Abstract
The distribution of cytosine methylation in 6.2 Mb of the mouse genome was tested using cohybridization of genomic representations from a methylation-sensitive restriction enzyme and its methylation-insensitive isoschizomer. This assay, termed HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR), allows both intragenomic profiling and intergenomic comparisons of cytosine methylation. The intragenomic profile shows most of the genome to be contiguous methylated sequence with occasional clusters of hypomethylated loci, usually but not exclusively at promoters and CpG islands. Intergenomic comparison found marked differences in cytosine methylation between spermatogenic and brain cells, identifying 223 new candidate tissue-specific differentially methylated regions (T-DMRs). Bisulfite pyrosequencing confirmed the four candidates tested to be T-DMRs, while quantitative RT-PCR for two genes with T-DMRs located at their promoters showed the HELP data to be correlated with gene activity at these loci. The HELP assay is robust, quantitative, and accurate and is providing new insights into the distribution and dynamic nature of cytosine methylation in the genome.
While cytosine methylation is a major component of epigenetic regulation of gene expression, many of the techniques used to test cytosine methylation at multiple loci are not suitable for comparing methylation levels at different loci within a genome. In contrast, analogous intragenomic profiling has been successfully developed for studying chromatin organization using chromatin immunoprecipitation with genomic microarrays (ChIP-chip) (Kim et al. 2005) and DNA copy number using array comparative genomic hybridization (aCGH) (Selzer et al. 2005). The establishment of a platform for intragenomic profiling is a prerequisite for integrating studies of cytosine methylation with other whole-genome studies of epigenetic regulation. Comprehensive reviews of cytosine methylation analytical techniques describe the various approaches used to date (Laird 2003; Ushijima 2005), not including several recent reports of note (Ching et al. 2005; Hu et al. 2005; Weber et al. 2005).
The use of restriction enzymes that are sensitive to cytosine methylation has allowed many of the early insights into the distribution of methylated CpG dinucleotides in the mammalian genome. For example, the use of HpaII revealed that most of the genome remains high molecular weight following digestion despite the short recognition motif (5′-CCGG-3′) at which the enzyme cuts (Singer et al. 1979). It was subsequently recognized that 55%–70% of HpaII sites in animal genomes are methylated at the central cytosine (Bird 1980; Bestor et al. 1984), which is part of a CpG dinucleotide. The minority of genomic DNA that cuts to a size of hundreds of base pairs was defined as HpaII Tiny Fragments (HTFs) (Bird 1986), revealing a population of sites in the genome at which two HpaII sites are close to each other and both unmethylated on the same DNA molecule. Cloning and sequencing of these HTFs revealed them to be (G+C)- and CpG dinucleotide-rich, allowing base compositional criteria to be created to predict presumably hypomethylated CpG islands (Gardiner-Garden and Frommer 1987). These criteria remain in use for genomic annotations today, defining sequences that tend to localize with transcription start sites, especially of genes active constitutively (Larsen et al. 1992) or during embryogenesis (Ponger et al. 2001).
Genome sequencing project data have revealed that <12% of HpaII sites in the human genome (and <9% in mouse) are located within annotated CpG islands (Fazzari and Greally 2004). This raised the question of whether a substantial proportion of HTFs is, in fact, derived from non-CpG island sequences and could be used to examine many non-CpG island sites in the genome for cytosine methylation status. We describe a technique that is based on HTF enrichment by ligation-mediated PCR, creating the acronym HELP that gives a name to this assay. We demonstrate that the HELP enrichment, used as part of comparative isoschizomer profiling and in combination with customized genomic microarrays, allows robust intragenomic profiling of cytosine methylation. We show that in primary mouse tissues 28%–34% of annotated CpG islands are categorized as methylated, that the technique reveals large numbers of tissue-specific differentially methylated regions (T-DMRs), and that some of the hypomethylated sites are located at repetitive sequences. These surprising patterns of cytosine methylation indicate that the intragenomic profiling capability of the HELP assay will allow insights into this major mediator of epigenetic regulation that were not possible with single-locus studies or intergenomic comparisons.
Results
Prior techniques that sample the methylated fraction of the genome (Frigola et al. 2002; Yan et al. 2002; Chen et al. 2003) have the disadvantage of generating a sample likely to be extremely enriched in repetitive sequences (Yoder et al. 1997), making it difficult to identify the genomic source of the sample using subsequent hybridization or sequencing techniques. Furthermore, the failure of one of these prior techniques to generate a representation for a specific genomic region could be due to a genuine difference in cytosine methylation, but it could also be due to a technical failure, deletion of the region (a major issue when studying cancer epigenetics), or base compositional differences (absence of CpGs or suitable restriction sites). With the HELP assay, an internal control allows these issues to be overcome. While HpaII profiling on its own is subject to exactly the same problems, a comparison between a HpaII profile and that of its CpG methylation-insensitive isoschizomer MspI should overcome these issues. The MspI profile can be considered to represent the total possible population of HTFs, of which the actual HTFs constitute a sample. The use of an internal reference allows every site from which HTFs originate to be analyzed in terms of the relative amounts generated by the HpaII and the MspI representations, creating a profile of cytosine methylation across the genome.
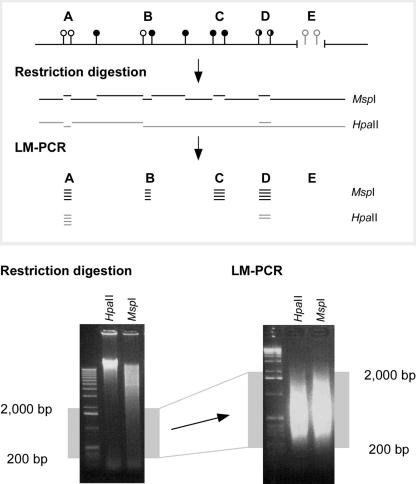
To create these HpaII and MspI representations, we isolate high molecular weight DNA, digest it to completion, and ligate an oligonucleotide pair that creates an end cohesive with that produced by the restriction enzymes. This serves as the template for a PCR primer to perform ligation-mediated PCR (LM-PCR). The PCR conditions create products ranging in size from 200 to 2000 bp (Fig. 1). The HpaII and MspI representations are labeled with different fluorophores using random priming and cohybridized to a customized genomic microarray.
Figure 1.
