminished in adaptively resistant bacteria (19), a finding which agrees well with the observation that surviving bacteria grow normally during the postexposure refractory phase (11, 19). These characteristics are opposite of those of another drug-recalcitrant subpopulation also selectable by aminoglycosides, composed of energy-deficient variants (also called small-colony variants) (7, 34).
Recently, several groups have almost simultaneously reported the identification in P. aeruginosa of a new multidrug efflux pump named MexXY (32, 35) or AmrAB (37). In addition to its ability to accommodate a wide range of antibiotics (e.g., tetracyclines, macrolides, quinolones, chloramphenicol, and β-lactams), the MexXY system has the distinctive property of being able to export aminoglycosides (29, 30, 35). This system is also involved in antagonism of aminoglycosides by divalent cations (28). MexXY proteins form a functional tripartite efflux machinery with outer membrane component OprM (32, 35) and contribute to the natural resistance of P. aeruginosa towards tetracyclines, macrolides, and aminoglycosides. Consideration of the inducible expression of MexXY by aminoglycosides (29) led us to examine the implication of the efflux system in adaptive resistance.
(A first account of this work was given at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 17 to 20 September 2000 [W. Mao, M. Warren, A. Lee, A. Mistry, and O. Lomovskaya, abstr. 1498]).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
P. aeruginosa PAO1 was used as the wild-type reference strain (B. W. Holloway). Mutant 11B is a mexX::Tn501 (Hgr [mercuric chloride-resistant]) insertion derivative of PAO1 showing hypersusceptibility to aminoglycosides, erythromycin, and tetracycline (35). PAO1T, an oprM::ΩHgr interposon mutant of PAO1, was kindly provided by T. Köhler (31). MICs of antibiotics were determined by the standard microdilution method in Mueller-Hinton broth (MHB) with adjusted concentrations of Ca2+ and Mg2+ (BBL, Cockeysville, Md.) by using inocula of 2.5 × 105 bacteria/ml (3). The resistance levels of PAO1, 11B, and PAO1T to gentamicin were 1.6, 0.4, and 0.25 mg/liter, respectively; MICs of ticarcillin for these strains were 14, 13.5, and 4 mg/liter, respectively. Escherichia coli JM105 (41) was the host strain in all DNA cloning experiments, while E. coli M15(pREP4) (Qiagen, Courtaboeuf, France) was used to produce recombinant peptides. Bacteria were cultured in Luria-Bertani broth (2), in MHB, or on Mueller-Hinton agar plates (Bio-Rad, Ivry sur Seine, France). When necessary, growth media were rendered selective by the addition of the following agents (final concentrations in milligrams per liter): amoxicillin (100), kanamycin (25), and mercuric chloride (15). All bacterial cultures were incubated at 37°C.
DNA methodology.
Chromosomal DNA suitable for PCR amplification was extracted and purified by following the procedure of Chen and Kuo (9). Plasmid DNA was prepared by the standard alkaline lysis method (2) or by using the Plasmid Midi Preps kit from Qiagen. Selected restriction fragments were purified from agarose electrophoresis gels with the JetSorb kit (Genomed Gmbh, Bad Oeynhauden, Germany). Other reagents for molecular biology were purchased from Invitrogen (Cergy Pontoise, France), Stratagene (La Jolla, Calif.), or Sigma-Aldrich (Saint Quentin, France). Transformation of strains of E. coli with plasmid DNA has been described in detail elsewhere (2).
Polyclonal MexY antiserum.
