Abstract
PDZ domain-containing proteins and their interaction partners are mutated in numerous human diseases and function in complexes regulating epithelial polarity, ion channels, cochlear hair cell development, vesicular sorting, and neuronal synaptic communication. Among several properties of a collection of documented PDZ domain–ligand interactions, we discovered embedded in a large-scale expression data set the existence of a significant level of co-regulation between PDZ domain-encoding genes and these ligands. From this observation, we show how integration of expression data, a comparative genomics catalog of 899 mammalian genes with conserved PDZ-binding motifs, phylogenetic analysis, and literature mining can be utilized to infer PDZ complexes. Using molecular studies we map novel interaction partners for the PDZ proteins DLG1 and CARD11. These results provide insight into the diverse roles of PDZ–ligand complexes in cellular signaling and provide a computational framework for the genome-wide evaluation of PDZ complexes.
The 90-amino-acid PDZ domain found in Caenorhabditis elegans, Drosophila melanogaster, and mammalian genomes takes its name from the first three PDZ-containing proteins identified: the post-synaptic density protein PSD-95/SAP90, the Drosophila tumor suppressor and septate junction protein Discs-large, and the mammalian epithelial tight junction protein zona-occludins-1 (ZO-1) (Kennedy 1995). The structural features of PDZ domains permit them to mediate specific protein–protein interactions, which assemble large protein complexes involved in polarity, vesicle transport, phototransduction, ion channel signaling, and synaptic signaling (Sheng and Sala 2001; Nourry et al. 2003; van Ham and Hendriks 2003; Macara 2004). A single PDZ protein may participate in different aspects of cell polarization, suggesting that developmental timing, cellular context, and multiple binding partners are critical regulators of its multidimensional usage (Betschinger et al. 2003; Betschinger and Knoblich 2004). The importance of understanding PDZ proteins is underscored by the fact that disrupting or deregulating PDZ domain-containing proteins or their ligands results in >20 human Mendelian diseases, while mutational screens suggest that PDZ proteins such as DLG1 may be critical in epithelial tumorgenesis (Bilder 2004; Fuja et al. 2004; Wang et al. 2004; Stephens et al. 2005).
PDZ domains bind to proteins via several mechanisms, the most common of which is the binding of PDZ domains to three classes of consensus carboxy-terminal binding motifs, although in a limited number of cases binding of PDZ domains to internal sites has been described (Songyang et al. 1997; Nourry et al. 2003; Penkert et al. 2004). Within a PDZ protein itself, the affinity of a particular PDZ domain for its corresponding ligand can be coupled to the engagement of protein partners located at neighboring PDZ or other domains, supporting complex temporal and hierarchical control of PDZ complexes in vivo (Penkert et al. 2004; Peterson et al. 2004).
To generate a resource to study the interaction between PDZ proteins and PDZ ligands, we sought to integrate the protein recognition code of PDZ domains with publicly available genomic data sets. Motivated by our observation that 96% of PDZ-binding motifs were conserved across three mammalian species in a collection of literature-curated PDZ–ligand interactions, we systematically discovered a genome-wide set of 899 genes encoding classical PDZ-binding motifs conserved across these three species (the PDZ Conserved Binding Motif proteome, or PDZCBM). Uniquely, we also considered the possibility that embedded in expression profiles exists the specific enrichment in co-expression between the set of genes encoding a particular domain and that set encoding for the respective cognate binding motif(s). Thus, we tested and found connectivity at the level of mRNA, reflected by co-regulation between PDZ domain proteins and PDZ ligands. As a result, we provide an integrated view of PDZ and the PDZCBM with respect to co-expression patterns, cellular localization, interologs, and literature co-citation profiles to enable the prediction of known and novel PDZ complexes.
Results
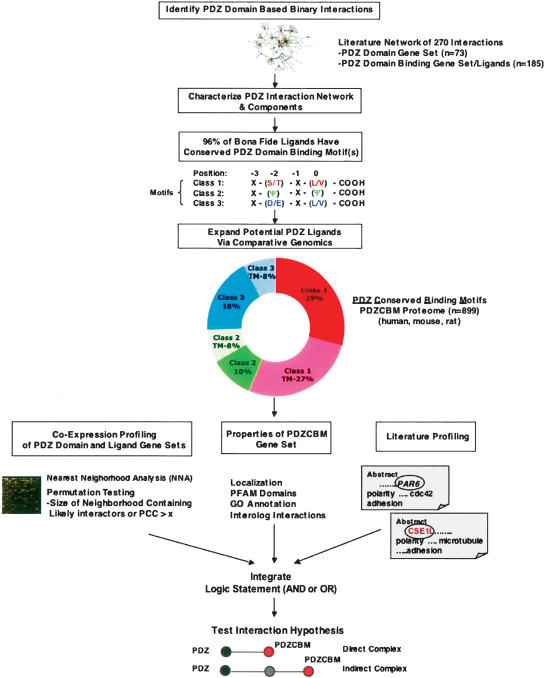
To gain insights into PDZ-mediated biological processes, we developed a schema outlined in Figure 1 to interrogate multiple types of genomic information in order to (1) generate focused experimental hypotheses as to potential PDZ complexes and (2) provide a resource for systematic study of PDZ domain–ligand interactions. To assemble an inventory of PDZ domain-encoding genes, we used the SCOP and SMART databases, which are derived from alignment profiles and Hidden Markov models of PDZ domain sequences. A set of 136 human genes encoding proteins with PDZ domains was compiled utilizing these databases along with the respective D. melanogaster and C. elegans orthologs by reciprocal best-hit BLAST searches. The human genes encode large proteins (994 amino acids as compared with the genome average of 478 amino acids) with multiple PDZ domains (136 genes encode a minimum of 237 PDZ domains along with 70 other domains), consistent with the presence of multiple interaction surfaces to function in the assembly of macromolecular complexes (Supplemental Table 1).
Figure 1.
Computational schema to identify PDZ ligands and PDZ complexes. Step (1): Created database of 270 binary PDZ domain-based ligand interactions from manual literature curation. Step (2): Analyzed characteristics of PDZ–ligand interaction network including expression correlation between PDZ-encoding gene and ligand gene sets, as well as properties of bona fide ligands (e.g., level of PDZ-binding motif conservation). Step (3): Identified that 96% of literature-identified ligands have conserved PDZ-binding motifs among three mammalian genomes (human, rat, mouse). Of these, 79% match one of three “canonical” consensus carboxy-terminal motifs. The canonical consensus motifs shown for each of the three classes were derived from reviews (Sheng and Sala 2001; Nourry et al. 2003). Step (4): Performed systematic comparative genomic analysis of three mammalian proteomes to reveal an 899-member gene set that encodes proteins with canonical PDZ Conserved Binding Motifs (PDZCBM). The motif class distribution and percentage of each class predicted to have a transmembrane (TM) domain by the TMHMM algorithm is shown (Krogh et al. 2001). Step (5): Pairwise relationships among PDZ domain-encoding genes and the PDZCBM gene set are determined from correlated mRNA levels, cellular localization, and common literature co-citation patterns between a PDZ gene and potential ligand. Criteria for significant co-expression levels are based on nearest neighborhood analysis (NNA) indexes and/or a Pearson’s correlation coefficient (PCC) threshold. Orthologous information from model organisms (e.g., interologs) is mined to derive potential PDZ–ligand interactions. Step (6): All data are integrated from these diverse data types employing logical operators AND or OR to provide testable hypothesis as to PDZ complexes. Step (7): The predicted interactions are tested in mammalian cells by co-immunoprecipitation, analyzing the effects of mutating interaction motifs/domains, and/or functional assays.
