Abstract
A 45-kb R plasmid, pRAS1, that confers resistance to tetracyclines, trimethoprim, and sulfonamides was isolated in 1989 from an atypical strain of the fish pathogen Aeromonas salmonicida. This plasmid could be transferred by conjugation to Escherichia coli with a high degree of efficiency (frequency, 0.48). The following year pRAS1 was isolated from A. salmonicida subsp. salmonicida in the same area. Incompatibility group U plasmid pRAS1 contained a drug resistance-determining region of 12 kb consisting of a class 1 integron similar to In4 of Tn1696 but with a dfrA16 gene cassette inserted. Close to IS6100 at the right end of Tn4 was a truncated Tn1721. Restriction enzyme analysis showed that R plasmid pAr-32, isolated from A. salmonicida in Japan in 1970, had the same backbone structure as pRAS1, while the drug resistance-determining region contained a complex class 1 integron with an aadA2 cassette; the chloramphenicol resistance gene catA2, as in In6 of pSa; and a duplicate of the 3′ conserved segment of the integron.
The intensity of farming of Atlantic salmon (Salmo salar L.) has increased rapidly since the 1970s in Norway (31). Aeromonas salmonicida subsp. salmonicida, the etiological agent of the salmonid disease furunculosis, was introduced into Norwegian salmon farms through smolts imported into mid-Norway from Scotland in 1985 (33). Until 1993, when effective vaccines were introduced, the use of antimicrobial drugs was essential for control of the disease. The quinolones oxolinic acid and flumequine were primarily used (13, 14); but decreased susceptibilities to these agents, often observed after only a few months of treatment, made the use of oxytetracycline or a combination of sulfadiazine and trimethoprim necessary (17).
Furunculosis has been treated with antimicrobial drugs in several countries worldwide, and at the end of the 1950s the type strain of A. salmonicida subsp. salmonicida (NCMB833), from the United States, was found to carry transferable antimicrobial drug resistance (7, 31). In Japan, sulfonamide-, chloramphenicol-, and streptomycin-resistant isolates of A. salmonicida subsp. salmonicida were detected in 1970. In one of these strains, a 29-MDa plasmid, pAr-32, was found to be responsible for resistance to all three drugs (6) and was later shown to contain a class 1 integron (20). Plasmid pAr-32 is identical to plasmid RA3, isolated from Aeromonas hydrophila in Japan (6), which is the reference plasmid of plasmid incompatibility group U (IncU) (12).
Class 1 integrons are commonly found in antibiotic-resistant clinical isolates of gram-negative bacteria. Each class 1 integron contains up to several gene cassettes encoding drug resistance, and the pool of such cassettes seems to be large. The cassettes are incorporated into the attI site of the integron by the site-specific enzyme integrase, encoded by the intI1 gene, which is identical in all class 1 integrons (26). The class 1 integron may have originated from the Tn402 transposon with conservation of the 5′ conserved segment (5′-CS), where the integrase gene and the attI site are located (25, 30). Downstream of the cassettes there is a 3′ conserved segment (3′-CS) encoding a truncated qacE gene, a sul1 gene, and sometimes, one or two open reading frames, orf5 and orf6. However, the 3′-CS may vary considerably between the class 1 integrons. The genes and open reading frames of the 3′-CS are considered to be trapped gene cassettes, while the original segment from Tn402 with transposition genes is often completely lost (15, 24).
In this study we have characterized the R plasmid pRAS1 and examined the resistance-determining regions of the two related R plasmids pRAS1and pAr-32.
MATERIALS AND METHODS
Bacterial isolates.
The first isolate of A. salmonicida subsp. salmonicida in Norway found to be resistant to tetracycline, sulfadiazine, and trimethoprim, in addition to quinolones, was strain NVI1995/91, also named strain NVH4133 and denoted strain 1995 here. Strain 1995 was isolated from diseased Atlantic salmon (Salmo salar L.) in 1991 at the Osterfjord of Hordaland County, on the west coast of Norway (32). Atypical A. salmonicida strain NVI2402/89, also denoted strain 718 (frozen stock number) and called strain 2402 here, was found in same narrow fjord in 1989 (32).