The alignment of amino acid sequences of Mex pumps in P. aeruginosa indicated the second periplasmic loop of MexY (238 residues located between transmembrane segments TM7 and TM8) as a potentially interesting peptide for the generation of specific MexY antibodies, as it does not share significant sequence homologies with other Mex pumps. A His tag was first added to the N terminus of the peptide to facilitate its purification by affinity chromatography. The 713-bp sequence encoding the periplasmic loop was amplified by PCR from the pBlueScript II KS(+) recombinant plasmid pJR49 (35). The sense (5′-GGGGATCCGAAGGCACGCCGATG-3′) and antisense (5′-GGAGATCTGTAGCGGGTCAGTTGCGG-3′) DNA primers used in the PCR were designed to add BamHI and BglII restriction sites (in bold), respectively, at the ends of the amplicon in order to help subsequent subcloning experiments. The reaction mixture (50 μl) contained 100 ng of pJR49, 1 μM (each) primers, 100 μM (each) deoxynucleoside triphosphates, 6% (vol/vol) dimethyl sulfoxide, 2.5 mM MgCl2, and 2 U of Taq polymerase (Perkin Elmer, Foster City, Calif.) in 1× amplification buffer. The mixture was heated at 94°C for 5 min and then subjected to 30 thermal cycles, each consisting of 45 s at 94°C, 45 s at 62°C, and 1 min at 72°C, before a final step at 72°C for 7 min. Once digested with BamHI and BglII, the amplification product was ligated to BamHI-linearized plasmid pQE-30 (Amxr) to yield pQY and introduced by transformation into competent cells of E. coli JM105. Plasmid pQY was subsequently transferred to E. coli M15(pREP4) (Kmr) for quantitative production of the selected peptide.
Overnight culture of the pQY-carrying M15 strain in Luria-Bertani medium supplemented with amoxicillin (100 mg/liter) and kanamycin (25 mg/liter) was diluted 1:59 into fresh medium and incubated at 37°C with shaking. Expression of the cloned sequence was induced by the addition of a 1 mM final concentration of isopropyl-β-thiogalactopyranoside (IPTG) to the exponentially growing cells (A650 of 0.5) and incubation for five additional hours at 37°C to reach peak levels. Bacteria were then collected by centrifugation (2,000 × g, 20 min, 4°C), resuspended in buffer A (50 mM Tris-HCl [pH 8], 100 mM NaCl, 0.2 mM β-mercaptoethanol, 0.05% [vol/vol] Tween 20) at 5 ml per g (wet weight) of pellet, and lysed by sonication (1 min, 20 W at 4°C) (Branson Ultrasonics, Danbury, Conn.). Inclusion bodies corresponding to aggregates of the recombinant peptide were recovered by centrifugation (8,000 × g, 10 min, 4°C), suspended in buffer B (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris-HCl [pH 8]), sonicated for 2 min, and left in the buffer for 1 h at room temperature under gentle shaking. The solution was cleared by another centrifugation (10,000 × g, 10 min, 4°C) and applied on a Ni-nitrilotriacetic acid column (1 by 10 cm; Qiagen S.A.) equilibrated with buffer B. Proteins bound to the column by nonspecific interactions were removed with buffer C (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris-HCl [pH 6.3], 10 mM imidazole), and the His-tagged peptide was finally eluted with buffer C in the presence of 250 mM imidazole. Fractions of interest were pooled and dialyzed at room temperature for 24 h against 5 liters of buffer consisting of 50 mM Tris-HCl (pH 6.3), 200 mM NaCl, and 0.1 mM EDTA. The purified peptide served to raise antibodies in two rabbits by multiple intradermal injections (36). Cross-reactivity of the MexY antiserum with other Mex pumps was checked by using mutants of PAO1 overproducing MexAB-OprM (strain 4098E), MexCD-OprJ (strain ERYR), and MexEF-OprN (strain PAO7H) efflux systems (21, 25, 31). No band in the 100- to 140-kDa range was detected by immunoblotting with the MexY antiserum in total membrane preparations from these strains (data not shown).
Membrane preparations.
The protocol followed to extract the outer membrane or the total (inner and outer) membrane of P. aeruginosa was adapted from that described by Michéa Hamzehpour et al. (31). Briefly, drug-exposed bacteria in MHB were collected by centrifugation, resuspended in 10 mM HEPES (pH 7.2), and lysed by two passages through a French pressure cell (SLM AMINCO, Rochester, N.Y.). Unbroken cells were removed by centrifugation at 4°C. Total bacterial membranes were then harvested at 100,000 × g for 1 h at 4°C and resuspended in a 15 mM MOPS (morpholinepropanesulfonic acid)-100 mM NaCl buffer (pH 8.0). The inner membrane components were subtracted from this preparation by differential solubilization in a 2% (wt/vol) final concentration of sodium N-lauroylsarcosinate. After a 30-min incubation at room temperature, insoluble materials corresponding to the outer membrane fraction were pelleted by centrifugation at 25,000 × g for 30 min at 4°C and resuspended in the same buffer. Proteins were quantified spectrophotometrically in each fraction by using the BCAprot reagent (Pierce Chemical, Rockford, Ill.) at 60°C and bovine serum albumin as a standard.