Expression analysis of the human PDZ gene complement
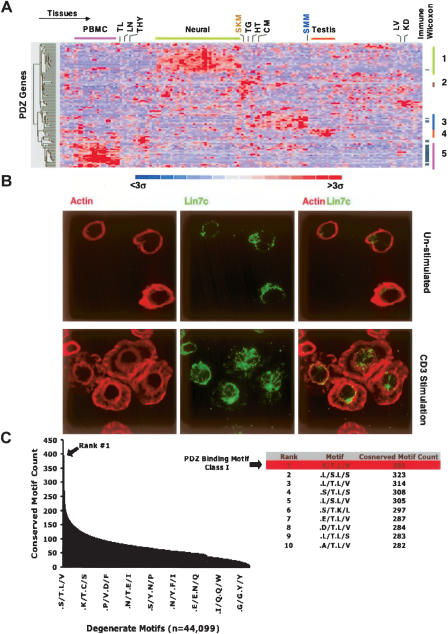
Next, using gene expression data from 79 human tissues, we examined the tissue/cell expression distribution of the PDZ domain-encoding genes, since, for many genes, detailed expression patterns had not been previously reported (Su et al. 2004). As shown in Figure 2A, two large clusters of PDZ genes in neuronal-related tissue and in immune cells/tissues are observed. The large number of PDZ genes expressed in neuronal tissues is consistent with their well documented function in neuronal signaling. In contrast, the number of PDZ genes with high expression in immune tissues was unexpected. In order to quantify the enrichment of PDZ genes in immune tissues in a systematic manner, the Wilcoxon rank-sum test was performed. To calculate the Wilcoxon rank-sum statistic, we fractionated the 79 tissue/cell types into those that are part of the immune system and all other tissue/cell types, thereby making two sample classes. In this regard, 35 PDZ genes were enriched in the immune system prior to Bonferroni correction, with 11 out of 35 significantly enriched after correction (P < 0.05). Several of the immune-enriched PDZ genes such as CARD11, DLG1, SCRIB, and SLC9A3R1 have previously been shown to redistribute to the immune synapse during T cell polarization, while others such as RAPGEF6 and LIN7C have not been characterized in immune signaling. (Hanada et al. 2000; Egawa et al. 2003; Hara et al. 2003, 2004; Ludford-Menting et al. 2005). To corroborate the mRNA expression patterns, we demonstrate the expression of the PDZ protein LIN7C in Jurkat cells by immunofluorescence, observing its redistribution upon TCR activation (Fig. 2B). These expression data provide a framework to further characterize PDZ proteins in immune cell signaling and other tissues.
Figure 2.
Systematic analysis of PDZ expression profiles and carboxy-terminal motifs. (A) mRNA expression profiles visualized across 79 tissue/cell types performed in duplicate from the human GNF compendium for 107 PDZ genes (Su et al. 2004). Genes and tissues were hierarchically clustered and visualized using dCHIP (Schadt et al. 2001). Selected samples aggregated into common tissue types are described at the top of the panel. (PBMC) Peripheral blood mononuclear cells, (TN) tonsil, (LN) lymph node, (THY) thymus, (SKM) skeletal muscle, (TG) tongue, (HT) heart, (CM) cardiac myocyte, (SMM) smooth muscle, (LV) liver, (KD) kidney. Four non-immune co-expression clusters among the various tissues are designated with corresponding genes in each cluster found in Supplemental Table 1. PDZ genes significantly enriched in the immune system (P < 0.05) as found by the Wilcoxon rank sum test are denoted with a tick mark on the right. (B) Endogenous expression of LIN7C in lymphocytes was confirmed by immunoflourescence in unstimulated or CD3-stimulated human Jurkat T cells. (C) The number of conserved instances for each of the observed 44,099 twofold generate motifs at positions −2 and 0 in the carboxyl terminus among the 11,044 aligned genes between human, dog, rat, and mouse. (Table) The top 10 ranked degenerate motifs showing that the Class I PDZ motif (–X-[S/T]-X-[L/V]-COOH) is the most conserved motif.
Characterization of PDZ–ligand interactions via a published literature survey
PDZ-binding motifs generally fall into three established motif classes, based on the residues at positions −2 and 0 with respect to the carboxyl amino acid of a protein (Sheng and Sala 2001; Nourry et al. 2003). In order to standardize definitions, we refer to a carboxy-terminal motif as “canonical” if the carboxy-terminal residues matched those shown in Figure 1. These motifs are taken from two reviews (Sheng and Sala 2001; Nourry et al. 2003). To further characterize PDZ–ligand interactions, we sought to identify a large set of interacting pairs from the published literature. Specifically, we searched the PubMed database using PDZ as keyword and identified 1262 articles published over a period of 10 yr. The review of the abstracts and then the texts of 525 publications reporting on interaction experiments in mammalian systems (human, mouse, rat) created a large, but not exhaustive, set of interactions. These articles reported on a total set of 270 binary interactions involving 179 unique ligands where the experimental evidence demonstrated that the interactions were via the PDZ domain (Supplemental Table 2). Further examination revealed that 76.5% (137/179) of the non-redundant ligands interact via canonical carboxy-terminal PDZ-binding motifs, 19.0% (34/179) via non-canonical carboxy-terminal binding motifs, and 4.5% (8/179) reflected the PDZ domain interacting with a non-carboxyl region. Although canonical motifs were observed in three-quarters of the literature-identified ligands, approximately a fifth of the interactions occurred through non-canonical motifs, with a consensus emerging at least for some ligands—12 out of 34 of such ligands followed the pattern X-(S/T)-X-(I/A)-COOH.
We next assessed the level of conservation of the PDZ-binding motif for each ligand in human, mouse, and rat via a reciprocal best-hit strategy. Among the 137 ligands harboring canonical carboxy-terminal motifs, the PDZ-binding motif was conserved in 95.6% (131/137) of the cases. In the remaining 4.4% (6/137) of cases, the ligand contained a canonical PDZ-binding motif, but only from the species in which the interaction was originally identified. Furthermore, among the 34 ligands containing non-canonical motifs, the PDZ motif was conserved in all cases. Taken together, these data would suggest a bias for bona fide interactions to occur via conserved carboxyl motifs.
Systematic discovery of carboxy-terminal motifs identifies PDZ ligand motifs
The functional importance of the carboxy-terminal motif in PDZ domain recognition and the high level of conservation observed among verified ligands led us to investigate whether the carboxy-terminal PDZ-binding motif was actually common or rare as compared with other conserved carboxy-terminal tail motifs in mammalian genomes. To address this question in a systematic fashion, we started with the collection of 22,737 human reference mRNAs from the RefSeq database (downloaded from UCSC Genome Browser, 04/2005 freeze) and, for these, constructed genome-wide alignments among three other mammalian species (mouse, rat, dog) as described previously (Xie et al. 2005). Ultimately, 13,913 mRNAs mapping to 11,044 unique genes were capable of being aligned among human, mouse, dog, and rat based on the requirement that upon translation, the stop codons for all four species occurred at the identical amino acid position. Since the three canonical PDZ-binding motifs have twofold degeneracy at the 0 and −2 position, we examined the number of conserved instances of all possible twofold degenerate motifs at these positions in our database of aligned reference sequences. Theoretically, there are 204 or 160,000 possible consensus motifs in this search space (i.e., two amino acid positions with twofold degeneracy), and we observed that there was at least one conserved instance of 44,099 out of the possible 160,000 motifs in our collection of sequences. For each of the 44,099 motifs, we counted the number of conserved instances, thereby generating a rank-ordered list of conserved twofold degenerate carboxy-terminal motifs. We called the carboxyl terminus conserved if sequences at positions −2 and 0 of all four species satisfied the consensus motif. In addition, we counted the frequency of all 399 observed conserved non-degenerate motifs at positions −2 and 0. Strikingly, the Class I motif was ranked number one among all twofold generate motifs possible, with (S/T) X (L/V) having 391 conserved instances. Furthermore, we observed that Class I tails occupied four out of the five top-ranked non-generate conserved motifs. These data imply that PDZ-binding motif Class I is likely to be the most strongly conserved carboxy-terminal binding motif in mammalian species (Fig. 2C).
Discovery of a genome-wide set of 899 potential PDZ ligands
Non-canonical motifs can interact with PDZ domains; however, the majority of interactions identified in our examination of the published PDZ–ligand interactions occurred via conserved canonical PDZ-binding motifs. We therefore chose to focus on this subset in order to identify a more complete set of potential PDZ ligands. Specifically, we searched the international protein index (IPI) database for proteins with carboxy-terminal sequences that contained one of the three canonical PDZ-binding consensus motifs and for which rat or mouse orthologous proteins also contained the motif. This approach identified 505, 165, and 229 genes that encode proteins containing conserved Class I, II, and III PDZ-binding consensus motifs, respectively. This will be referred to as the PDZ Conserved Binding Motif (PDZCBM) gene set (n = 899) (Fig. 1; Supplemental Table 3). We next examined if any of the literature-defined PDZ ligand carboxy-terminal motifs overlapped with another motif described in the cell biology knowledge base. This analysis revealed that the carboxy-terminal KDEL ([KRHQSA]-[DENQ]-E-L-COOH) ER retrieval sequence overlapped with the Class III PDZ-binding motif (X-[D/E]-X-[L/V]-COOH). Only a subset of the Class III carboxyl termini (52/229) match the ER consensus motif, suggesting that the remaining Class III potential ligands are not likely to be the result of ER resident protein contribution. The PDZCBM catalog contains 92.1% (116/126) of the literature ligands with conserved canonical binding motifs used in our search. However, in the remaining cases (10/126), IPI was either missing that particular protein and/or the database did not contain those splice isoforms with a consensus motif for some ligands (e.g., NF2 and SYNJ2). Nevertheless, our ability to capture 92.1% of the literature-verified ligands confirms our IPI database search algorithm was sensitive.