Principle of the HELP assay. The HELP assay is based on a comparison of representations from the genome following digestion by HpaII or its methylation-insensitive isoschizomer MspI. The representations are limited to a size range of 200–2000 bp by the use of ligation-mediated PCR. The MspI representation is the total potential population of sites that could be generated by the HpaII representation were none of these sites to be methylated. However, as 55%–70% of these sites are methylated in animal genomes (Bird 1980; Bestor et al. 1984), the HpaII representation will always represent a subset of the MspI representation. By comparing the relative representation at individual loci, assignment can be made of cytosine methylation status. While loci such as A should be amplified in both the HpaII and MspI representations, the failure of HpaII to digest both sites at loci B and C will yield a representation from MspI alone, while the partial methylation depicted at locus D should generate a lower HpaII/MspI ratio than at locus A. If a locus is deleted (or has a sequence change at the enzyme cleavage sites) as shown at E, neither representation will generate the locus.
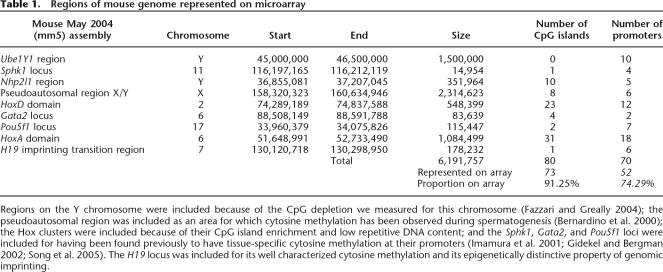
We designed the genomic microarray to represent some of the diversity within the mouse genome, including autosomes and sex chromosomes, constitutively active and tissue-specific genes, regions we found to be CpG-depleted (the Y chromosome; Fazzari and Greally 2004), CpG island-rich regions (Hox gene clusters), and an imprinted locus (H19) (Table 1). The HpaII-amplifiable fragments from these regions were identified in silico as loci where two HpaII sites are located 200–2000 bp apart with at least some unique sequence between them. Each HpaII-amplifiable fragment was represented on the microarray by 10 oligonucleotides, each 50 nucleotides in length. A genome-wide bioinformatic analysis was performed to ensure that each oligonucleotide used was unique in the genome. In Supplemental Figure 1, we show that the range of sizes represented is as low as 200 bp and that the distribution of sizes in the genomic regions studied is skewed toward smaller fragments. The resolution of the HELP assay is therefore in the range of hundreds of base pairs. A total of 1339 sites representing the HpaII-amplifiable fragments from ∼6.2 Mb of the mouse genome was represented on a microarray with >13,000 oligonucleotides.
Table 1.
Regions of mouse genome represented on microarray
Regions on the Y chromosome were included because of the CpG depletion we measured for this chromosome (Fazzari and Greally 2004); the pseudoautosomal region was included as an area for which cytosine methylation has been observed during spermatogenesis (Bernardino et al. 2000); the Hox clusters were included because of their CpG island enrichment and low repetitive DNA content; and the Sphk1, Gata2, and Pou5f1 loci were included for having been found previously to have tissue-specific cytosine methylation at their promoters (Imamura et al. 2001; Gidekel and Bergman 2002; Song et al. 2005). The H19 locus was included for its well characterized cytosine methylation and its epigenetically distinctive property of genomic imprinting.
The cell samples chosen for analysis were spermatogenic cells and whole brains from young adult mice, since we had generated preliminary data (not shown) revealing differences in cytosine methylation at certain loci between these tissues. To reduce the influences of genotype, sex, age, diet, or other environmental influences on epigenetic organization, identically housed male littermates were used for these experiments. To measure the variability due to the experimental protocol alone, digested DNA from one of the mice was subjected to three separate experimental preparations.
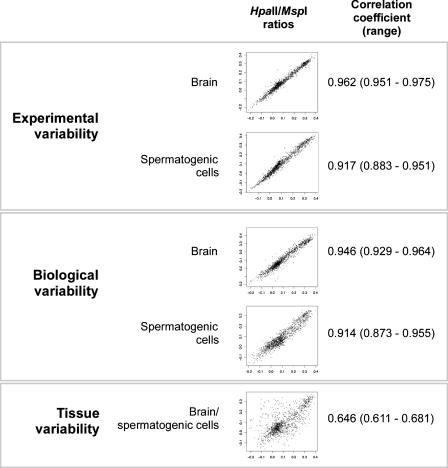
Our first goal was to explore the sources of variability in the assay. We represent these findings in two ways, by calculating the mean and ranges of correlation coefficients for HpaII/MspI log ratios (illustrated with representative scattergrams in Fig. 2) and for HpaII and MspI individually (Supplemental Fig. 2A). We also depict the same data by means of branching dendrograms (Supplemental Fig. 2B). The amount of variability due to differences in cytosine methylation between tissues is greatly in excess of the biological and experimental variability.
Figure 2.
The variability due to tissue-specific differences in cytosine methylation greatly exceeds all other sources of variability. We show the mean and range of correlation coefficients for all of the experimental replicates (three independent HELP assays performed on DNA from a single mouse) and biological replicates (three different mice) for the HpaII/MspI ratios. An illustrative scattergram is shown for each situation. The variability involving HpaII/MspI representations across tissues greatly exceeds any of the other sources of variability in the experimental system.
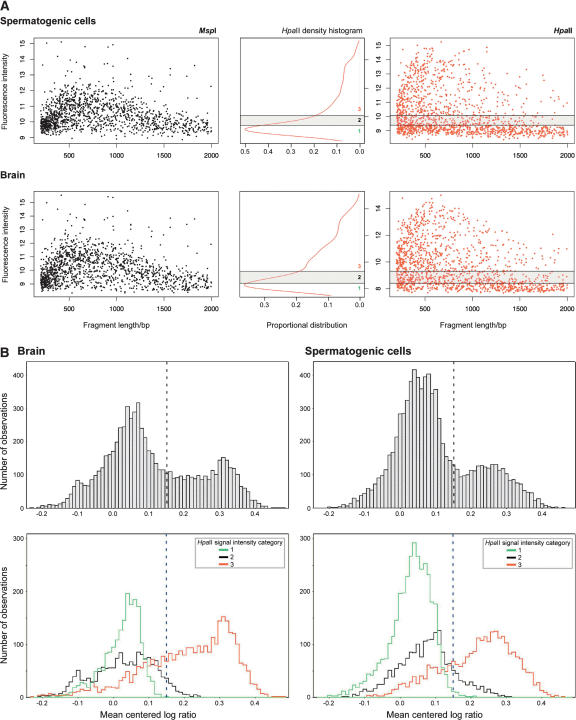
When we studied the microarray signal characteristics (Fig. 3A), we found that the median fluorescence intensity for each of the 1339 loci varies as a function of the size of the HpaII fragment. This was the expected consequence of heterogeneity in PCR amplification efficiency, resulting in differences in signal intensities across the range of sizes represented for each isoschizomer. However, it was also strikingly apparent that the HpaII representation is distinctive for having a population of loci throughout the size range that shows little signal. These loci result from the failure of the HpaII digestion to create a fragment for PCR, the outcome expected for methylated loci. A frequency histogram demonstrates this population, while a mixture model allows us to categorize each locus in terms of where it falls within this distribution of signal intensities. The category with the lowest signal intensities allowed us to normalize signal intensities across microarrays, as described in the Methods section.