Immunoblotting experiments.
Purified extracts of total membrane (20 μg of protein) and outer membrane (10 μg of protein) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (with 15% [wt/vol] acrylamide in the running gel) according to the method of Laemmli (23) and transferred electrophoretically to nitrocellulose filters as previously described (43). These filters were subsequently blocked with 3% (wt/vol) gelatin and hybridized for 1 h with MexY, OprM (43), or MexB (17) antiserum diluted 1:20,000, 1:5,000, or 1:1,000, respectively, in phosphate-buffered saline. The development of membranes was carried out with alkaline phosphatase conjugated to an anti-rabbit secondary antibody by using the AP color reagent kit from Bio-Rad.
Adaptive resistance in vitro.
Adaptive resistance was induced as previously described (11). Briefly, an overnight culture was diluted 10-fold in prewarmed, cation-adjusted MHB and incubated with gentamicin at a fraction or multiple of the MIC for 2 h at 37°C (first exposure). In parallel, a sample of the same culture was simultaneously incubated in drug-free broth and used as a control. After 2 h, the bacteria were harvested by centrifugation at 1,350 × g for 10 min and washed twice with drug-free medium to remove cell-bound antibiotic. Previous studies have demonstrated that this procedure reduces the extracellular concentrations of aminoglycosides to inactive levels (about 1,000-fold) (20). The pellet was resuspended in drug-free broth to yield a postexposure suspension of 104 to 105 CFU/ml (determined in preliminary experiments) and then reincubated at 37°C. Aliquots of this culture were removed every 2 h for 6 h and reexposed to gentamicin (at the MIC for 2 h). Colony counts were determined in duplicates before and after the second exposure on Mueller-Hinton agar plates (supplemented with 15 ng of mercuric chloride/ml for strain PAO1T) with a Spiral Plater apparatus (AES Laboratoire, Combourg, France). Experiments were repeated at least twice to ensure the reproducibility of the data. Adaptive resistance was characterized by the reduced bactericidal effect of the second gentamicin exposure compared with that of the first exposure.
RESULTS AND DISCUSSION
Medical experience shows that the treatment of P. aeruginosa infections with antibiotics otherwise found efficacious by in vitro tests may be unsuccessful (10, 22). How drug-susceptible P. aeruginosa may survive and persist in patients under appropriate chemotherapy is a long-standing question. This complex, multifactorial phenomenon actually involves host-related and bacterium-related factors (8). In the present work, we attempted to identify some of the mechanisms by which initially susceptible bacteria may transiently develop resistance to aminoglycosides and evade chemotherapy.
Role of efflux system MexXY-OprM in adaptive resistance.
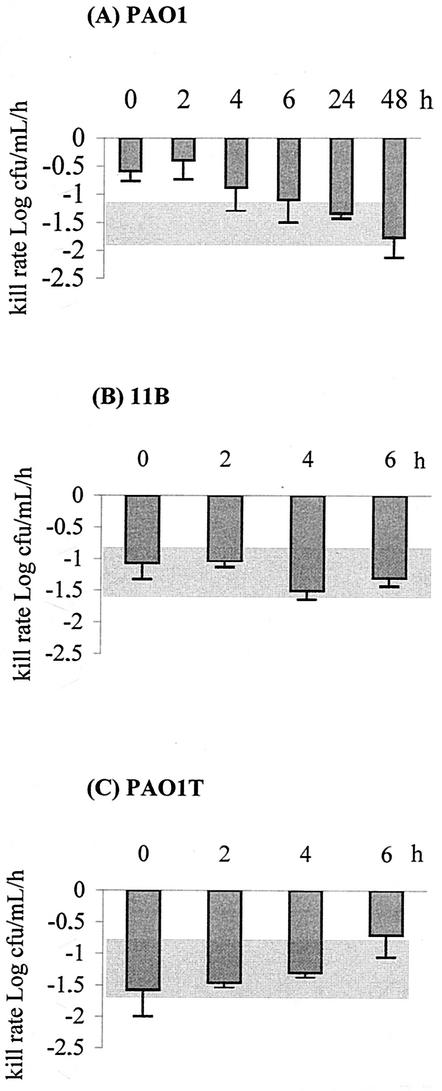
The implication of MexXY-OprM was investigated in wild-type strain PAO1 and its mexXY- and oprM-defective mutants 11B and PAO1T, respectively. The conditions used in the pioneering work of Daikos et al. (11) to demonstrate and characterize adaptive resistance were reproduced. Figure 1A shows that a single exposure of strain PAO1 to the MIC of gentamicin (1.6 mg/liter) for 2 h resulted in significant bacterial killing averaging −1.4 ± 0.3 log10 CFU/ml/h over the growth period tested. As observed elsewhere (11, 19), reexposure of PAO1 to gentamicin (at the MIC for 2 h) at different time intervals after the removal of the aminoglycoside demonstrated the appearance and recession of bacterial refractoriness to drug killing. This so-called adaptive resistance was maximal 2 h and reversed 6 h after the initial exposure. Interestingly, mutants 11B and PAO1T did not develop such a resistance when treated under the same conditions, that is, with the MICs of gentamicin (0.4 and 0.25 mg/liter, respectively) (Fig. 1B and C). This finding strongly suggested that the MexXY-OprM system is, at least in part, responsible for the resistance process.