The PDZCBM gene set: Cellular localization, functional annotation, and tissue distribution
Next, characterization of the PDZCBM gene set in parallel with the known literature ligands was undertaken, analyzing binding motif distributions, cellular localization, and functional annotation using the Gene Ontology classification system. Such an analysis would allow us to evaluate the extent to which our comparative genomics strategy recovered a set of genes that followed the characteristics of the literature PDZ ligands.
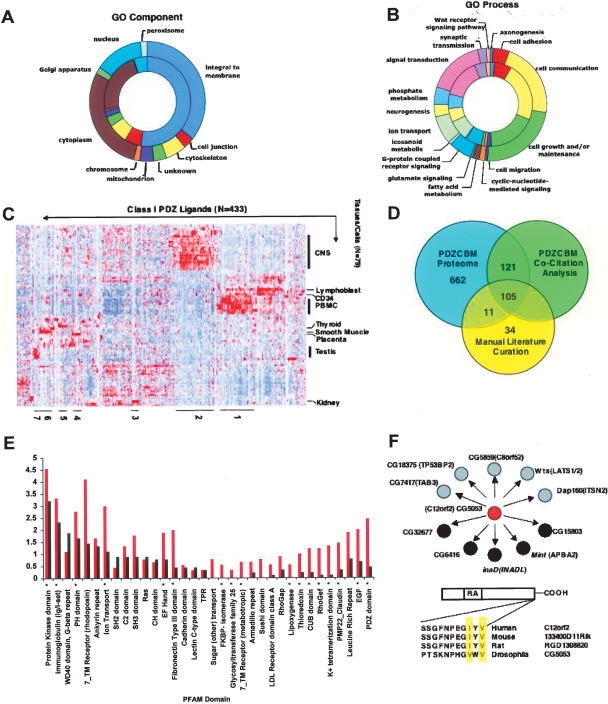
Like those ligands previously reported in the literature, the majority of the PDZCBM gene set was found to have Class I motifs (56% and 72%, respectively), with a similar number of Class II motifs (21% vs. 18%). Class III genes were not compared directly, with only five ligands of this class reported in the literature to date (Itoh et al. 1999; Mancini et al. 2000; Jelen et al. 2003). Structurally, two-thirds (66%) of Class I literature ligands are predicted by the TMHMM algorithm to harbor transmembrane domains, along with 48% of the proteins encoded by the PDZCBM gene set. The number of predicted transmembrane domains among Class II motif genes in the literature versus PDZCBM sets was 19% and 43%, respectively. Aggregating all motif classes, both gene sets are significantly enriched in membrane localization in comparison with the human proteome based on Gene Ontology compartment annotation (GO: Integral to Membrane P-value = 8.64 × 10−29 [literature] vs. 4.12 × 10−23 [PDZCBM]; Bonferroni-corrected Fischer’s exact test; Fig. 3A).
Figure 3.
Characteristics of the PDZCBM. Distribution of cellular component (A) and biological process (B) GO categories. (Inner rings) Proteins encoded by the literature ligands (174 with GO component and 172 with GO biological process identifiers); (outer rings) proteins encoded by the PDZCBM (728 with GO component and 733 with GO biological process identifiers). Each section represents the number of proteins assigned to a given GO category. Both the literature and the PDZCBM gene sets are significantly enriched for integral to membrane (P < 0.001). The distribution between GO biological process categories did not change between the literature and the PDZCBM gene set, with 12 out of the 16 categories enriched in both gene sets (P < 0.05, Fischer’s exact test-hypergeometric probability distribution using a background set of 13,802 GO annotated human genes). (C) Identification of neuronal and immune tissue expression modules in the PDZCBM gene set. For illustrative purposes, the mRNA expression patterns after hierarchical clustering of both tissue and genes are shown for 433 Class I members of the PDZCBM gene set (433 out of 505 Class I genes had probes meeting filtering criteria). Although there exists strong expression in smooth muscle and testes for several ligands, for instance, the profiles are dominated by the prominent clusters of ligands in neural and immune tissues, paralleling the expression of subsets of PDZ genes. Similar dominant immune and neural expression clusters were observed for Class II and III subsets. The identity of the individual PDZCBM constituents in the numbered tissue expression clusters can be found in Supplemental Table 3. (D) Assessment of previous experimentally confirmed PDZ ligands within the PDZCBM set versus potential novel ligands. The automated MILANO literature mining software package revealed that 219 of the 899 genes were co-cited (as of Jan. 15, 2006) with the term PDZ, with an additional 18 genes found to be co-cited by manual examination of PubMed. The 237 overlapped with 116 of the unique ligands in our manually curated literature PDZ–ligand interaction data set (Rubinstein and Simon 2005). Eleven out of 116 genes that are verified PDZ ligands with conserved canonical binding motifs were not found by the automated co-citation software to be linked to the term PDZ. An additional 34 genes with non-canonical binding motifs were identified by manual literature curation. In summary, there are 662 genes that have not been shown to bind to a PDZ protein or be co-cited with the term PDZ with conserved canonical PDZ-binding motifs. (E) Frequency distribution and enrichment of PFAM domains in the PDZCBM proteome. The PFAM database was used to identify domains for the reference protein sequence of each member of the 899 PDZCBM gene set. For illustrative purposes, PFAM domain frequency of the PDZCBM is shown compared with an equivalently sized random gene set. Domains marked with asterisks are significantly (P-value < 0.01, Fischer’s exact test) enriched across the entire PDZCBM proteome compared with the domain composition of the human proteome. (F) Interolog data of PDZ–ligand interactions. We identified Drosophila orthologs for the 29.9% (269/899) PDZCBM members. Of these Drosophila proteins, 31.6% (93/269) had conserved the mammalian PDZ-binding motif. We subsequently interrogated the large-scale Drosophila Y2H screen of Giot et al. (2003) for this subset of proteins looking for interactions with PDZ domain-containing proteins. For example, C12orf2 is an RA domain-containing protein whose fly ortholog, CG5053, was found to interact with four PDZ domain-encoding proteins (black circles) as well several other proteins (gray circles) with high confidence. Thus, comparative genomics coupled with annotation transfer strongly suggest that C12orf2 interacts with PDZ proteins.
To examine in an unbiased fashion the extent to which the literature and PDZCBM gene sets shared similar functions, GO biological process criteria were applied. We show in Figure 3B the distribution for the literature ligands among 16 biological processes, with 13 of these processes showing significant enrichment (e.g., cell adhesion, synaptic transmission, and ion transport function) when compared with annotation of the entire human proteome (all P-value < 0.05; Bonferroni-corrected Fischer’s exact test). In the larger PDZCBM gene set, these same 13 biological process categories were enriched by Fischer’s exact test (P-value < 0.05); while some categories remained significant after multiple hypothesis testing corrections (for example, ion transport: 2.74 × 10−16), others did not maintain significance (for example, cell adhesion). Overall, the literature and PDZCBM gene sets exhibit similar size fractions of proteins annotated to different GO functional categories. At the same time, a distinct group of biological processes, not found in the literature gene set, emerged as significantly enriched after correcting for multiple hypothesis testing in the PDZCBM gene set, such as icosanoid (P < 0.005) and fatty acid metabolism (P < 0.01), suggesting a potential role for PDZ proteins in these metabolic processes.
Finally, we assessed the expression pattern of the PDZCBM gene set across the GNF tissue compendium. Interestingly, modules of strong neuronal and immune expression were observed for the PDZ genes themselves, suggesting sub-networks of PDZ–ligand genes having specific function in these tissues (Fig. 3C). To gauge the potential number of novel PDZ ligands in the PDZCBM gene set, we systematically searched the PubMed literature database for occurrence of co-citation(s) between the term PDZ and the 899 PDZCBM members as of Jan. 15, 2006, employing the MILANO software tool (Rubinstein and Simon 2005). The MILANO co-citation algorithm is a sensitive metric as to the extent the PDZCBM has previously linked to PDZ complexes, since it was capable of tagging 90.5% (105/116) ligands in the PDZCBM identified manually as confirmed ligands (Fig. 3D). Notably, only 26% (237/899) of the PDZCBM genes have been co-cited with the term PDZ, suggesting in conjunction with similarity of gene properties of known ligands that the remaining 74% of genes not co-cited with the term PDZ represents a large pool of potential PDZ ligands.