Figure 3.
(A) Microarray signal characteristics for MspI (black) and HpaII (red). The signal intensities mirror the relative amount of product across the range of sizes in the PCR amplification. The HpaII representation shows the additional characteristic of a population of loci with low signal intensities, likely to represent the more methylated loci in the sample. A mixture model applied to the distribution allows loci to be categorized according to their relative signal intensities (center panel). (B) When the normalized mean HpaII/MspI log ratios are plotted as density histograms for each tissue, two populations of loci are apparent: the majority have low ratios, with a minority falling into a higher distribution. The breakdown of the log ratios by HpaII signal intensity category shows the low ratio peak to be mostly composed of those loci with lower HpaII signal intensities. In both tissues, a threshold of ∼0.15 serves to distinguish these two distributions.
Our starting hypothesis was that the comparison with MspI would create a valuable internal control for our HpaII representation. When we create a HpaII/MspI log ratio density plot averaged over the three biological replicates for each tissue (Fig. 3B), we see that the distribution of values creates what resembles a bimodal distribution. It is apparent that the majority of loci tested falls into a group with little HpaII representation compared with the MspI representation. This fits with the known methylation of most HpaII sites in the genome (Bird 1980; Bestor et al. 1984) and indicates a group of loci for which a methylated status can be assigned with confidence. When we break down the distribution by the categories of signal intensities that we observed for the HpaII representation, we find that the peak of lower ratio values is mostly generated by loci with low HpaII signal intensities, with the peak of higher ratios generated by the correspondingly higher HpaII signal intensities. The data indicate a log ratio value of 0.15 to be a reasonable threshold for this experiment, discriminating the majority methylated population from the range of less methylated loci in each tissue. The proportion of loci below this threshold is greater in spermatogenic cells than in brain (72% vs. 65%).
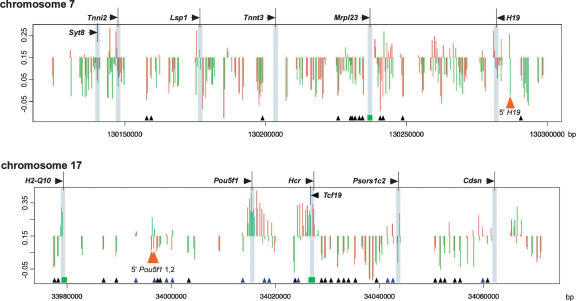
The HpaII/MspI log ratio is plotted in Figure 4 as a function of genomic position for each tissue in two representative regions. We represent each locus in terms of its deviation from the 0.15 threshold value; loci with greater methylation generate a negative deflection, while relatively hypomethylated loci generate a positive value. The intragenomic profiles of cytosine methylation in regions of chromosomes 7 and 17 are illustrated. Some striking patterns are evident: The majority of the genome exists as contiguous methylated blocks, interspersed by hypomethylated clusters that are mostly located at annotated transcription start sites. Most but not all repetitive elements are methylated, and there exist some hypomethylated sites that are located neither at promoters nor CpG islands.
Figure 4.
Intragenomic profiling and intergenomic comparisons for two chromosomal regions. The HpaII/MspI log ratios are depicted in terms of their deviation from the threshold value of 0.15. A downward deflection is indicative of a methylated locus, and an upward deflection is a relatively hypomethylated locus. (Red) Brain cells, (green) spermatogenic cells. Transcription start sites are shared, CpG islands are represented (green rectangles), and loci for which the fragment has one or both flanking HpaII sites located in a repetitive element are depicted (small triangle at the bottom of the graph). (Black triangles) Methylated, (blue) hypomethylated in at least one of the tissues tested. (Orange) Loci subsequently tested by bisulfite pyrosequencing. The red and green histograms are slightly shifted relative to each other for clarity. The intragenomic profile of cytosine methylation shows that methylation is the predominant pattern in these regions, with short blocks of hypomethylation located mostly at promoters and CpG islands, although some non-promoter, non-CpG island hypomethylation is also apparent. The intergenomic comparison between tissues shows overall concordance but some clear tissue-specific differences, such as those at the orange arrowheads.
The data not only reveal the intragenomic profile of cytosine methylation, they also illustrate the intergenomic comparison that is possible. Some loci are clearly distinctive in terms of cytosine methylation between brain and spermatogenic cells. Such tissue-specific differentially methylated regions (T-DMRs) have been identified by means of challenging techniques such as RLGS, resulting in the publication of several hundred to date (Shiota et al. 2002; Song et al. 2005); in this study, we demonstrate that T-DMRs are frequent and not confined to gene promoter regions.
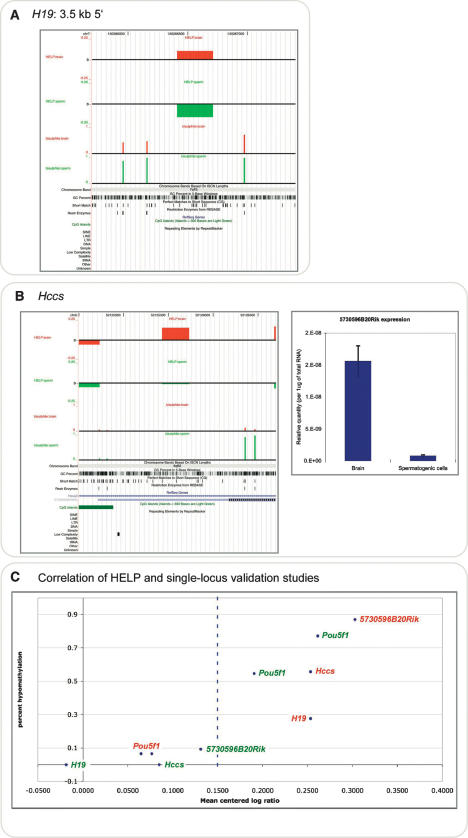
We performed bisulfite pyrosequencing (Dupont et al. 2004) at four loci to validate the HELP results indicating differences in cytosine methylation between these tissues. Each was chosen for a specific reason: two were located at promoters of genes where we could perform correlative gene expression studies, and two represented the intriguing class of hypomethylated loci that are located at intergenic, non-promoter, non-CpG island sites. Of the latter, one is the region upstream of H19 known to be methylated in spermatogenic cells and allele-specifically methylated in somatic tissues (Davis et al. 1999). Other than this locus, none of the others was previously recognized to exhibit tissue-specific methylation. In Figure 5, we show these results integrated with the HELP data and genomic annotations (available as custom tracks for the UCSC Genome Browser using the links below). The HELP data indicating the site 5′ to H19 to be methylated in spermatogenic cells and relatively hypomethylated in brain were confirmed. The sites ∼20 kb upstream from Pou5f1 (Oct3/4) showed the opposite pattern of methylation, with relative hypomethylation in spermatogenic cells. The most telomeric HpaII site is located within a B2 SINE but is only partially methylated by bisulfite pyrosequencing in each tissue, confirming for this locus what is indicated to occur at multiple loci using the HELP assay: A small proportion of interspersed repetitive elements is hypomethylated in the genome (Supplemental Fig. 3). We tested to see whether the cytosine methylation results correlated with gene expression, choosing two loci exhibiting differential methylation at gene promoters. In both cases, we found that the methylation of these loci in spermatogenic cells was associated with the near-complete silencing of expression from each locus, while hypomethylation in brain was associated with much higher levels of expression (Fig. 5; Supplemental Fig. 3). These results indicate that the HELP assay may provide information that correlates with genome-wide gene expression data.