FIG. 1.
Bactericidal effects on P. aeruginosa of a second exposure to the MIC of gentamicin for 2 h. (A) Wild-type PAO1. (B) MexXY-defective mutant 11B. (C) OprM null mutant PAO1T. Error bars show standard deviations. The test cultures were preexposed for 2 h to the MIC. The grey areas indicate the bactericidal rates in control cultures of a single exposure to gentamicin for 2 h at the MIC (−1.40 ± 0.30, −1.27 ± 0.38, and −1.25 ± 0.42 log10 CFU/ml/h for the PAO1, 11B, and PAO1T strains, respectively).
MexY is overproduced during the adaptation phase.
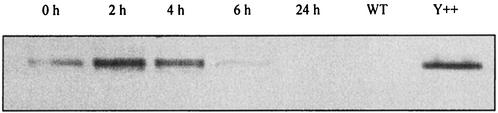
To determine whether the development of adaptive resistance correlates with enhanced production of MexXY in gentamicin-exposed bacteria, total membrane extracts from strain PAO1 were analyzed by Western blotting with a MexY-specific antiserum (obtained as described in Materials and Methods). MexY was almost undetectable on immunoblots prepared from bacteria grown in antibiotic-free medium (Fig. 2, lane WT), a result that confirms the very low level of expression of mexXY efflux genes in the absence of aminoglycosides (29). On the other hand, the exposure of PAO1 cells to the MIC of gentamicin for 2 h triggered the rapid production of MexY, which reached a maximum 2 h and started to recess 4 h after drug removal (Fig. 2, lanes 0 h to 24 h). Thus, the increase in MexY amounts was concomitant to the adaptive period (compare with Fig. 1).
FIG. 2.
Amounts of MexY produced by P. aeruginosa PAO1 immediately (0 h) and 2, 4, 6, or 24 h after a single gentamicin exposure of 2 h at the MIC. Total membranes were extracted from the exposed bacteria and electrophoresed (20 μg of protein per lane) in sodium dodecyl sulfate-15% polyacrylamide gels. Western blot analyses were performed with MexY-specific antiserum. MexXY-overproducing strain PAO1(pAGH97) was used as a positive control (Y++) for MexY detection, and PAO1(pAK1900) was used as a wild-type control (WT) (35).
These data show quite conclusively that the efflux system MexXY is rapidly overproduced in a subpopulation of bacteria surviving the first contact with the aminoglycoside (less than 1% of the initial inoculum). Whether these bacteria have features distinctive from those of the rest of the population that could account for their stronger intrinsic resistance to drug killing is not known. In terms of survival, however, fast activation of MexXY production (within 2 h) is likely to be an advantage since it prevents intracellular accumulation of aminoglycosides, which are efficient inhibitors of protein synthesis. Consistent with this notion, adaptive resistance was reported to develop only in metabolically active, growing cells, to require a certain time to appear (1 to 4 h), and to be suppressed by DNA polymerase inhibitor rifampin (5, 11, 19, 38). De novo protein synthesis is therefore necessary to increase resistance to aminoglycosides during the adaptation period. The observation that the inactivation of MexXY or OprM abolishes the resistance process strongly supports the notion that adaptive resistance is due to aminoglycoside efflux rather than decreased drug uptake. As shown by Fig. 2, the activation of mexXY expression is turned down when P. aeruginosa is no longer in contact with an aminoglycoside. Recent data obtained in our laboratory with a plasmid-borne mexX::lacZ fusion have provided evidence that MexZ, a member of the TetR family, exerts a strong repression on efflux operon mexXY (C. Vogne, D. Hocquet, J. Ramos Aires, F. El Garch, P. Plésiat, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-434, 2002).