Protein domains of PDZCBM: Enrichment in RhoGEFs and RhoGAPs
The protein domain composition of the PDZCBM was next examined to identify domains that functionally cooperate with PDZ proteins (see Supplemental Table 3 for annotation of PFAM domains for each PDZCBM gene). This analysis was performed by comparing the distribution of PFAM domains found in the PDZCBM with that of the human proteome using a one-tailed Fischer’s exact test (cumulative hyper-geometric probability distribution) to calculate the P-values. For the 12.1% (116/899) of the PDZCBM proteome not characterized by a PFAM domain, BLASTp analysis was performed to the non-redundant (nr) proteome database at NCBI, revealing homology with known proteins in some instances. The PDZCBM was enriched (P < 0.05) in several PFAM domains such as ion transport, immunoglobulin, and PH domains, many of which are typically found in proteins known to participate in PDZ-mediated processes and in the literature set of ligands (Supplemental Table 7). Interestingly, there was also a set of domains that was enriched in the PDZCBM but not in the literature subset, suggesting that the larger data set may yield additional insights into the functionality and mechanism of PDZ complexes (Fig. 3E). For instance, both RhoGEF and RhoGAP domains are enriched in the PDZCBM (P-value = 9.1 × 10−5 and P-value = 1.7 × 10−2; Fischer’s exact test), respectively. Intriguingly, the balance between RhoGEFs versus RhoGAPs appears to control the epithelium-to-mesenchyme transition and corresponding establishment of polarity in C. elegans, suggesting the 19 identified RhoGEF/GAPs with PDZ-binding motifs, of which 5/19 are known to bind PDZ proteins, may shape similar decisions in vertebrate organisms (Labouesse 2004; Supplemental Fig. 3).
Gene family analysis of proteins encoding conserved PDZ-binding motifs
The previously reported literature suggests that PDZ proteins often interact with multiple members of a gene family to execute their functions, as in the case of membrane conductance (e.g., Ca2+ and K+ ion channels), neuronal synaptic communication (e.g., glutamate receptors), and epithelial adhesion (e.g., claudins) (Balda and Matter 2000; Kim and Sheng 2004). Conversely, the presence of multiple gene family members with conserved PDZ-binding motifs may suggest that such a family functions as PDZ ligands. We define a gene family as a group of genes that share significant sequence similarity with common domain architectures. We therefore examined the extent to which novel ligands in the PDZCBM belonged to known PDZ-binding families. To this end, multiple protein sequence alignment was performed on the PDZCBM proteome using CLUSTALW and phylogenetic trees derived by neighbor-joining analysis. Interestingly, 48% (435/899) of the PDZCBM genes fall into 163 gene families with two or more members, with 49% (80/163) having at least one family member as a known PDZ ligand based on literature curation and/or co-citation with term PDZ (see Supplemental Table 3 for full gene family annotation).
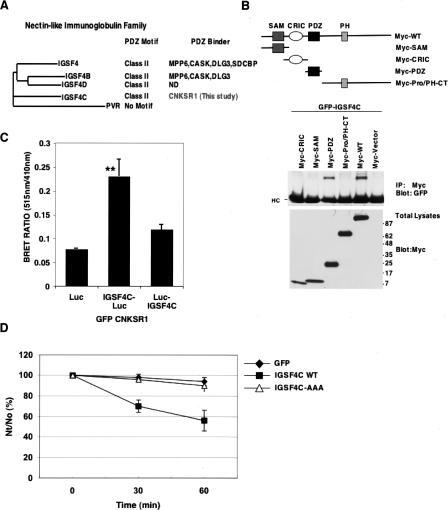
As a result of phylogenetic analysis, ligands were categorized into (1) those occurring in a family, but where perhaps the extent of potential PDZ-binding ligands in a family is less appreciated; (2) published gene families, but where no member had been suggested or shown in vivo to be involved in PDZ complexes to our knowledge; and (3) novel gene families with members containing PDZ-binding motifs or (4) ligands not falling into gene families. As an example of the first category, the nectin-like immunoglobulin gene family contains five members, with four out of five harboring conserved Class II PDZ-binding motifs. Two of the four nectin-like proteins (IGSF4 and IGSF4D) with motifs have been shown to bind PDZ proteins (Fig. 4A). We now show that the PDZ domain of CNKSR1 interacts with a third family member, IGSF4C, identified via a two-hybrid screen (Fig. 4B). In addition, we confirmed the in vivo interaction of CNKSR1 and IGSF4C by bioluminescence resonance energy transfer (BRET) in intact cells (Fig. 4C). The functions for the nectin-like molecules are incompletely understood; however, several have been implicated in cell adhesion (Sakisaka and Takai 2004). We also demonstrate the PDZ-binding motif of IGSF4C is functionally important, since mutation of the motif leads to a reduction in cell adhesion (Fig. 4D). Therefore, in an unbiased manner, these observations support our findings that existence of a conserved PDZ-binding motif in a gene family may predict a PDZ–ligand interaction.
Figure 4.
The CNKSR1 PDZ domain binds IGSF4C, whose PDZ motif mediates cell adhesion. (A) Dendogram of the five-member nectin-like immunoglobulin family based on CLUSTALW protein sequence alignment showing conserved Class II PDZ-binding motifs and annotation of those family members documented to interact with PDZ domain-encoding proteins. (B) CNKSR1 interacts with the PDZ domain of IGSF4C. (Upper panel) Schematic representations of Myc-tagged CNKSR1 constructs used to verify and map interaction between CNKSR1 and IGSF4C; (middle panel) along with GFP-tagged IGSF4C, HEK293T cells were transfected with constructs encoding Myc-tagged wild-type CNKSR1 (Myc-WT) or Myc-tagged CNKSR1 constructs encoding various CNKSR1 domains. Cell lysates were immunoprecipitated with anti-Myc antibody and then blotted with anti-GFP to detect interactions. In the immunoprecipitates, heavy chains are indicated (HC); (bottom panel) total lysates were immunoblotted with anti-Myc antibody to confirm equal expression of constructs. (C) BRET assays confirm the interaction of CNKSR1 with IGSF4C in intact cells. GFP-CNKSR1 was transfected into HEK293T cells with one of three constructs: luciferase alone, luciferase fused to the IGSF4C cytoplasmic domain (IGSF4C-Luc), or luciferase fused to the IGSF4C extracellular domain (Luc-IGSF4C). Forty-eight hours after transfection, the cells were detached and subjected to BRET analysis. Shown are the results of three independent experiments with average bioluminescence resonance energy transfer and standard error. (Asterisks) Significant increase (t-test; P < 0.01) in BRET ratio upon expression of cytoplasmic domain-tagged IGSF4C compared with luciferase alone. (D) Cell aggregation activity of IGSF4C. HEK293 cells transfected with GFP (black diamonds), IGSF4C-GFP (black squares), or IGSF4C (AAA)-GFP (lacking the PDZ motif, white triangles) were treated with trypsin in the presence of EDTA and then dispersed by pipetting to obtain a single-cell suspension. Each single-cell suspension was rotated in Ca2+- and Mg2+-free HBSS containing 5 mM EDTA for 30 and 60 min. The degree of aggregation of cells was represented by the ratio of the total particle number at time t of incubation (Nt) to the initial particle number (No). Similar results were obtained when cell suspensions were rotated in HBSS containing Ca2+ and Mg2+ or Ca2+- and Mg2+-free HBSS.
As an example of the third category, novel gene families were discovered based on the observation of phylogenetically related proteins among the PDZCBM, as was the case for XKR4, XKR6, and XKR7. Although these proteins do not contain recognizable PFAM domains, all three share homology with several additional mammalian proteins, CG32579 of D. melongaster, and ced-8 in C. elegans. In fact, XKR6 is the ortholog of ced-8 (BLASTp 6 × 10−26), and iterative searching of databases revealed an extended gene family with nine members in total, including the gene (XK) responsible for X-linked McLeod Neuroacanthocytosis Syndrome (Ho et al. 1994; Supplemental Fig. 2). The function and mechanism of action of XK and ced-8 are unclear, but both have characteristics of membrane transport proteins, and ced-8 appears to regulate the timing of apoptosis (Stanfield and Horvitz 2000). As a subfamily of a larger ced-8–like gene family, XKR4, XKR6, and XKR7 suggest PDZ complexes may be linked to execution of ced apoptotic pathways.