Figure 5.
Validation studies using bisulfite pyrosequencing. The H19 locus indicated by the orange arrowhead in Fig. 4 was tested for cytosine methylation using bisulfite pyrosequencing of the original DNA samples used for the HELP assays. The samples from each of the three mice were tested individually. The primary data are provided in Supplemental Table 1, and are represented here as the median value of the three generated. The figures were generated by creating custom tracks for the UCSC Genome Browser and can be browsed in detail at http://greallylab.aecom.yu.edu/~greally/wiggle_tracks/HELP_data.htm. The degree of cytosine methylation is plotted as a percentage for spermatogenic cells (green) and brain (red). The HELP data are shown for reference using the same color scheme. (A) The complete methylation of the HpaII sites at the H19 locus in spermatogenic cells is consistent with the methylated categorization in the HELP assay and with prior studies of this region in spermatogenic cells (Davis et al. 1999). The downstream HpaII site is more methylated in brain than the upstream site. As the proportion of molecules digested and available for amplification in the HELP assay is dependent on the digestion of both flanking sites, the site with the greater degree of methylation determines this proportion. We conclude that the 72.4% methylation of this site allowed the HELP representation categorized as hypomethylated. (B) The Hccs promoter shows clear differences in cytosine methylation between tissues at all HpaII sites tested, with a corresponding change in gene expression levels. (C) Correlation of all of the loci plotted by hypomethylation (bisulfite pyrosequencing data) against normalized HpaII/MspI ratios, with results from brain samples (red) and spermatogenic cell samples (green) shown.
The validation studies allow us to correlate a number of results from individual loci with the corresponding HpaII/MspI log ratio values. The amount of HpaII product that can be formed at a locus depends on the flanking HpaII site with the greater amount of methylation, as both sites need to be digested to generate a template for PCR. This allows us to calculate the values shown in Figure 5C, in which the proportional hypomethylation is plotted against the HpaII/MspI log ratio. Those loci with log ratios <0.15 are almost fully methylated, whereas those loci with log ratios exceeding this threshold have methylation levels ranging from 13% to 72%. With further single-locus validation studies, we will be able to test more rigorously whether the HpaII/MspI log ratio predicts intermediate degrees of cytosine methylation, as suggested by the current data.
T-DMRs in the context of these data could be said to be ubiquitous, as no locus has identical HpaII/MspI ratios in both tissues. However, the practical use of this categorization is in defining loci at which methylation is markedly different between tissues. We could rank every locus by the degree of divergence of HpaII/MspI ratios, but a simple analysis uses our threshold value of 0.15 to discriminate loci that are likely to be highly methylated from less methylated loci. This categorization defines 223 T-DMRs, 167 of which are hypomethylated in brain and methylated in spermatogenic cells; the remainder (56 loci) show the opposite pattern. As 223 represents one-sixth of all of the loci on the microarray, we can appreciate that T-DMRs are frequent in the genome.
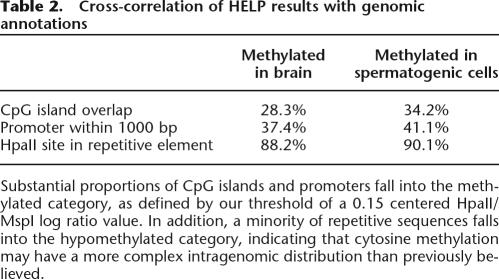
Because the validation studies showed the HELP assay to be generating reliable data, we performed data mining of genomic sequence annotations from the UCSC Genome Browser (Kent et al. 2002) to determine how the cytosine methylation was distributed in the heterogeneous regions represented on the microarray. We identified the loci represented on the microarray that had any overlap with CpG islands and those within 1.0 kb upstream of transcription start sites of RefSeq genes. We also identified those loci on the microarray for which one or both HpaII sites were located within an annotated repetitive element. When we studied the methylation categorization of each of these elements, we found the results depicted in Table 2. Approximately 37%–41% of the sites at promoters were in the methylated category, while 28%–41% of sites overlapping CpG islands were categorized in the methylated category. In addition, the proportion of repetitive elements classified as hypomethylated was 10%–12%, a small minority of the total repetitive element content of the genome, but on a genomic scale potentially representing as many as tens of thousands of hypomethylated repetitive sequences that are of potential functional significance.
Table 2.
Cross-correlation of HELP results with genomic annotations
Substantial proportions of CpG islands and promoters fall into the methylated category, as defined by our threshold of a 0.15 centered HpaII/MspI log ratio value. In addition, a minority of repetitive sequences falls into the hypomethylated category, indicating that cytosine methylation may have a more complex intragenomic distribution than previously believed.
Discussion
This comparative isoschizomer profiling approach allows two types of comparisons of cytosine methylation to be made. The genomic DNA from two cell types can be compared (intergenomic comparison), revealing the presence of T-DMRs in this study. Simultaneously, the intragenomic cytosine methylation profile is generated for each cell type. The key to intragenomic comparisons is the use of an internal control to account for variables that would otherwise confound analysis. A difference in the HpaII representation from two loci in the genome could be due to a number of variables other than cytosine methylation. Chief among these is base composition, since CpG dinucleotide and HpaII site distributions are highly heterogeneous (Saccone et al. 1999). The internal control in the HELP assay is the MspI representation, controlling for base composition, DNA copy number (a major issue when studying cancer cells, in which amplifications and deletions are frequent [Lengauer et al. 1998]), and difficulty amplifying these frequently (G+C)-rich sites when generating the representations, which should affect the HpaII and MspI representations for individual loci to comparable extents. As the MspI representation remains relatively invariant in situations of differential methylation (for example, the brain and spermatogenic cell comparisons of Fig. 2), the MspI representation provides a robust control with which to compare the HpaII representation.