Plasmid pAr-32 was isolated from A. salmonicida strain Ar-32 in 1970 from diseased biwamasu (Oncorhynchus rhodurus f. rhodurus) in the Samegai trout hatchery in Shiga, Japan (7), transferred to Escherichia coli W3102 (str), and kindly supplied by T. Aoki.
E. coli DH5α (Life Technologies) was used as the recipient in the conjugation experiments.
Media and antimicrobial susceptibility testing.
A. salmonicida and E. coli were grown on either 5% bovine blood agar or Luria-Bertani agar (LA) plates at 15°C for 2 days and at 37°C for 1 day, respectively. They were stored in Luria-Bertani broth with 18% (vol/vol) glycerol at −80°C.
Susceptibility testing was performed on Mueller-Hinton (MH) agar (Difco, Detroit, Mich.) with antimicrobial discs containing tetracycline (80 μg), sulfamethoxazole (240 μg), trimethoprim (5.2 μg), streptomycin (100 μg), chloramphenicol (60 μg), or kanamycin (100 μg) (NeoSensitabs; Rosco, Taastrup, Denmark). A. salmonicida was incubated for 48 h at 15οC and E. coli was incubated for 24 h at 37°C before the inhibition zones were measured and compared with those of susceptible control strains.
Conjugation experiments and plasmid profiling.
Growing colonies of the donor strains were mixed with approximately equal amounts of growing recipient strain E. coli DH5α, Salmonella enterica serovar Enteritidis (1139), or S. enterica serovar Typhimurium (882/87) on LA plates incubated at room temperature from 1 h to 2 days. Mating was also performed in Luria-Bertani broth by mixing equal numbers of donor and recipient bacteria (measured by optical densitometry) in the exponential phase of growth.
The frequency of transfer was measured as the proportion of the recipients receiving an R plasmid; the total number of recipients was estimated after overnight incubation of MH agar without antibiotics at 37°C, a temperature that inhibits the growth of the donor strain, while selection of transconjugants was performed on MH agar plates with either 10 μg of tetracycline, 10 μg of trimethoprim, or 200 μg of sulfadiazine ml−1 after incubation under conditions similar to those described above. The transfer frequencies were calculated on the basis of the average number of colonies from three consecutive plates in a dilution series.
Plasmids were isolated by the procedure of Kado and Liu (18) or the mini procedure of Birnboim and Doly (9) before separation in 1% agarose gels by vertical gel electrophoresis with Tris-borate-EDTA (TBE) buffer (0.089 M boric acid, 0.0025 M EDTA, 0.089 M Tris [pH 8.0]) at 120 V for 3 h before staining with ethidium bromide and photography. Donor and recipient strains were used as controls in the plasmid profiling procedure.
Restriction endonuclease analysis and construction of pRAS1 restriction map.
The DNA of plasmid pRAS1 from strain 2402 was digested with 31 different restriction enzymes according to the instructions of the manufacturers. Double digestions were systematically used when they were found to be useful. pRAS1 from strain 1995 was digested with the eight enzymes (PstI, BamHI, HindIII, EcoRI, ClaI, AccI, XbaI, and SalI) and combinations of enzymes (SalI-BamHI and SalI-HindIII). The restricted plasmid DNA was separated in 0.7% (wt/vol) agarose gels (SeaKem GTG; FMC Bioproducts, Rockland, Maine) by electrophoresis in TBE buffer at 25 V for 15 h in a horizontal gel apparatus before staining with ethidium bromide and photography. The fragment lengths were calculated, and the pRAS1 restriction map was constructed manually. Subcloning and secondary digestion of fragments of plasmid DNA were not performed.
Hybridization studies.