Mapping the PDZCBM gene set to Drosophila: Phenotypic profiling and interologs
Several recent studies have demonstrated the value of large-scale protein interaction maps and phenotypic screens in model organisms to understand complex cellular processes and identify human disease genes. In some instances, studies have sought to identify conserved orthologous interactions, which are referred to as “interologs.” Nevertheless, the transfer of interaction information from model organisms to mammalians systems, although powerful, is imperfect (Bork et al. 2004; Ramani et al. 2005). In this regard, we propose that information transfer is likely to be more robust if the two proteins observed to interact in model organisms had conserved interaction surfaces in mammalian species, as might be observed between a protein with a PDZ domain interacting with a protein harboring a conserved PDZ-binding motif.
Thus, we sought to further annotate the PDZCBM gene set members and their potential involvement in PDZ protein complexes by analyzing the orthologous set of proteins in Drosophila at the sequence, interaction, and functional level. We chose D. melanogaster since many functions of PDZ complexes were originally identified in this model organism, coupled with the availability of sequence, large-scale yeast-two-hybrid maps, and phenotypic screens (RNAi or mutagenesis). To accomplish our goal we first identified fly orthologs of the PDZCBM by reciprocal best-hit strategy (E-cutoff 10−10). We found Drosophila orthologs for 30% (149/505), 28% (46/165), and 32% (74/229) of Class I, II, and III genes, respectively, with 34.5% (93/269) having a conserved mammalian PDZ-binding motif (see Supplemental Table 3 for list of fly orthologs). For all 269 fly orthologs, RNAi or mutant phenotypes were assigned based on manual curation of GO biological process annotation, review of individual articles, and a large-scale RNAi screen performed to examine cell morphology and create annotation profiles (Kiger et al. 2003) (see Supplemental Table 4 for complete annotation profiles).
In those Drosophila orthologs with mammalian motifs and phenotypic profiles available, a higher percentage are associated with polarity, adhesion, ion transport, or neuronal synaptic processes (manifested by mutants causing abnormal bristle polarity, myoblast fusion, and dorsal closure, for example) than not: 43% (40/93) compared with 26% (24/93), respectively. These results highlight that, in some instances, known PDZ ligands important in the regulation of polarity and adhesion have evolutionarily conserved PDZ-binding motifs, as is the case for the mutants Van Gogh (VANGL1), yurt (EPB4IL5), and rolling pebbles (TANC2) (Wolff and Rubin 1998; Menon and Chia 2001; Rau et al. 2001; Hoover and Bryant 2002). We also identified novel proteins in the PDZCBM set with highly conserved PDZ-binding motifs, including but not limited to CG31534 (DKFZp434I0312), CG7323 (PLEKHG5), and CG56987 (RASSF8, previously known as C12orf2) (Supplemental Table S1). Based on a Drosophila yeast-two-hybrid (Y2H) interaction map and interolog concepts, CG7323 (PLEKHG5) interacts with PDZ protein l(2)02045 (GIPC1), while CG56987 (C12orf2) is capable of interacting with multiple PDZ-domain-containing proteins, such as MINT (APBA2) (Fig. 4F; Giot et al. 2003; Kiger et al. 2003). Integrating Y2H interactions and sequence data therefore suggests these interactions may rely on conserved interaction surfaces (both PDZ domain and PDZ-binding motif), thereby increasing the probability that such PDZ complexes may be found in mammalian cells in vivo.
PDZ genes are co-expressed with ligands
For PDZ complexes to form and accomplish their biological functions, their components must be temporally and spatially co-localized. Some of this control should be contributed at the level of gene expression. Thus, we sought to determine whether co-expression patterns could be used as a tool to help predict the connectivity map between PDZ proteins and their ligands. To test this hypothesis, using the set of 270 interactions we identified in reviewing the current literature (see above), we examined whether the known PDZ proteins and their ligands were co-expressed more than expected by chance alone.
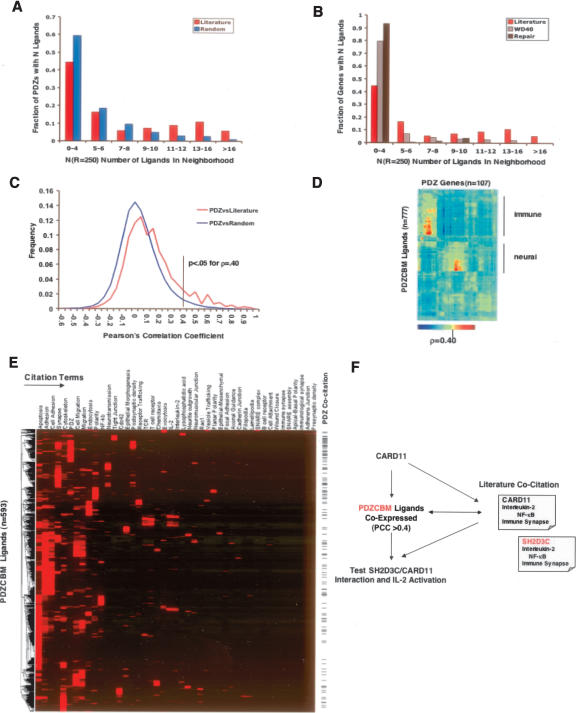
In order to assess the level of co-regulation between PDZ and ligand gene sets, we applied the previously described nearest neighborhood analysis (NNA) methodology to a large-scale, publicly available, mRNA expression atlas of human samples (79 normal or transformed tissues/cells; 16,684 genes, 117 PDZ genes) (Mootha et al. 2003; Owen et al. 2003). We found an enrichment of known ligands in PDZ neighborhoods, as compared with equally sized sets of randomly selected genes (permutation testing, P < 0.001). Specifically, we observed 31 PDZ proteins that have ≥10 literature ligands in the top 250-gene neighborhood. Examining 1000 random ligand sets equal in size to literature ligands, we observe an average of 0.3 (stddev 0.22, maximum of three) PDZ proteins having ≥10 random ligands in the 250-gene neighborhood (Fig. 5A). In contrast, we did not observe any co-regulation between known PDZ ligands with a set of 230 WD40 domain-encoding genes or with a set of 120 DNA repair genes by neighborhood analysis using NNA analysis, suggesting specificity in terms of our observed co-regulation (Fig. 5B).
Figure 5.
Co-expression and co-citation profiling. (A) The PDZ domain-containing proteins and literature confirmed PDZ’s ligand gene sets are co-regulated by nearest neighborhood analysis (Mootha et al. 2003). Briefly, the PDZ neighborhood index N(R = 250) is defined as the number of unique ligands previously reported in the literature to bind PDZ proteins that occur within the nearest 250 genes ranked by Euclidean distance to a PDZ gene. The distribution is plotted for random distribution of ligands and for the literature-curated list of ligands (n = 192). For comparison, a set of random ligands of equal size to the literature ligands was generated and the distribution was calculated 1000 times with the average value shown (P < 0.001; permutation testing). (B) No significant co-regulation between known PDZ ligands and a set of 230 WD40 domain-encoding genes or with a set of 120 DNA repair genes was detected by neighborhood analysis. (C) Determination of pairwise Pearson’s correlation (PCC) cutoff of ρ > 0.40 suggests a potential direct or indirect interaction between the protein products of the PDZ gene and PDZCBM ligand. Two distributions were examined consisting of the PCCs of the 270 binary PDZ–ligand interactions documented in the literature survey and another distribution comparing PDZ genes correlations with a random set of ligands. The random distribution was calculated by generating 1000 permutations of random ligands of the PDZCBM gene set size (n = 899). A significant pairwise correlation value for potential PDZ–ligand interaction was derived based on the fact that <5% of the random distribution had (ρ or PCC) > 0.40. (D) Hierarchical clustering of correlation values (ρ) between PDZ genes and PDZCBM (n = 107 × 777) showing modules of co-expression between PDZ and PDZCBM ligands in immune and neuronal tissues. (E) Five hundred and ninety-three genes of PDZCBM clustered by similarity of PubMed literature co-citation profiles with 43 terms describing location, functionality, and interacting proteins of known PDZ ligands and complexes. (Note: 306/899 did not have citations with any of these 43 terms.) The tick marks on the side indicate those PDZCBM genes that are co-cited with the term PDZ. (F) Flow of information used to identify CARD11–SH2D3C interaction combining PDZCBM comparative genomics with co-expression profiling and co-citation profiling.