The HELP assay can be used on any genomic microarray and with any combination of restriction enzyme isoschizomers differing in sensitivity to cytosine methylation of the target sequence. By using a customized oligonucleotide microarray, we achieved the maximum resolution of 200 bp permitted by our LM-PCR conditions. The HpaII/MspI isoschizomer pair allowed most of the 82 CpG islands present in the genomic regions encompassed by the microarray to be represented (73 represented, 89.0%), and most of the 71 annotated RefSeq transcription start sites, presumably representing true promoters, had HpaII-amplifiable fragments within 1.0 kb upstream (52 represented, 73.2%). Unexpectedly, we found that a number of repetitive elements were also represented using this unbiased design (4.2% of the total number of repetitive elements in the regions studied). The assay is readily scalable to the entire genome by expanding the representation on customized microarrays. Our in silico analysis demonstrates that the mouse genome has ∼600,000 loci that can be tested using HELP, with a corresponding 750,000 loci in the human genome. With sufficient microarray representation, the entire genome can be tested at once.
The number of techniques published for testing cytosine methylation genome-wide is now sizable. This topic has been extensively reviewed (Laird 2003; Ushijima 2005) prior to the recent publication of several new approaches (Ching et al. 2005; Hu et al. 2005; Weber et al. 2005). Each technique has clear strengths and weaknesses, depending on the application. As mentioned earlier, some techniques selectively enrich the methylated fraction of the genome (examples being differential methylation hybridization [DMH] [Yan et al. 2002] and methylated DNA immunoprecipitation [meDIP] [Weber et al. 2005]), which tends to enrich repetitive sequences, potentially causing difficulties with hybridization-based approaches. Set against this concern is the publication of successful outcomes of such experiments (Paz et al. 2003; Keshet et al. 2006), indicating that the problem is surmountable. Other techniques are inherently limited in terms of the number of loci that can be tested, including RLGS and other techniques involving rare-cutting restriction enzymes (Ching et al. 2005). RLGS has the additional problem of difficulty proceeding from the identification of a gel electrophoresis difference to a genomic locus for validation studies, a problem to some extent addressed by virtual image RLGS (Matsuyama et al. 2003). Base composition heterogeneity is another source of intragenomic variability that is not usually addressed. If one region has many CG dinucleotides and another has very few, the former region is likely to generate stronger signals indicating methylation or its absence than the latter whatever their relative methylation levels, especially with techniques that survey CG dinucleotides in bulk, including affinity-based techniques such as meDIP (Weber et al. 2005), the mcrBC-based cleavage of methylated DNA (Lippman et al. 2005), or methylation-sensitive restriction enzyme cocktails (Rollins et al. 2006; Schumacher et al. 2006). While these techniques are proving useful in intergenomic comparisons, normalization for base composition is essential before they can be used for accurate intragenomic profiling. Finally, the difficulty of validating results from these techniques is generally inversely related to the number of sites they survey. Techniques such as NotI profiling on BAC microarrays (Ching et al. 2005) generate small numbers of defined sites for analysis, whereas any technique that generates a signal based on bulk CG dinucleotide surveys requires the investigator to test the entire group of CGs in loci of interest. The HELP assay falls between the extremes, surveying fewer sites than the bulk techniques but generating fewer sites for validation. The customized microarray design means that the results obtained for each fragment are individually independent of base composition. Base composition affects the HELP assay only in terms of the density of loci that can be tested, with more dense representation in (G+C)-rich regions where the CCGG motif is more abundant. The signals from individual HpaII-amplifiable fragments in (G+C)-rich and -poor regions are otherwise directly comparable. In addition, the use of a methylation-insensitive isoschizomer controls for copy number, relative difficulty with PCR amplification of different loci, and polymorphisms at the restriction enzyme cleavage site, as was demonstrated in a recent publication (Hatada et al. 2006).
The HELP assay is technically quite straightforward and associated with very little experimental variability (Fig. 2; Supplemental Fig. 2). We identified 223 loci that are strong candidates for being T-DMRs using the HELP assay, confirming the five candidates by testing using bisulfite pyrosequencing. Our cross-correlation studies found a substantial proportion of sites annotated as promoters or as CpG islands to be methylated, and a minority of repetitive elements to be hypomethylated. The data in Figure 4 suggest that hypomethylation is discretely organized and restricted to small clusters within the genome that can extend into immediately flanking repetitive sequences. Any process that is restricted in terms of its physical extent in the genome should fail to involve most sequence features, including repetitive elements. Whether these “hypomethylated islands” of DNA non-randomly spare repetitive sequences or are influenced by the base compositional criteria used in the definition of CpG islands remains to be tested in a more extensive study.
An immediate disease-related application of genome-wide cytosine methylation assays is to study cancer, in which epigenetic regulation is profoundly disturbed (Jones and Baylin 2002). The use of a methylation-insensitive isoschizomer controls for a common variable in cancer, that of changes in copy number (Lengauer et al. 1998). By reporting the ratio of isoschizomer representations, amplified or deleted regions will generate a measure of cytosine methylation that can be used for intragenomic comparisons with normal, diploid regions, making the HELP assay exceptionally suited to cancer epigenomic studies. However, as we begin to recognize the role of epigenetics in other processes and diseases, such as aging (Fraga et al. 2005), mediation of dietary influences (Wolff et al. 1998), and possibly the sequelae of assisted reproductive technology (Maher et al. 2003), genome-wide cytosine methylation assays are likely to find applications beyond the cancer focus.
Methods
Sample preparation
C57Bl/6J male littermate mice were housed and fed identically and sacrificed using humane techniques at ∼8 wk of age. DNA was isolated from the whole brain and from the tubular contents of the testes using proteinase K digestion, phenol-chloroform-isoamyl alcohol extraction, dialysis against 0.2× SSC, and concentration by surrounding the dialysis bag with PEG 20,000 to reduce water content by osmosis. The quality of the DNA was checked by gel electrophoresis and spectrophotometry.