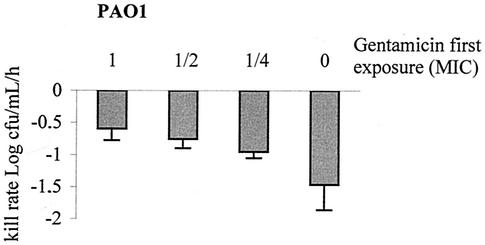
Previous studies (5, 39) have established that bacterial refractoriness is greater in intensity (resistance of survivors to killing) and in duration (persistence of resistance after the original exposure) when P. aeruginosa cells are initially confronted with higher doses of aminoglycosides. We thus compared the inductive effects of different concentrations of gentamicin on MexY production. As expected, amounts of MexY (Fig. 3) and adaptive resistance (Fig. 4) proved to be greater in cells treated with higher concentrations of gentamicin, thereby demonstrating that the degree of adaptive resistance depends on MexXY levels.
FIG. 3.
Amounts of MexY produced by strain PAO1 after a single gentamicin exposure at 1.6 mg/liter (MIC; lane 1a), 0.8 mg/liter (half the MIC; lane 2a), or 0.4 mg/liter (one-fourth the MIC; lane 3a) for 2 h. Western blots were prepared as described in the legend for Fig. 2 and developed with a MexY antiserum. In each case, exposed bacteria were subsequently cultured for 24 h in drug-free MHB (lanes 1b, 2b, and 3b).
FIG. 4.
Bactericidal effects on P. aeruginosa PAO1 of a second exposure to gentamicin at the MIC for 2 h. The bacteria were preexposed for 2 h to no gentamicin (0), one-fourth or one-half the MIC, or the MIC (1) of gentamicin. Error bars show standard deviations.
Production of OprM and MexB.
Since MexXY proteins are functional with outer membrane component OprM (32, 35), we investigated whether amounts of OprM and MexXY increase in parallel in adaptively resistant bacteria. OprM appears to be produced constitutively in P. aeruginosa cells grown under standard laboratory conditions (26). Its structural gene, oprM, has been found to be cotranscribed with mexA and mexB from two promoters located upstream of mexA (13). However, an expression of oprM independent of that of mexAB has also been demonstrated, involving a third promoter located within gene mexB (42). In agreement with other results (29), immunoblotting analysis of outer membrane preparations from strain PAO1 developed with an OprM-specific antiserum (43) failed to reveal an increased production of OprM in postexposure resistant cells (data not shown). The same result was obtained with strain 11B (defective in MexXY) examined 4 h after an initial exposure to the MIC of gentamicin (the time of peak of adaptive resistance in PAO1) (data not shown). This indicates that, while required by MexXY for aminoglycoside export in drug-challenged bacteria, OprM is not upregulated with MexXY. Alternatively, one could imagine that the expression of mexAB is repressed during the induction of mexXY, as some efflux pumps appear to be inversely coregulated in P. aeruginosa (24). Semiquantitative determination of MexB on total membrane immunoblots developed with a MexB-specific antiserum demonstrated that it is not the case (data not shown). Given that OprM forms a tripartite drug efflux complex with MexA-MexB and that MexB amounts do not decrease in adaptively resistant cells, it is reasonable to assume that MexXY competes with MexAB to bind OprM. If this assumption is correct, resistance to aminoglycosides (substrates for MexXY but not for MexAB) will develop to the detriment of resistance to ticarcillin (a substrate for MexAB but not for MexXY) (30). If confirmed, such a competition between the two pump systems for OprM could, at least in part, account for the synergistic interactions frequently observed between aminoglycosides and β-lactams in P. aeruginosa.