Although certainly many of the PDZ proteins and their ligands are constitutively expressed and/or controlled at the level of translation or post-translational modification, we were interested in determining the fraction of known PDZ–ligand interactions that can be detected using co-expression patterns as measured by individual expression neighborhood indexes (see Methods). Examining individual PDZ gene expression neighborhoods, we detected 14% and 21% of the published PDZ–ligand interaction pairs in the 250- and 500-gene neighborhood radius, respectively.
Given that there was significant co-expression of the known PDZ and ligand gene sets, it was of interest to be able to assess the chance any specific PDZ and potential ligand pair interacted based on co-regulation. The challenge of examining any given PDZ expression neighborhood for potential ligands is to know what correlation threshold will be the most sensitive and specific for identifying true ligands. One approach may be to consider only those ligands in the nearest neighbors of a PDZ in the radius of 250 or 500 based on the above gene set results. Another approach, however, is to examine the distribution of Pearson’s pairwise correlation values between known PDZ–ligand interactions compared with that between PDZ and random sets of genes. Based on this analysis, we determined a pairwise Pearson’s correlation cutoff (PCC) of ρ > 0.40, since it corresponds to <5% of the random distribution (P-value < 0.05) (Fig. 5C). Specifically, we examined the expression neighborhood of each PDZ gene and identified all of those PDZCBM genes that were correlated at level of ρ > 0.40. Those PDZCBM genes that met this threshold were considered candidates for functional and/or physical interactions with the examined PDZ gene.
Identification of novel PDZ complexes
Given the ability of correlation thresholds and NNA to capture known PDZ complexes, we next sought to test novel predictions based on NNA/PCC, the PDZCBM comparative genomics catalog, cellular localization, and literature parsing.
The DLG family of PDZ proteins can interact with overlapping and different sets of PDZ ligands (Montgomery et al. 2004; Supplemental Fig. 1). Previous studies have shown that DLG1 is broadly expressed and plays a central role in neuronal synapse assembly, cell growth, polarity determination, and T cell signaling (Hanada et al. 2000; Kim and Sheng 2004). We examined the neighborhood indexes of DLG1 and pairwise correlation values for known ligands, given our interest in understanding the underpinnings of Discs large actions in various biological processes. Mammalian discs large homolog 1 is the ortholog of the fly tumor suppressor dlg. Loss of dlg causes overproliferation of imaginal discs epithelium in a cell-autonomous fashion as well as aberrant cellular morphology and organization in the nervous system (Perrimon 1988; Woods and Bryant 1991; Bilder 2004). The mechanism by which fly or mammalian DLG1 might regulate proliferation has remained elusive. Examination of the correlation values of DLG1 showed that 18 Class I ligands (the preferred binding motif Class I of known DLG1 interactors) had ρ-values >0.40, including DGKζ (ρ = 0.56), BCR (ρ = 0.48), GRIK2 (ρ = 0.48), and KIF1 (ρ = 0.61). GRIK2 and KIF1 were previously identified as interacting partners of DLG1, whereas Breakpoint Cluster Region (BCR) has not been reported to associate with DLG1 (Garcia et al. 1998; Mok et al. 2002).
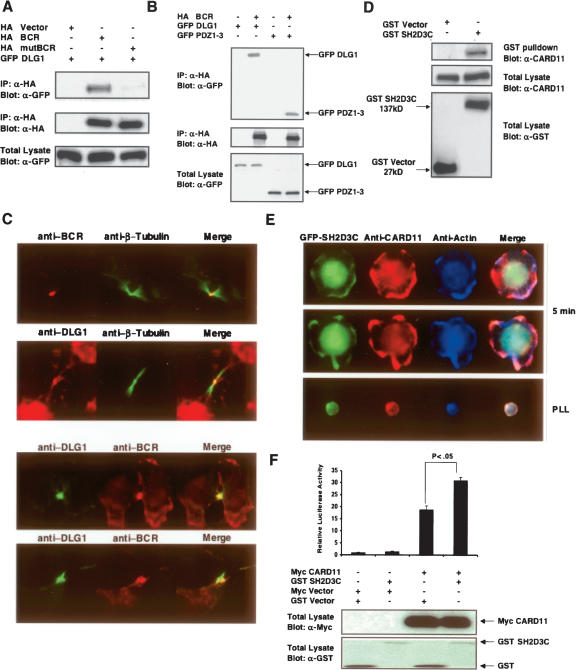
Figure 6A shows that BCR co-immunoprecipitates with DLG1 in 293T cells. Furthermore, mutation of the Class I binding motif of BCR abolishes the interaction with full-length DLG1. Having established that the PDZ-binding motif of BCR is necessary for BCR–DLG1 interaction, we mapped the binding site of BCR onto DLG1. As shown in Figure 6B, PDZ domains of DLG1 are sufficient for binding to BCR. We next examined the localization of endogenous BCR and DLG1. We observed a striking co-localization of the endogenous proteins during cytokinesis, with both proteins localized at the midbody (Fig. 6C). It has previously been shown that DLG1 localizes to the midbody, but BCR localization to this structure has not been previously reported in mammalians cells (Massimi et al. 2003). However, the ortholog of BCR has been implicated in Dictyostelium cytokinesis (Knetsch et al. 2001). These findings suggest that co-expression analysis enables the detection of novel PDZ protein–ligand interactions.
Figure 6.
Experimentally verified novel PDZ complexes. (A) DLG1 binds the break point cluster region (BCR) protein. 293T cells were co-transfected with wild-type HA-tagged BCR or PDZ-binding motif mutant (mutBCR) and GFP-tagged DLG1. Anti-HA antibody immunoprecipitates were resolved by SDS-PAGE and immunoblotted for GFP-DLG1. (Upper panel) Proteins retained on Ha beads were probed with anti-GFP antibody; (middle panel) cell extracts probed with anti-Ha antibody; (lower panel) cell extracts probed with anti-GFP. (B) Co-immunoprecipitation demonstrating that DLG1 PDZ domains are sufficient for binding to BCR. Co-transfection of HA epitope-tagged BCR along with full-length wild-type GFP-DLG1 or the isolated DLG1 PDZ domains 1–3. (Upper panel) Proteins retained on anti-HA beads probed with anti-GFP antibody; (middle panel) cell extracts probed with anti-HA antibody; (bottom panel) cell extracts probed with anti-GFP antibody. (C) Staining of endogenous BCR-DLG1 shows their co-localization at the midbody in U20S cells during cytokinesis. (D) SH2D3C associates with CARD11. Interaction between GST-tagged SH2D3C with endogenous CARD11 in Jurkat cells. Expression constructs encoding GST vector and GST-SH2D3C were electroporated into Jurkat cells, and 24 h post-transfection lysates were prepared and subjected to GST pull-down. The immunoprecipitates (IP) and total lysates (TL) were resolved by SDS-PAGE and immunoblotted with anti-CARD11 and anti-GST. (E) GFP-SH2D3C and CARD11 co-localize upon cell spreading in response to T-cell stimulation. Jurkat cells expressing GFP-SH2D3C (green) were activated to induce spreading for 5 min at 37°C on glass slides coated with anti-CD3 and CD28 antibodies (two examples shown in two upper panels). As a control, cells were also stained on poly-lysine-coated slides (PLL) lacking anti-CD3 and anti-CD28. After fixation, cells were stained with rabbit anti-CARD11 followed by anti-rabbit IgG conjugated with Alexa-Flour 568 (red) and phalloidin conjugated with Alexa-Flour 350 (blue–actin). These results represent one of three independent experiments, which gave similar results. (F) SH2D3C activates the CARD11-mediated IL-2 activation. Jurkat E6 cells (5 × 106) were electroporated with GST-Vector (8 μg) or GST-SH2D3C (8 μg), Myc-vector (5 μg), or Myc-CARD11 (5 μg), along with two reporter plasmids (7 μg of IL-2-luc and 0.2 ng of renilla-luc). After 24 h of electroporation, cells were harvested, lysed, and examined for luciferase activity, showing a significant increase in IL-2 promoter activation upon co-transfection of both genes (student’s t-test P < 0.05). (Lower panel) Expression levels of the transfected proteins.