Isoschizomer representations
Ten micrograms of each DNA sample were digested to completion overnight using HpaII or MspI. The quality of digestion was assessed using gel electrophoresis. The digested DNA was cleaned by phenol-chloroform extraction, isopropanol-precipitated in the presence of glycogen, and re-dissolved in TE pH 8.0. One-tenth of the digested sample was added to T4 DNA ligase and the following oligonucleotide pair:
 |
The reaction mix was placed in a thermocycler for 5 min at 55°C then ramped over 1 h to 4°C, at which time 1 unit of T4 DNA ligase was added for overnight ligation at 16°C. One of the digested DNA samples from each mouse was divided into three separate reactions to measure the amount of experimental variability. To perform LM-PCR, 1/50 of the MspI or 1/25 of the HpaII sample was amplified using the 24-mer oligonucleotide shown above. An initial extension for 10 min at 72°C filled in the 5′ overhang, followed by 20 cycles of PCR using 30 sec at 95°C and 3 min at 72°C, with a final 10 min extension at 72°C. The quality of the LM-PCR reaction was tested using gel electrophoresis, looking for the size range of products shown in Figure 1, and the product was cleaned using the Qiagen PCR purification kit and quantified using spectrophotometry.
Microarray design and use
The microarrays were designed to represent loci amplified by the LM-PCR reaction. The size range of product was 200–2000 bp (Fig. 1), so an in silico digest was conducted with HpaII (CCGG), and all sequence fragments of the appropriate size range were retained. An initial probe set was generated by selecting a 50mer oligo every 10 bp, avoiding repeat-masked regions and sequence containing ambiguities. A measure of small oligo frequency was determined by sliding a 15mer window along the length of each 50mer oligo and determining the average frequency. The uniqueness of each 50mer was determined by looking for perfect matches in the entire mouse genome using SSAHA2 (Ning et al. 2001). Ten 50mer oligonucleotides were selected to represent each HpaII fragment using a score based selection algorithm based on three primary parameters: average 15mer frequency, 50mer count, and base pair composition rules. The base pair composition rules add penalties for homopolymer runs: Stretches of more than three Gs or Cs, or more than five As or Ts, are penalized, with larger penalties for longer stretches. After the first oligo is selected, an additional positional parameter is added to encourage uniform distribution of subsequent oligos along the length of the fragment.
A microarray of ∼13,500 oligonucleotides was used to represent the ∼6.2 Mb of the mouse genome in this study. These were printed using maskless array synthesis (Nuwaysir et al. 2002) in the NimbleScreen 12 format (NimbleGen Systems Inc). The LM-PCR products were labeled for microarray analysis as previously described (Selzer et al. 2005) using Cy3 or Cy5-conjugated oligonucleotides and random primers. The HpaII and MspI representations were cohybridized to the microarray in the NimbleGen Service Laboratory and scanned to quantify the 532 and 635 nm fluorescence at each oligonucleotide on the microarray.
Data analysis
Each cohybridization was analyzed by visual inspection of the image file to ensure that the signals were uniform. Each fragment represented on the microarray consists of 10 separate oligonucleotide probes, each with an associated signal intensity. We calculated the median signal intensity for each fragment to define the fragment’s signal intensity. The HpaII and MspI signal intensities were correlated and plotted against each other or fragment length using the R statistical package (http://www.r-project.org/) to generate the data in Figures 2 and 3. The branching dendrograms of Supplemental Figure 2 were generated based on an epigenomic distance measurement of (1 − correlation coefficient) and plotted using MatLab. The frequencies of loci with different HpaII signal intensities were modeled using a mixed Gaussian model (one variant) to separate loci into groups with 90% or 10% probabilities of being in the group of low intensity signals, defining categories 1 and 2 in Figure 3; the remainder of the loci with higher signal intensities were categorized as group 3. The range of intensities for group 1 was used as a measure of variability between arrays. Normalization was performed by subtracting the mean log ratio of this group of signal intensities in order to center log ratios over the entire array. The normalized HpaII/MspI log ratios for the three biological replicates in one array were used to generate the data in Figures 3 and 4.
Validation studies using bisulfite pyrosequencing
Four loci showing tissue-specific cytosine methylation were chosen for validation studies. The chromosome 7 sequence 5′ to H19 appeared to be methylated in spermatogenic cells and hypomethylated in brain, while a site 20 kb 5′ to Pou5f1 had the opposite pattern, as did an immediately adjacent site representing a HpaII site present in a B2 SINE. Two promoter sequences also exhibited differential methylation, at the Hccs and the 5730596B20Rik loci. We used a technique published for microdissected cells (Kerjean et al. 2001) for bisulfite conversion, digesting DNA from the same brain and spermatogenic cell samples used for the HELP assay overnight, denaturing the DNA using heat and NaOH, and embedding it in agarose beads. The DNA was then treated with sodium bisulfite for 4 h at 50°C and then washed in TE pH 8.0, followed by desulfonation with 0.2 M NaOH. The beads were dialyzed in water prior to amplification using PCR. The PCR and pyrosequencing primers were designed using Biotage’s proprietary software. Amplification using the primer pairs specific for each HpaII site was performed. Pyrosequencing was performed in a shared resource at this institution, performing the assays on the three samples of DNA from different animals. The primer sequences for each of these loci are shown in Supplemental Table 2.
Quantitative analysis of gene expression
RNA was extracted from the same tissues used for the HELP assays and analyzed for quality and concentration using electrophoresis and spectrophotometry using Nanodrop (NanoDrop Technologies). RT-PCR primers were designed (primer sequences in Supplemental Table 2) for quantitative PCR using SYBR Green (SYBR Green PCR Master mix, Applied Biosystems) and the DNA Engine OPTICON 2 (BioRad). As well as oligo-dT, gene-specific primers were used for reverse transcription given the location of 5730596B20Rik within an intron of Hoxa3. We were concerned that the analysis might be influenced by amplification of the unprocessed Hoxa3 transcript, but we found near-identical results for both reverse transcription primers (not shown). Neither Gapdh nor Hprt was suitable as a control locus, as each is substantially silenced in spermatogenic cells. However, the more robust approach in this case was to calculate relative levels of expression (mean of triplicate C(t) values) as a simple function of RNA concentration to generate the data in Figure 5.
Mining and analysis of genomic sequence annotations
Genome sequence annotations were mined from the mm5 version of the mouse genome at the UCSC Genome Browser (NCBI build 33, http://genome.ucsc.edu/), the same database used to design the microarrays. The coordinates for CpG islands, transcription start sites of RefSeq genes, and repetitive elements were downloaded for the genomic regions represented. Perl scripts were used to identify the colocalization of loci on the microarray with these genomic sequence features.