The plasmid DNA in agarose gels was transferred to nylon membranes by Southern blotting (34) and fixed to the nylon membranes with UV light. Probe DNA (DNA specific for IncU, Tet A, Tet B, Tet C, Tet D, Tet E, sul1, sul2, dfrA1, dfrA2, dfrA3, drfA4, dfrA5, dfrA7, dfrA10, and dfrA12) was prepared from the appropriate plasmids after separation of the restricted fragments in low-melting-point agarose gels (SeaPlaque GTG agarose; FMC Bioproducts) and was labeled with 32P by using an oligonucleotide labeling kit (Bethesda Research Laboratories, Inc., Gaithersburg, Md.). Hybridization was performed at 65°C overnight after 3 h of prehybridization. The membranes were washed at 65 or 70°C by standard procedures. X-ray films (XR film; Kodak, Rochester, N.Y.) were exposed to the membranes at −70°C by using intensifier screens.
PCR primers.
On the basis of the restriction maps of pRAS1 (this study) and pAr-32 (6), areas of similarity to other R plasmids that have been studied were recognized; and PCR primers were designed from the sequences of Tn1696 (EMBL accession number U12338), Tn1721 (EMBL accession number X61367), pSa (EMBL accession number L06822), and catA2 of E. coli (EMBL accession number X53796).
PCR amplification and sequencing of amplified products.
PCRs were performed according to the instructions of the manufacturer of the Taq DNA polymerase (Qiagen GmbH, Hilden, Germany). Template DNA either was isolated plasmid DNA or was from boiled bacterial suspensions. The PCR protocol had an initial denaturation step of 94°C for 3 min and 30 cycles of 94°C for 1 min, 65 to 43°C for 1 to 3 min (depending on the primer set), and 72°C for 2 min, before a final step of 72°C for 5 min. The PCR products were separated by electrophoresis in 0.7% agarose gels in a horizontal gel apparatus.
PCR amplicons were purified by use of the Qiaquick PCR purification kit (Qiagen GmbH), the reaction mixtures for the sequencing PCR were prepared, and the products were purified as recommended in the protocol of the BigDye Terminator Ready Reaction kit (Applied Biosystems, Warrington, United Kingdom). The products from the sequencing PCR were separated with an ABI Prism 377 automated sequencing machine (Perkin-Elmer Cetus Corp., Foster City, Calif.), and the DNA sequences were analyzed by using the GCG Sequence Analysis software package (version 8; Genetics Computer Group, Madison, Wis.) and BLAST search analysis (4).
Nucleotide sequence accession numbers.
The nucleotide sequence from the pRAS1 resistance-determining region has been assigned EMBL accession number AJ517790, and the nucleotide sequence from the pAr-32 resistance-determining region has been assigned EMBL accession number AJ517791.
RESULTS AND DISCUSSION
Antimicrobial susceptibility and plasmids.
Both atypical A. salmonicida 2402 and A. salmonicida subsp. salmonicida 1995 were resistant to tetracycline, trimethoprim, and sulfamethoxazole. Conjugation experiments and plasmid profiling showed that a plasmid of about 25 MDa encoded the drug resistance. The R plasmid from strain 2402 was found to be identical to that from strain 1995, and the common R plasmid of the two strains was named pRAS1 (for plasmid resistance A. salmonicida number 1).
Restriction map of pRAS1 and pAr-32.
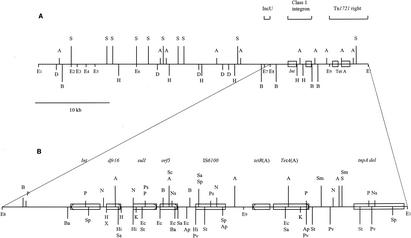
The size of pRAS1 was found to be approximately 45 kb, based on the sum of the sizes of the fragments from suitable restriction digests. No differences were found between digests of pRAS1 from strains 1995 and 2402. It was possible to map the various restriction sites to a resolution of a few hundred base pairs; sites from the same enzyme closer than this were mapped as one (Fig. 1). Of the 94 mapped restriction sites recognized by 23 enzymes, 57 sites (60%) were concentrated to an area of 11.5 kb, which is about 25% of the plasmid (Fig. 1B). Only 7 of the 23 enzymes digested regions in the remaining larger (75%) part of pRAS1. These seven enzymes digested none (DraI) or only a few sites that mapped to the 25% part of the plasmid with a high density of restriction sites, while they digested regions at several positions evenly distributed over the remaining 75% of the plasmid. This uneven distribution of restriction sites may be related to the different G+C contents of the two parts of the plasmid; however, this was not determined. Six enzymes, BclI, ClaI KpnI, NotI, ScaI, and XbaI, did not digest pRAS1.