Coupling expression patterns to co-citation profiles
To gain further insights into the relationship of expression patterns between PDZ genes and ligands, we performed hierarchical clustering of pairwise correlation values between PDZ proteins and ligands (PDZCBM) in the GNF tissue compendium. Figure 5D reveals distinct PDZ and ligand gene co-regulation clusters in neuronal and immune tissues, suggesting sub-networks of interactions that may function in these tissues. These results suggested that potential interactions relevant to immune function may be embedded in the immune module. To enhance our ability to predict biologically relevant interactions within the immune signaling, we coupled these expression profiles with orthogonal information (Jansen et al. 2002; Troyanskaya et al. 2003; Lee et al. 2004). Here, we reasoned that subcellular location and pathway knowledge inferred from literature citations would permit us to further refine our approach. In order to provide an additional mechanism to systematically organize the involvement of a particular PDZ or PDZCBM gene into pathways, we created co-citation expression vectors, which could be analyzed by hierarchical clustering algorithms as shown in Figure 5E. Each component of a gene co-citation vector represents the absolute number of times the gene was cited in an article with each of 43 citation terms. These co-citation terms were selected to describe the location, functionality, and interacting proteins of known PDZ ligands and complexes (terms such as adherens junction, polarity, and Cdc42). Illustration of our proposed approach to identify and experimentally test predicted PDZ complexes utilizing such literature profiles is shown in Figure 5F, employing our results concerning CARD11 and SH2D3C (referred to as Chat-H in Sakakibara et al. 2003) as an example.
Recent biochemical and genetic studies have demonstrated a central role for CARD11 as a positive regulator of antigen receptor signaling (Egawa et al. 2003; Hara et al. 2003; Jun et al. 2003; Newton and Dixit 2003). In this case, we applied the AND rule of logic operators to predict candidate CARD11 interactions with members of the PDZCBM, requiring that a candidate ligand be co-expressed at ρ > 0.40 AND have literature citations supporting a positive role in antigen receptor signal transduction. As a result, six out of 899 PDZCBM genes (RAC2, SH2D3C, FYN, SCAP2, TBC1D10A, and PKCα) emerged as potential CARD11 interactors (Liu et al. 1998; Marie-Cardine et al. 1998; Black et al. 2000; Reczek and Bretscher 2001; Yu et al. 2001; Itoh et al. 2002; Sakakibara et al. 2003; Sugie et al. 2004; Matsumoto et al. 2005; Rahmouni et al. 2005). Among the six candidate ligands, previous studies have demonstrated that SH2D3C over-expression enhances IL2 production.
As shown in Figure 6D, we confirmed that GST-tagged SH2D3C interacts with endogenous CARD11 in T cells. Given that CARD11 and SH2D3C form a complex, we next tested if CARD11 and SH2D3C co-localize upon activation of T cells by anti-CD3 and CD28 co-stimulation. As shown in Figure 6E, GFP-SH2D3C co-localizes with CARD11 after 5 min following TCR stimulation. As a functional readout, we measured the effect of SH2D3C on CARD11-mediated induction of IL-2 promoter activity. As shown in Figure 6F, low levels of ectopic expression of SH2D3C enhanced CARD11-mediated IL-2 promoter induction. These results demonstrate that integration of motif conservation, co-expression, and literature annotation can be utilized to gain insight into biologically relevant novel PDZ protein complexes.
Discussion
In this study, we present a strategy for the discovery of novel PDZ complexes based on the integration of co-expression, comparative genomics, and citation profiles. As an integral part of this strategy, the identification and expression patterns of the human PDZ domain-containing gene complement as well as that of 899 genes that encode for proteins with conserved PDZ-binding motifs are reported. Using publicly available tissue expression profiles, we show that a distinct subset of PDZ genes and ligands are preferentially expressed in the immune system, a number comparable to the subset of PDZ genes/PDZ ligands enriched in the nervous system. Inspection of the tissue compendium revealed that PDZ genes such as RAPGEF6 and paralogs of evolutionarily conserved polarity components such as APBA2 and LIN7C are prominently expressed in lymphocytes. (Fig. 1; Supplemental Table 1; Butz et al. 1998). These predictions are consistent with the recent demonstration that PDZ domain proteins coordinate T cell polarity (Ludford-Menting et al. 2005).
Similar to the PDZ gene expression catalog, the PDZCBM catalog serves as a resource. The significance of the conservation rate of the PDZ-binding motif(s) was evaluated by conducting a systematic interrogation of all possible twofold carboxy-terminal degenerate motifs in four mammalian species. Importantly, the Class I motif is the most conserved carboxy-terminal motif in the four species examined. Further, sequence alignments and domain annotation suggested that PDZ ligands (PDZCBM) fall into gene families and, when coupled with information derived from literature parsing, interolog data and co-expression enables placement in biological pathways.
In terms of integrative strategies, co-expression profiles have previously been employed to infer functional or physical interactions. However, few experimentally verified interactions have been documented prospectively, especially in mammalian systems. In addition, it has not been reported whether embedded in such profiles exists the specific enrichment in co-expression between the set of genes encoding a particular domain and that set encoding for the respective cognate binding motif(s). Using the GNF expression atlas of human tissues, we demonstrated enrichment of known PDZ ligand genes in the expression neighborhoods of PDZ domain-encoding genes. In support of this observation, a retrospective mining of co-expression patterns between PDZ domains and known ligands enabled the detection of 21% of 239 PDZ–ligand interactions published over the past 10 yr. This level of sensitivity between co-expression and physical interaction is consistent with that seen in a systems view of the C. elegans interactome (Li et al. 2004). Prospectively, and as proof of principal, the co-expression patterns in this data set suggested a novel interaction between DLG1 and BCR, which was then experimentally verified. DLG1 is a tumor suppressor involved in the regulation of cell cycle progression through not entirely clear mechanisms, but it interacts with proteins such as PBK (a mitotic Ser/Thr kinase) and APC (Matsumine et al. 1996; Gaudet et al. 2000). Here, we demonstrated that DLG1 interacts and co-localizes with BCR at the mitotic spindle. Recent proteomic surveys have also reported that PDZ ligands such as ACTN4, HAPIP, and NADRIN as well as potential PDZ ligands by our analysis (e.g., CGI-23) are localized to the midbody (Skop et al. 2004). These observations suggest that PDZ proteins, mirroring the role of PDZ complexes in other synaptic processes, may regulate the synaptic connection between cells at cytokinesis.
Our approach is complementary to other approaches such as yeast two-hybrid screens (Y2H) and proteomic surveys, since no one approach will identify all known PDZ protein complexes. This is most directly demonstrated by the modest level of overlap between identified protein complexes based on mass spectrometry or Y2H screens in interaction maps published to date (Uetz et al. 2000; Ito et al. 2001; Stanyon et al. 2004). One strength of correlative profiling compared with Y2H screens lies in that our methodology extracts information from the status of cells/tissues in their endogenous state as opposed to the environment of yeast cells. On the other hand, mass spectrometry-based approaches certainly account for post-translational modifications, which are not reflected in co-expression patterns. Extending our approach to the analysis of gene expression data sets derived from mucosal epithelial surfaces, solid tumors, and embryonic tissues may provide additional insights into co-expression patterns of PDZ genes with PDZ ligands. In addition, the current PDZCBM catalog is likely to under-represent the number of potential PDZ ligands. An expanded PDZCBM should be discovered by designing algorithms to detect conserved PDZ-binding motifs in splice variants of genes and by utilizing the entire spectrum of observed literature PDZ-binding motifs to broaden the comparative genomics searches. Lastly, the impact on expression and the composition of PDZ complexes by microRNAs provides an important avenue for future exploration, given that 42 of the PDZ-encoding genes and numerous ligands are predicted to be miRNA targets (John et al. 2004).
In summary, we have developed a systematic computational platform, based on the integrative analysis of the biological properties of PDZ domain-encoding genes and ligands, to facilitate further understanding of the PDZ complexes in health and disease.