Clinical aspects.
It has long been known that the exposure of P. aeruginosa to inhibitory concentrations (near the MICs) of aminoglycosides easily selects for drug-resistant subpopulations (1, 33). Some of these bacteria, designated small-colony variants because of their reduced growth rate on solid media, are deficient in energy-dependent uptake of aminoglycosides (7, 14, 16). Over the past decade, evidence has accumulated that such variants may emerge in vivo, resulting in bad clinical response or failure of therapy (7, 15, 18, 34). A second recalcitrant subpopulation can also be selected by aminoglycosides. The resistance process developing in these bacteria does not alter their growth rates and has been qualified as adaptive resistance (5, 11, 19). There is now little doubt that the two aforementioned resistant populations reflect distinct phenotypic adaptations to the lethal action of aminoglycosides and that these populations may coexist at the infection site, such as the cystic fibrotic lung (6, 18). Adaptive resistance is one of the major arguments for once-daily aminoglycoside therapy (4). Interestingly, a MexXY-OprM efflux pump inhibitor enhancing the activity of aminoglycosides has recently been discovered (A. Lee, D. Lofland, D. Madsen, M. S. Warren, P. Plésiat, O. Lomovskaya, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-433, 2002). According to our results, such an inhibitor could prevent the manifestation of adaptive resistance and represent a valuable approach to improve the clinical efficacy of aminoglycoside antibiotics.
Acknowledgments
C.V. and F.E.G. were sponsored by the French Cystic Fibrosis Association “Vaincre la mucoviscidose.”
REFERENCES
- 1.Annear, D. I. 1975. Unstable gentamicin resistance with linkage to colony size in Pseudomonas aeruginosa. Pathology 7:281-283. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Balows, A., W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy. 1991. Manual of clinical microbiology, 5th ed. ASM Press, Washington, D.C.
- 4.Barclay, M. L., and E. J. Begg. 2001. Aminoglycoside adaptive resistance: importance for effective dosage regimens. Drugs 61:713-721. [DOI] [PubMed] [Google Scholar]
- 5.Barclay, M. L., E. J. Begg, and S. T. Chambers. 1992. Adaptive resistance following single doses of gentamicin in a dynamic in vitro model. Antimicrob. Agents Chemother. 36:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barclay, M. L., E. J. Begg, S. T. Chambers, P. E. Thornley, P. K. Pattemore, and K. Grimwood. 1996. Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J. Antimicrob. Chemother. 37:1155-1164. [DOI] [PubMed] [Google Scholar]
- 7.Bayer, A. S., D. S. Norman, and K. S. Kim. 1987. Characterization of impermeability variants of Pseudomonas aeruginosa isolated during unsuccessful therapy of experimental endocarditis. Antimicrob. Agents Chemother. 31:70-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan, L. E. 1989. Microbial persistence or phenotypic adaptation to antimicrobial agents: cystic fibrosis as an illustrative case, p. 411-418. In L. E. Bryan (ed.), Microbial resistance to drugs. Springer-Verlag, Berlin, Germany.
- 9.Chen, W. P., and T. T. Kuo. 1993. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 21:2260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig, W. A., and S. C. Ebert. 1994. Antimicrobial therapy in Pseudomonas aeruginosa infections, p. 441-517. In A. L. Baltch and R. P. Smith (ed.), Pseudomonas aeruginosa infections and treatment. Marcel Dekker, Inc., New York, N.Y.