FIG. 1.
(A) Restriction enzyme map of 45-kb plasmid pRAS1 from A. salmonicida. The restriction enzyme sites for EcoRI (E) are numbered from 1 to 9. (B) Restriction enzyme map of the antibiotic resistance-determining region of pRAS1. The restriction enzyme sites are abbreviated as follows: A, AccI; Ap, ApaI; Ba, BamHI; B, BglII; D, DraI; Ec, EcoRV; Hi, HindIII; H, HpaI; K, KpnI; N, NcoI; Ns, NsiI; Ps, PstI; Pv, PvuI; P, PvuII; Sa, SalI; Sc, SacI; Sm, SmaI; Sp, SphI; S, SspI; St, StuI; and X, XhoI. Int, integrase gene of the class 1 integron.
Conjugation.
E. coli DH5α mated with atypical strain A. salmonicida 2402 became resistant to tetracycline, trimethoprim, and sulfamethoxazole at a frequency that indicated that half of the E. coli cells received pRAS1 after 24 h of mating on LA (Table 1). Under similar conditions, E. coli DH5α mated with A. salmonicida subsp. salmonicida 1995 received the R plasmid at a frequency of 6.5 × 10−3. The frequency of transfer of pRAS1 from the two A. salmonicida subspecies increased by 3 to 4 logs when the mating period was prolonged from 1 to 48 h, but for all times the frequency of transfer from atypical strain A. salmonicida 2402 remained approximately 103 times higher than that from A. salmonicida subsp. salmonicida 1995 (Table 1). This donor-dependent transfer frequency was also observed when the two S. enterica serovars were used as recipients. Furthermore, pRAS1 could be transferred from both donor strains to Vibrio cholerae, Vibrio parahaemolyticus, and Yersinia ruckeri but could not be transferred to Pseudomonas aeruginosa (ATCC 27853) or Staphylococcus aureus (ATCC 25923) (data not shown).
TABLE 1.
Conjugative transfer of pRAS1 measured by transfer frequencies (proportion of E. coli recipient cells receiving the plasmid) after mating at 22°C for various periods either on a solid surface or in liquid medium
| Strain | Transfer frequency on the following type of medium after the indicated mating time (h):
|
Broth, 24 | Transfer frequency ratio for plate mating/ broth mating | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Agar
| ||||||||||
| 1 | 2 | 4 | 6 | 8 | 14 | 24 | 51 | |||
| 2402 | 1.2 × 10−3 | 4.0 × 10−3 | 3.5 × 10−3 | 0.13 | 0.18 | 0.25 | 0.48 | 0.59 | 1.1 × 10−3 | 436 |
| 1995 | 1.6 × 10−6 | 2.5 × 10−6 | 3.3 × 10−5 | 1.1 × 10−4 | 4.1 × 10−4 | 6.7 × 10−4 | 6.5 × 10−3 | 6.8 × 10−2 | 8.7 × 10−5 | 75 |
The conjugative transfer of pRAS1 in broth was in general lower than that on agar (Table 1). Plasmids of the IncU group express short rigid pili that are characteristic of conjugative plasmids with a broad host range (11). Conjugation by such structures is surface dependent, making conjugation 2,000 to 300,000 times faster on a solid surface than in broth (12). The results of the conjugation experiments with pRAS1 support this, and even though the differences between broth and agar in this study are less extreme, the tendency is clear.
The conjugation experiments demonstrate the large potential of the effective spread of pRAS1 and, supposedly, other IncU plasmids. The more successful transfer of pRAS1 from the atypical A. salmonicida strain than from A. salmonicida subsp. salmonicida suggests that atypical strains are particularly important in the spread of R plasmids. pRAS1 has also been shown to be highly transferable under conditions mimicking natural environments, as Kruse and Sørum (19) transferred pRAS1 from A. salmonicida subsp. salmonicida to E. coli DH5α in raw salmon and in pig feces at frequencies comparable to the ones detected in this study.