Fluorescence in situ hybridization
The MspI LM-PCR product was labeled by nick translation and hybridized to a normal mouse metaphase, with fluorophores imaged separately by fluorescence microscopy and merged using Photoshop (Adobe).
Acknowledgments
The following shared resources at Albert Einstein College of Medicine were used to generate data for this study: the DNA Sequencing Facility (Christina Lowes), the Bioinformatics Shared Resource, the Genome Imaging Facility, and the Analytical Imaging Facility, as well as resources from the Albert Einstein Cancer Center. This work is supported by a grant from the National Institutes of Health (NCI) R03 CA111577 to J.M.G.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5273806
References
- Bernardino J., Lombard M., Niveleau A., Dutrillaux B., Lombard M., Niveleau A., Dutrillaux B., Niveleau A., Dutrillaux B., Dutrillaux B. Common methylation characteristics of sex chromosomes in somatic and germ cells from mouse, lemur and human. Chromosome Res. 2000;8:513–525. doi: 10.1023/a:1009271706488. [DOI] [PubMed] [Google Scholar]
- Bestor T.H., Hellewell S.B., Ingram V.M., Hellewell S.B., Ingram V.M., Ingram V.M. Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol. Cell. Biol. 1984;4:1800–1806. doi: 10.1128/mcb.4.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Chen C.M., Chen H.L., Hsiau T.H., Hsiau A.H., Shi H., Brock G.J., Wei S.H., Caldwell C.W., Yan P.S., Huang T.H., Chen H.L., Hsiau T.H., Hsiau A.H., Shi H., Brock G.J., Wei S.H., Caldwell C.W., Yan P.S., Huang T.H., Hsiau T.H., Hsiau A.H., Shi H., Brock G.J., Wei S.H., Caldwell C.W., Yan P.S., Huang T.H., Hsiau A.H., Shi H., Brock G.J., Wei S.H., Caldwell C.W., Yan P.S., Huang T.H., Shi H., Brock G.J., Wei S.H., Caldwell C.W., Yan P.S., Huang T.H., Brock G.J., Wei S.H., Caldwell C.W., Yan P.S., Huang T.H., Wei S.H., Caldwell C.W., Yan P.S., Huang T.H., Caldwell C.W., Yan P.S., Huang T.H., Yan P.S., Huang T.H., Huang T.H. Methylation target array for rapid analysis of CpG island hypermethylation in multiple tissue genomes. Am. J. Pathol. 2003;163:37–45. doi: 10.1016/S0002-9440(10)63628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching T.T., Maunakea A.K., Jun P., Hong C., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., Maunakea A.K., Jun P., Hong C., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., Jun P., Hong C., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., Hong C., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., Fridlyand J., Mao J.H., Shchors K., Mao J.H., Shchors K., Shchors K., et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat. Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- Davis T.L., Trasler J.M., Moss S.B., Yang G.J., Bartolomei M.S., Trasler J.M., Moss S.B., Yang G.J., Bartolomei M.S., Moss S.B., Yang G.J., Bartolomei M.S., Yang G.J., Bartolomei M.S., Bartolomei M.S. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics. 1999;58:18–28. doi: 10.1006/geno.1999.5813. [DOI] [PubMed] [Google Scholar]
- Dupont J.M., Tost J., Jammes H., Gut I.G., Tost J., Jammes H., Gut I.G., Jammes H., Gut I.G., Gut I.G. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Fazzari M.J., Greally J.M., Greally J.M. Epigenomics: Beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., Ropero S., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., Cigudosa J.C., Urioste M., Benitez J., Urioste M., Benitez J., Benitez J., et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J., Ribas M., Risques R.A., Peinado M.A., Ribas M., Risques R.A., Peinado M.A., Risques R.A., Peinado M.A., Peinado M.A. Methylome profiling of cancer cells by amplification of inter-methylated sites (AIMS). Nucleic Acids Res. 2002;30:e28. doi: 10.1093/nar/30.7.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gidekel S., Bergman Y., Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 2002;277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- Hatada I., Fukasawa M., Kimura M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Fukasawa M., Kimura M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Kimura M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Yamanaka S., Endo C., Sakurada A., Sato M., Endo C., Sakurada A., Sato M., Sakurada A., Sato M., Sato M., et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25:3059–3064. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- Hu M., Yao J., Cai L., Bachman K.E., van den Brule F., Velculescu V., Polyak K., Yao J., Cai L., Bachman K.E., van den Brule F., Velculescu V., Polyak K., Cai L., Bachman K.E., van den Brule F., Velculescu V., Polyak K., Bachman K.E., van den Brule F., Velculescu V., Polyak K., van den Brule F., Velculescu V., Polyak K., Velculescu V., Polyak K., Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat. Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- Imamura T., Ohgane J., Ito S., Ogawa T., Hattori N., Tanaka S., Shiota K., Ohgane J., Ito S., Ogawa T., Hattori N., Tanaka S., Shiota K., Ito S., Ogawa T., Hattori N., Tanaka S., Shiota K., Ogawa T., Hattori N., Tanaka S., Shiota K., Hattori N., Tanaka S., Shiota K., Tanaka S., Shiota K., Shiota K. CpG island of rat sphingosine kinase-1 gene: Tissue-dependent DNA methylation status and multiple alternative first exons. Genomics. 2001;76:117–125. doi: 10.1006/geno.2001.6607. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjean A., Vieillefond A., Thiounn N., Sibony M., Jeanpierre M., Jouannet P., Vieillefond A., Thiounn N., Sibony M., Jeanpierre M., Jouannet P., Thiounn N., Sibony M., Jeanpierre M., Jouannet P., Sibony M., Jeanpierre M., Jouannet P., Jeanpierre M., Jouannet P., Jouannet P. Bisulfite genomic sequencing of microdissected cells. Nucleic Acids Res. 2001;29:E106. doi: 10.1093/nar/29.21.e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Pikarski E., Young R.A., Niveleau A., Cedar H., Young R.A., Niveleau A., Cedar H., Niveleau A., Cedar H., Cedar H., et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;426:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Larsen F., Gundersen G., Lopez R., Prydz H., Gundersen G., Lopez R., Prydz H., Lopez R., Prydz H., Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K.W., Vogelstein B., Kinzler K.W., Vogelstein B., Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Colot V., Martienssen R., Gendrel A.V., Colot V., Martienssen R., Colot V., Martienssen R., Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- Maher E.R., Brueton L.A., Bowdin S.C., Luharia A., Cooper W., Cole T.R., Macdonald F., Sampson J.R., Barratt C.L., Reik W., Brueton L.A., Bowdin S.C., Luharia A., Cooper W., Cole T.R., Macdonald F., Sampson J.R., Barratt C.L., Reik W., Bowdin S.C., Luharia A., Cooper W., Cole T.R., Macdonald F., Sampson J.R., Barratt C.L., Reik W., Luharia A., Cooper W., Cole T.R., Macdonald F., Sampson J.R., Barratt C.L., Reik W., Cooper W., Cole T.R., Macdonald F., Sampson J.R., Barratt C.L., Reik W., Cole T.R., Macdonald F., Sampson J.R., Barratt C.L., Reik W., Macdonald F., Sampson J.R., Barratt C.L., Reik W., Sampson J.R., Barratt C.L., Reik W., Barratt C.L., Reik W., Reik W., et al. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART). J. Med. Genet. 2003;40:62–64. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T., Kimura M.T., Koike K., Abe T., Nakano T., Asami T., Ebisuzaki T., Held W.A., Yoshida S., Nagase H., Kimura M.T., Koike K., Abe T., Nakano T., Asami T., Ebisuzaki T., Held W.A., Yoshida S., Nagase H., Koike K., Abe T., Nakano T., Asami T., Ebisuzaki T., Held W.A., Yoshida S., Nagase H., Abe T., Nakano T., Asami T., Ebisuzaki T., Held W.A., Yoshida S., Nagase H., Nakano T., Asami T., Ebisuzaki T., Held W.A., Yoshida S., Nagase H., Asami T., Ebisuzaki T., Held W.A., Yoshida S., Nagase H., Ebisuzaki T., Held W.A., Yoshida S., Nagase H., Held W.A., Yoshida S., Nagase H., Yoshida S., Nagase H., Nagase H. Global methylation screening in the Arabidopsis thaliana and Mus musculus genome: Applications of virtual image restriction landmark genomic scanning (Vi-RLGS). Nucleic Acids Res. 2003;31:4490–4496. doi: 10.1093/nar/gkg488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z., Cox A.J., Mullikin J.C., Cox A.J., Mullikin J.C., Mullikin J.C. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir E.F., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Richmond T., Gorski T., Berg J.P., Ballin J., Gorski T., Berg J.P., Ballin J., Berg J.P., Ballin J., Ballin J., et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz M.F., Fraga M.F., Avila S., Guo M., Pollan M., Herman J.G., Esteller M., Fraga M.F., Avila S., Guo M., Pollan M., Herman J.G., Esteller M., Avila S., Guo M., Pollan M., Herman J.G., Esteller M., Guo M., Pollan M., Herman J.G., Esteller M., Pollan M., Herman J.G., Esteller M., Herman J.G., Esteller M., Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- Ponger L., Duret L., Mouchiroud D., Duret L., Mouchiroud D., Mouchiroud D. Determinants of CpG islands: Expression in early embryo and isochore structure. Genome Res. 2001;11:1854–1860. doi: 10.1101/gr.174501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins R.A., Haghighi F., Edwards J.R., Das R., Zhang M.Q., Ju J., Bestor T.H., Haghighi F., Edwards J.R., Das R., Zhang M.Q., Ju J., Bestor T.H., Edwards J.R., Das R., Zhang M.Q., Ju J., Bestor T.H., Das R., Zhang M.Q., Ju J., Bestor T.H., Zhang M.Q., Ju J., Bestor T.H., Ju J., Bestor T.H., Bestor T.H. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S., Federico C., Solovei I., Croquette M.F., Della Valle G., Bernardi G., Federico C., Solovei I., Croquette M.F., Della Valle G., Bernardi G., Solovei I., Croquette M.F., Della Valle G., Bernardi G., Croquette M.F., Della Valle G., Bernardi G., Della Valle G., Bernardi G., Bernardi G. Identification of the gene-richest bands in human prometaphase chromosomes. Chromosome Res. 1999;7:379–386. doi: 10.1023/a:1009220131225. [DOI] [PubMed] [Google Scholar]
- Schumacher A., Kapranov P., Kaminsky Z., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Kapranov P., Kaminsky Z., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Kaminsky Z., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Virtanen C., Winegarden N., Cheng J., Gingeras T., Winegarden N., Cheng J., Gingeras T., Cheng J., Gingeras T., Gingeras T., et al. Microarray-based DNA methylation profiling: Technology and applications. Nucleic Acids Res. 2006;34:528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Nair P., Brothman A.R., Stallings R.L., Brothman A.R., Stallings R.L., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Shiota K., Kogo Y., Ohgane J., Imamura T., Urano A., Nishino K., Tanaka S., Hattori N., Kogo Y., Ohgane J., Imamura T., Urano A., Nishino K., Tanaka S., Hattori N., Ohgane J., Imamura T., Urano A., Nishino K., Tanaka S., Hattori N., Imamura T., Urano A., Nishino K., Tanaka S., Hattori N., Urano A., Nishino K., Tanaka S., Hattori N., Nishino K., Tanaka S., Hattori N., Tanaka S., Hattori N., Hattori N. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells. 2002;7:961–969. doi: 10.1046/j.1365-2443.2002.00574.x. [DOI] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Riggs A.D., Roberts-Ems J., Riggs A.D., Riggs A.D. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979;203:1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Song F., Smith J.F., Kimura M.T., Morrow A.D., Matsuyama T., Nagase H., Held W.A., Smith J.F., Kimura M.T., Morrow A.D., Matsuyama T., Nagase H., Held W.A., Kimura M.T., Morrow A.D., Matsuyama T., Nagase H., Held W.A., Morrow A.D., Matsuyama T., Nagase H., Held W.A., Matsuyama T., Nagase H., Held W.A., Nagase H., Held W.A., Held W.A. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc. Natl. Acad. Sci. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat. Rev. Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Haase M., Lam W.L., Schubeler D., Lam W.L., Schubeler D., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wolff G.L., Kodell R.L., Moore S.R., Cooney C.A., Kodell R.L., Moore S.R., Cooney C.A., Moore S.R., Cooney C.A., Cooney C.A. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- Yan P.S., Chen C.M., Shi H., Rahmatpanah F., Wei S.H., Huang T.H., Chen C.M., Shi H., Rahmatpanah F., Wei S.H., Huang T.H., Shi H., Rahmatpanah F., Wei S.H., Huang T.H., Rahmatpanah F., Wei S.H., Huang T.H., Wei S.H., Huang T.H., Huang T.H. Applications of CpG island microarrays for high-throughput analysis of DNA methylation. J. Nutr. 2002;132:2430S–2434S. doi: 10.1093/jn/132.8.2430S. [DOI] [PubMed] [Google Scholar]
- Yoder J.A., Walsh C.P., Bestor T.H., Walsh C.P., Bestor T.H., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]