- 11.Daikos, G. L., G. G. Jackson, V. T. Lolans, and D. M. Livermore. 1990. Adaptive resistance to aminoglycoside antibiotics from first-exposure down-regulation. J. Infect. Dis. 162:414-420. [DOI] [PubMed] [Google Scholar]
- 12.Daikos, G. L., V. T. Lolans, and G. G. Jackson. 1991. First-exposure adaptive resistance to aminoglycoside antibiotics in vivo with meaning for optimal clinical use. Antimicrob. Agents Chemother. 35:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber, A. U., and W. A. Craig. 1982. Aminoglycoside-selected subpopulations of Pseudomonas aeruginosa: characterization and virulence in normal and leukopenic mice. J. Lab. Clin. Med. 100:671-681. [PubMed] [Google Scholar]
- 15.Gerber, A. U., P. A. Vastola, J. Brandel, and W. A. Craig. 1982. Selection of aminoglycoside-resistant variants of Pseudomonas aeruginosa in an in vivo model. J. Infect. Dis. 146:691-697. [DOI] [PubMed] [Google Scholar]
- 16.Gilleland, L. B., H. E. Gilleland, J. A. Gibson, and F. R. Champlin. 1989. Adaptive resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. J. Med. Microbiol. 29:41-50. [DOI] [PubMed] [Google Scholar]
- 17.Gotoh, N., H. Tsujimoto, M. Tsuda, K. Okamoto, A. Nomura, T. Wada, M. Nakahashi, and T. Nishino. 1998. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häuβler, S., B. Tümmler, H. Weiβbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 19.Karlowsky, J. A., M. H. Saunders, G. A. Harding, D. J. Hoban, and G. G. Zhanel. 1996. In vitro characterization of aminoglycoside adaptive resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:1387-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlowsky, J. A., G. G. Zhanel, R. J. Davidson, and D. J. Hoban. 1994. Postantibiotic effect in Pseudomonas aeruginosa following single and multiple aminoglycoside exposures in vitro. J. Antimicrob. Chemother. 33:937-947. [DOI] [PubMed] [Google Scholar]
- 21.Köhler, T., M. Michéa Hamzehpour, U. Henze, N. Gotoh, L. Kocjancic Curty, and J. C. Pechère. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 22.Korvick, J., and V. Yu. 1991. Antimicrobial agent therapy for Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:2167-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Li, X. Z., N. Barré, and K. Poole. 2000. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 46:885-893. [DOI] [PubMed] [Google Scholar]
- 25.Li, X. Z., D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 38:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacArthur, R. D., V. Lolans, F. A. Zar, and G. G. Jackson. 1984. Biphasic, concentration-dependent and rate-limited, concentration-independent bacterial killing by an aminoglycoside antibiotic. J. Infect. Dis. 150:778-779. [DOI] [PubMed] [Google Scholar]
- 28.Mao, W., M. S. Warren, A. Lee, A. Mistry, and O. Lomovskaya. 2001. MexXY-OprM efflux pump is required for antagonism of aminoglycosides by divalent cations in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michéa Hamzehpour, M., J. C. Pechère, P. Plésiat, and T. Köhler. 1995. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:2392-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson, L., L. Sörén, and G. Rådberg. 1987. Frequencies of variants resistant to different aminoglycosides in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 20:255-259. [DOI] [PubMed] [Google Scholar]
- 34.Parr, T. R., Jr., and A. S. Bayer. 1988. Mechanisms of aminoglycoside resistance in variants of Pseudomonas aeruginosa isolated during treatment of experimental endocarditis in rabbits. J. Infect. Dis. 158:1003-1010. [DOI] [PubMed] [Google Scholar]
- 35.Ramos Aires, J., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaitukaitis, J. L. 1981. Production of antisera with small doses of immunogen: multiple intradermal injections. Methods Enzymol. 73:46-52. [DOI] [PubMed] [Google Scholar]
- 37.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong, Y. Q., J. Caillon, H. Drugeon, G. Potel, and D. Baron. 1996. The effect of rifampicin on adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. J. Antimicrob. Chemother. 37:993-998. [DOI] [PubMed] [Google Scholar]
- 39.Xiong, Y. Q., J. Caillon, H. Drugeon, G. Potel, and D. Baron. 1996. Influence of pH on adaptive resistance of Pseudomonas aeruginosa to aminoglycosides and their postantibiotic effects. Antimicrob. Agents Chemother. 40:35-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong, Y. Q., J. Caillon, M. F. Kergueris, H. Drugeon, D. Baron, G. Potel, and A. S. Bayer. 1997. Adaptive resistance of Pseudomonas aeruginosa induced by aminoglycosides and killing kinetics in a rabbit endocarditis model. Antimicrob. Agents Chemother. 41:823-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanisch-Perron, C., J. Vieria, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 42.Zhao, Q., X. Z. Li, R. Srikumar, and K. Poole. 1998. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob. Agents Chemother. 42:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziha-Zarifi, I., C. Llanes, T. Köhler, J. C. Pechère, and P. Plésiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]