Hybridization to pRAS1 incompatibility and drug resistance genes.
The IncU probe hybridized to an EcoRI fragment of 0.95 kb, i.e., a fragment of the same size as the probe which originated from pAr-32. The hybridization of the restriction digests showed that the IncU region of pRAS1 is located 2 kb outside of the region of the 25% of the plasmid with a high density of restriction sites. The Tet A-specific probe hybridized to fragments that mapped close to the middle of this 25% region. The sul1-specific probe hybridized to the same region and was localized between Tet A and IncU (Fig. 1). The sul2-specific probe did not hybridize to the plasmid.
Drug resistance-determining region of pRAS1 and pAr-32 DNA.
The sizes of all fragments amplified from pRAS1 and pAr-32 by PCR were those expected from the restriction maps of pRAS1 and pAr-32 and the sequences of Tn1696, pSa, and catA2.
The drug resistance-determining region in pRAS1 comprises two main elements: a complete class 1 integron and a fragment of the Tet A transposon Tn1721. A copy of IS6100 is located between these two elements (Fig. 1).
The Tet A determinant in pRAS1 is part of a fragmented form of transposon Tn1721. A complete Tn1721 consists of two halves, in which one half includes complete tnpA and tnpR genes, while the other half includes the Tet A gene and a truncated tnpA gene (3). The transposon has three inverted repeats (IRs): the bordering left IR (IRL) and right IR (IRR) and an additional central IRR. The left part of Tn1721, called Tn1722, is able to transpose itself. All of this minitransposon except the 344 bp adjacent to the central IRR is absent from pRAS1. The complete Tn1721 was present in pRAS1-related IncU plasmids from aeromonads isolated from effluents in the United Kingdom, and these plasmids could have represented a former version of pRAS1 which later experienced a loss of Tn1722, as discussed by Rhodes et al. (27). However, the persistence of the central IRR in pRAS1 and remnants of tnpA from Tn1722 indicate that this has not happened and that the missing part of Tn1721 is due to another deletion mechanism. Due to this loss of both IRL and the transposition genes, the truncated Tn1721 in pRAS1 is not able to transpose, in opposition to Tn1720, a variant of Tn1721 with an internal deletion found in bacteria from apple orchards, which still has this ability (29).
A dfrA16 cassette is the only cassette in the integron of pRAS1; and the 3′-CS includes qacEΔ1, sul1, orf5, and orf6 (Fig. 2). The sequence of the pRAS1 integron shows that it is identical to the In4 integron in Tn1696 from plasmid R1033 in P. aeruginosa (10) in both the 5′-CS and the 3′-CS and in the localization of IS6100 after orf6 (23, 24). However, a partial copy of IS6100 that is present in In4, in addition to a complete copy, is absent from pRAS1; but flanking segments of the right-hand end of Tn402 on either side of IS6100, found in several In4-like integrons, are present.
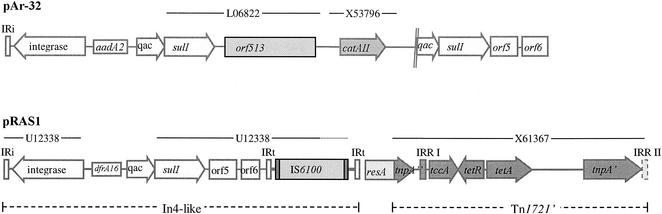
FIG. 2.
Comparison of the resistance-determining regions of pRAS1 and pAr-32. The areas of the plasmids that are the same as the sequences in Tn1696 (EMBL accession number U12338), Tn1721 (EMBL accession number X61367), pSa (EMBL accession number L06822), and catA2 of E. coli (EMBL accession number X53796) are indicated by the lines at the top. The areas of pRAS1 with similarities to In4 and Tn1721 are indicated at the bottom.
A short segment of 466 bp that does not relate to either of the elements involved in the part of pRAS1 involved with resistance separates the In4-like integron and the truncated Tn1721 in pRAS1. This area does not show any similarities to known sequences on a nucleotide basis, but a protein search revealed 56% identity to DNA invertases belonging to the site-specific recombinase resolvase family. The origin of this deleted novel resolvase, resA, which is present in pRAS1 (Fig. 2), and a possible former function of this gene are unknown. However, it has been shown that integrons may be inserted in the res site of transposons or plasmids (21). Resolvase genes are often located close to the integrated integron on the left side, and this is the first time that a resolvase gene has been found to the right of an integron (24). Possibly integrons are preferably integrated into res sites of transposons or plasmids by site-specific integration.
In pAr-32, aadA2 is the only cassette in the integron (20), and the sequences of the conserved segments in pAr-32 are identical to those of other class 1 integrons, except for the lack of orf5 and orf6. The pAr-32 integron is very similar to the In6 integron of pSa, except for the additional aacA4 cassette in In6. As in pSa, pAr-32 has an open reading frame originally called ORF341 (35), later renamed orf513, followed by the chloramphenicol resistance gene catA2. There are duplications of orf513 in both pAr-32 and pSa, but the duplicated area is larger in pSa. This has also been observed and discussed by Valentine et al. (37), who suggest that the duplication may have a relation to the mechanism by which catA2 was inserted in In6 of pSa. Downstream of catA2 is a deleted second copy of the 3′-CS of the integron, including the last two-thirds of the qacEΔ1 gene, the sul1 gene, and also orf5 and orf6 (Fig. 2). We have not been able to identify IRt (tni) downstream of orf6.
Dissemination of IncU plasmids.
IncU R plasmids have been isolated from Aeromonas species from aquatic environments throughout the world. In Japan, several R plasmids isolated from A. hydrophila and A. salmonicida strains from various freshwater districts were identical to pAr-32 and RA3 (2, 5, 8). In the United Kingdom, such R plasmids were isolated from both A. salmonicida strains from diseased Atlantic salmon in Scotland and A. hydrophila and A. caviae strains from effluents of a hospital in Cumbria, England (Table 2) (1, 27). Schmidt et al. (28) found class 1 integrons on R plasmids of the same size as other IncU plasmids in A. salmonicida strains from the Faroe Islands, and L'Abée-Lund and Sørum (20) found a pRAS1-related R plasmid in an A. salmonicida strain originating from the United States. Hedges et al. (16) found IncU R plasmids from A. hydrophila strains isolated from a hospital in Paris, France (22), and from A. hydrophila strains isolated from a fish hatchery on the west coast of Ireland (Table 2). However, IncU plasmids have also been isolated from bacteria other than Aeromonas spp.; in a study by Tschäpe et al. (36), five R plasmids from enteric bacteria from humans with clinical infections and from hospital sewage were characterized as IncU plasmids. These plasmids were also compared to RA3 by single digestions with EcoRI, BamHI, and PstI; and the investigators concluded that there are no similarities between them or to RA3. However, the dissimilarities may be concentrated on the variable part of the plasmids, the part encoding resistance, as the endonucleases used in their study frequently cut in such areas. One of these R plasmids, pIE420, which was isolated from an E. coli strain that caused pyelonephritis in Germany in the late 1970s, has already been shown to be identical to pRAS1 (27).
TABLE 2.
IncU plasmids, their origins and resistance features, and presence of resistance elements and determinants, when known
| IncU plasmid | Original host/ environment | Country or region | Phenotypic resistancea | Known resistance elements | Known resistance determinants | Comments | Source or reference(s) |
|---|---|---|---|---|---|---|---|
| pRAS1 | A. salmonicida/diseased fish | Norway | SUL, TMP, | Tn1721 right part, In4-like integron | dfrA16, Tet A, sulI | Identical to pIE420 | This paper; 20 |
| pAr-32 | A. salmonicida/diseased fish | Japan | SUL, STR, CHL | In6-like integron | aadA2, sul1, catA2 | Identical to RA3 | This paper; 6, 20 |
| RA3 | A. hydrophila/ diseased fish | Japan | SUL, STR, CHL | IncU reference plasmid; identical to pAr-32 | 6 | ||
| pASOT | A. salmonicida/diseased fish | Scotland | SUL, TET, CHL | Complete Tn1721, integron | aadA2 or dfrIIc (dfrB3), Tet A | 1, 20, 27 | |
| pASOT2 | A. salmonicida/diseased fish | Scotland | SUL, TET | Integron | aadA2, Tet A | 1, 20, 27 | |
| pASOT3 | A. salmonicida/diseased fish | Scotland | SUL, TET, STR | Integron | aadA2, Tet A | 1, 20, 27 | |
| pIE420 | E. coli/human pyelonephritis | Germany | SUL, TMP, TET, STRb | Identical to pRAS1 | 27, 36 | ||
| pFBAOT 6 | A. caviae/hospital effluent | England | TMP, STR | Complete Tn1721 | Tet A | 27 | |
| pUG1001 | A. hydrophila/fish hatchery | Ireland | SUL, TMP, TET, STR | EcoRI digests similar to those of pIE420 | 16 |
SUL, sulfonamides; TMP, trimethoprim; TET, tetracycline; STR, streptomycin; CHL, chloramphenicol.
pIE420 is reported to confer streptomycin resistance. Strains of A. salmonicida with pRAS1 are phenotypically intermediately resistant to streptomycin, but we do not consider pRAS1 as conferring streptomycin resistance.
Conclusions.
IncU plasmids pRAS1 and pAr-32 have two different integrons, an In4-like integron and an In6-like integron, respectively, that are inserted at what appears to be the same spot. Whether this point of insertion has features that make it a hot spot for the insertion of integrons is unknown. Thus, plasmids pRAS1 and pAr-32 may have arisen from two separate integrons inserted at the same site of a universal IncU plasmid and may not have evolved from a common ancestor by two separate lines of development.
It seems apparent that all plasmids typed to be IncU plasmids so far appear to be modifications of the same plasmid. This means that the same plasmid is isolated from aeromonads in various parts of the world, either from freshwater or marine water related to disease in farmed fish or from hospital effluents. In addition, it has also been isolated from enterobacteria pathogenic for humans. Hence, despite their wide distribution, IncU plasmids seem to be an evolutionarily narrow group of plasmids with the same backbone structure in which variations are restricted to the region containing resistance-determining genes.
Acknowledgments
We thank Takashi Aoki for the gift of plasmid pAr-32, Kevin Towner for all the dfrA-specific probes except dfrA9, and Ola Sköld for the dfrA9- and sul-specific probes.
REFERENCES
- 1.Adams, C. A., B. Austin, P. G. Meaden, and D. McIntosh. 1998. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl. Environ. Microbiol. 64:4194-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akashi, A., and T. Aoki. 1986. Characterization of transferable R plasmids from Aeromonas hydrophila. Bull. Jpn. Soc. Sci. Fish. 52:649-655. [Google Scholar]
- 3.Allmeier, H., B. Cresnar, M. Greck, and R. Schmitt. 1992. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111:11-20. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoki, T., S. Egusa, C. Yada, and T. Watanabe. 1972. Studies of drug resistance and R factors in bacteria from pond-cultured salmonids. I. Amago (Oncorhynchus rhodurus macrostomus) and Yamame (Oncorhynchus masou ishikawae). Jpn. J. Microbiol. 16:233-238. [DOI] [PubMed] [Google Scholar]
- 6.Aoki, T., Y. Mitoma, and J. H. Crosa. 1986. The characterization of a conjugative R-plasmid isolated from Aeromonas salmonicida. Plasmid 16:213-218. [DOI] [PubMed] [Google Scholar]
- 7.Aoki, T., S. Egusa, T. Kimura, and T. Watanabe. 1971. Detection of R factors in naturally occurring Aeromonas salmonicida strains. Appl. Microbiol. 22:716-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki, T., T. Kitao, T. Ando, and T. Arai. 1979. Incompatibility grouping of R plasmids detected in fish pathogenic bacteria, Aeromonas salmonicida, p. 219-222. In S. Mitsuhashi (ed.), Microbial drug resistance. University Park Press, Baltimore, Md.
- 9.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bissonnette, L., S. Champetier, J. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradley, D. E. 1980. Morphological and serological relationships of conjugative pili. Plasmid 4:155-169. [DOI] [PubMed] [Google Scholar]
- 12.Bradley, D. E., T. Aoki, T. Kitao, T. Arai, and H. Tschäpe. 1982. Specification of characteristics for the classification of plasmids in incompatibility group U. Plasmid 8:89-93. [DOI] [PubMed] [Google Scholar]
- 13.Grave, K., A. Lillehaug, B. T. Lunestad, and T. E. Horsberg. 1999. Prudent use of antibacterial drugs in Norwegian aquaculture? Surveillance by the use of prescription data. Acta Vet. Scand. 40:185-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grave, K., A. Markestad, and M. Bangen. 1996. Comparison in prescribing patterns of antibacterial drugs in salmonid farming in Norway during the periods 1980-1988 and 1989-1994. J. Vet. Pharmacol. Ther. 19:184-191. [DOI] [PubMed] [Google Scholar]
- 15.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedges, R. W., P. Smith, and G. Brazil. 1985. Resistance plasmids of aeromonads. J. Gen. Microbiol. 131:2091-2095. [Google Scholar]
- 17.Høie, S., B. Martinsen, S. Sohlberg, and T. E. Horsberg. 1992. Sensitivity patterns of Norwegian clinical isolates of Aeromonas salmonicida subsp. salmonicida to oxolinic acid, flumequine, oxytetracycline, and sulphadiazine/trimethoprim. Bull. Eur. Assoc. Fish Pathol. 12:142-144. [Google Scholar]
- 18.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse, H., and H. Sørum. 1994. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60:4015-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L'Abée-Lund, T. M., and H. Sørum. 2001. Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microb. Drug Resist. 7:263-272. [DOI] [PubMed] [Google Scholar]
- 21.Minakhina, S., G. Kholodii, S. Mindlin, O. Yurieva, and V. Nikiforov. 1999. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33:1059-1068. [DOI] [PubMed] [Google Scholar]
- 22.Mizon, F. M., G. R. Gerbaud, H. Leclerc, and Y. A. Chabbert. 1978. Occurrence of R plasmids belonging to incompatibility group incC in Aeromonas hydrophila strains isolated from sewage water. Ann. Microbiol. 129B:19-26. (In French.) [PubMed] [Google Scholar]
- 23.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rådström, P., O. Sköld, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundström. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes, G., G. Huys, J. Swings, P. McGann, M. Hiney, P. Smith, and R. W. Pickup. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant Tet A. Appl. Environ. Microbiol. 66:3883-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt, A. S., M. S. Bruun, J. L. Larsen, and I. Dalsgaard. 2001. Characterization of class 1 integrons associated with R-plasmids in clinical Aeromonas salmonicida isolates from various geographical areas. J. Antimicrob. Chemother. 47:735-743. [DOI] [PubMed] [Google Scholar]
- 29.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro, J. A., and P. Sporn. 1977. Tn402: a new transposable element determining trimethoprim resistance that inserts in bacteriophage lambda. J. Bacteriol. 129:1632-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sørum, H. 1999. Antibiotic resistance in aquaculture. Acta Vet. Scand. Suppl. 92:29-36. [PubMed] [Google Scholar]
- 32.Sørum, H. 2000. Farming of Atlantic salmon—an experience from Norway. Acta Vet. Scand. Suppl. 93:129-134. [PubMed] [Google Scholar]
- 33.Sørum, H., J. H. Kvello, and T. Håstein. 1993. Occurrence and stability of plasmids in Aeromonas salmonicida subsp. salmonicida isolated from salmonids with furunculosis. Dis. Aquat. Org. 16:199-206. [Google Scholar]
- 34.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 35.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 36.Tschäpe, H., E. Tietze, and C. Koch. 1981. Characterization of conjugative R plasmids belonging to the new incompatibility group IncU. J. Gen. Microbiol. 127:155-160. [DOI] [PubMed] [Google Scholar]
- 37.Valentine, C. R., M. J. Heinrich, S. L. Chissoe, and B. A. Roe. 1994. DNA sequence of direct repeats of the sulI gene of plasmid pSa. Plasmid 32:222-227. [DOI] [PubMed] [Google Scholar]