Methods
Identification of PDZ domain-containing proteins
A reference catalog of genes that encode PDZ domain-containing proteins was obtained by searching the SMART ([http://smart.embl-heidelberg.de/] [IPR001478]) and SCOP superfamily (release 1.65 [http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?ipid=SSF50156]) databases. These databases recorded 479 and 145 PDZ-encoding proteins, respectively, in the human genome. The protein-encoding sequence and gene IDs were obtained and mapped to a LocusLink (currently known as Entrez gene) or UniGene identifiers to reveal a non-redundant set of 136 genes encoding proteins with PDZ domains (Supplemental Table 1). In addition, the protein sequences of five PDZ domain proteins were used to perform a TBLASTN search of ESTs identifying no further PDZ-encoding genes. For each of the 136 genes, the reference protein sequence (for those with multiple isoforms, the longest isoform) was used to search against the conserved domain database (CDD; http://www.ncbi.nih.gov/Structure/cdd/cdd.shtm) to identify the number of PDZ domains (E-cutoff 10−5) encoded by a particular gene. These 136 genes were found to encode 237 unique PDZ domains, based on the longest reference sequence.
Literature PDZ–ligand interactions
In order to curate a set of ligands confirmed by experimental data in the literature, we queried the PubMed database using PDZ as keyword and identified 1262 articles from the period of January 1995 to November 2004. Based on review of the abstract and then of the text when appropriate, 525 publications regarding PDZ proteins in mammalian systems (human, rat, mouse) were analyzed. These articles reported on a total set of 270 unique binary interactions involving 185 ligands, where the experimental evidence demonstrated that the interactions were via a PDZ domain and/or the carboxyl terminus of the ligand (see Supplemental Table 2 for the list of interactions). Our recorded set of interactions is equivalent to the set of non-redundant mammalian PDZ–ligand interactions in PDZbase (as of July 2005), which was published after the initiation of our work (Beuming et al. 2005). In our data set, there were fewer interactions than publications reviewed, since some publications reported identical interactions as others, experimental evidence was lacking that interaction was mediated by PDZ domain and/or cytoplasmic tail, or reported interactions mapped to a non-PDZ domain, although we recorded a small subset of the latter (n = 7). Further examination of these 185 carboxy-terminal non-redundant ligands revealed that 145 were interacting via canonical PDZ carboxy-terminal binding motifs: Class I (X-[S/T]-X-[L/V]-COOH); Class II (X-Y-X-V; X-F-X-A; X-Y-X-I; X-V-X-I; X-V-X-V; X-I-X-V-COOH); and Class III (X-[D/E]-X-[L/V]-COOH). Motifs were defined as “canonical” based on the reviews of Sheng and Sala (2001) and Nourry et al. (2003). For the 270 interactions, we identified probe sets that met our filtering criteria (see below) in the current data set for both the PDZ and ligand in 88% (239/270) of interactions. All references for the 270 interactions are available on request.
PDZ ligand identification
A stand-alone Perl script was written such that for each human protein (reference sequence or predicted) in the International Protein Database (IPI) (June 2004 build), the carboxy-terminal 10 amino acids were extracted and checked for the presence of consensus Class I, Class II, or Class III PDZ-binding motifs described above. To identify redundant IPI protein entries with consensus positive motifs encoded by the same gene, pairwise BLASTp analysis utilizing the entire protein sequence coupled with assignment of each protein unique identifiers including UniGene and/or LocusLink ID (Entrez gene ID) was performed. Next, we identified the subset of genes that had a reference protein sequence (either NP or XP accessions) confirming that the carboxyl terminus indeed matched a consensus PDZ-binding motif. If no reference protein sequence was available for a given database entry (e.g., for some UniGene entries), we manually examined all available evidence such as gene predictions, EST alignments, and comparative genomics to ascertain if a gene model or a cDNA sequence encoded for a carboxy-terminal PDZ-binding motif. We employed a combination of NCBI functions BLASTp, TBLASTN, and/or BLINK analysis at the proteome level and genomic alignments via BLAT functions of the UCSC Genome Browser to find mouse/rat orthologs and to crosscheck human gene models. This level of stringency was taken since in some instances the protein sequence in the IPI database represented in actuality a partial fragment, which by chance had a consensus motif, but the full-length protein did not have a motif. Next, the HomolGene and InParanoid databases were used to identify the reciprocal best hit in the mouse and rat proteome (E-value < 10−10). If no HomolGene or InParanoid entry was available, manual curation was performed as above identifying the reciprocal best mouse and/rat sequence by BLASTp analysis of a non-redundant protein database (NCBI). Perfect conservation of the consensus motif in either the rat or mouse ortholog was required to be included in subsequent analyses. The above criteria and filtering lead to the identification of a total of 899 proteins with conserved PDZ-binding domains in their carboxy-terminal ends (505 Class I, 165 Class II, and 229 Class III genes; see Supplemental Table 3).
Carboxy-terminal motif search
We started with 22,737 reference mRNA sequences from the RefSeq database (downloaded from UCSC Genome Browser, 04/2005 freeze). Upon translation, we were able to align 13,913 of these RefSeqs to orthologous mouse/dog/rat reference sequences based on the requirement that all four species had aligned stop codons. The alignments were extracted from nucleotide whole-genome alignments between human/mouse/rat/dog generated by the UCSC Genome Browser and translated to amino acid sequences as described previously (Xie et al. 2005). If a RefSeq accession, when translated, had a different stop codon in human versus mouse, the RefSeq was not used in the analysis. In other words, if a stop codon in human was amino acid 555 and mouse was 554, these were not used for further analysis. Further mapping revealed that these 13,913 reference sequences mapped to 11,044 unique genes based on Entrez/LocusLink IDs. To ascertain the level of conservation of the known consensus PDZ-binding motifs (I, II, and III) that exhibit twofold degeneracy at positions −2 and 0 (e.g., class I: X-[S/T]-X-[L/V]-COOH), we conducted an unbiased search for all possible twofold degenerate carboxy-terminal binding motifs at positions −2 and 0 in the 11,044 aligned reference sequences. In theory, there are 160,000 possible consensus motifs in a twofold degenerate motif search space. Of the 160,000 theoretically possible consensus motifs, we observed 44,099 of such motifs upon examination of the translated reference sequences among all species in our collection of 11,044 aligned genes. We called a site “conserved” if the sequences at positions −2 and 0 of all four species (human/dog/mouse/rat) satisfied one of the possible 44,099 motifs. Subsequently, we counted the total number of genes with conserved instances for each of these 44,099 twofold degenerate motifs. As a result, we have generated a ranked ordered list, with the most conserved twofold generate motif at positions −2 and 0 to the least conserved. We observed that the PDZ Class I motif ranked number one among twofold degenerate motifs with the largest number of conserved instances. In addition, we derived counts for all 399 conserved non-degenerate motifs at positions −2 and 0 with the 4/5 top-ranked motifs corresponding to individual Class I motifs.
Domain, GO, co-citations, and phylogenetic analysis of ligands
Transmembrane domains were predicted by the TMHMM algorithm (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al. 2001). We used the published DAVID 2.0 program to compute enrichments of both the transmembrane and GO biological processes by using Fischer’s exact probability with Bonferroni corrections, which identifies functional categories over-represented in a gene list relative to the representation within the proteome of a given species (http://david.niaid.nih.gov/david/version2/index.htm) (Dennis Jr. et al. 2003; Hosack et al. 2003). Domains of ligands were identified by querying the PFAM database against the reference protein accession of each potential ligand in the PDZCBM proteome. Supplemental Table 3 contains the PFAM domain for each potential ligand. To identify gene families in the PDZCBM proteome, CLUSTALW (http://align.genome.jp/sit-bin/clustalw) was employed using default parameters to build a phylogenetic tree derived by neighbor-joining analysis applied to pairwise sequence distances. If the distance between any two pairs of proteins was <0.40, which empirically corresponded to known gene families and was consistent with common domains as a crosscheck, we considered such proteins a family. Co-citation profiles for the ligands with the term PDZ were performed by the MILANO (Microarray Literature Based Annotation) software (http://milano.md.huji.ac.il/) (Rubinstein and Simon 2005; Supplemental Table 3). Similarly, co-citation profiles for each of the PDZ and PDZCBM genes were identified for a common set of 40 citation terms representing biological processes in which PDZ genes have been implicated in the literature (Fig. 6). MILANO reports the absolute number of times a gene, including all known name aliases, was cited in articles containing the user-defined terms. Therefore, for each gene we created a co-citation expression vector, which could be analyzed by hierarchical clustering algorithms as described below.
Mendelian disease identification
To identify PDZ or PDZCBM genes known to result in Mendelian or complex diseases, the morbidmap was downloaded from (ftp://ftp.ncbi.nih.gov/repository/OMIM/morbidmap). In Supplemental Table 5, we provide a list of human PDZ genes and known or potential PDZCBM ligands associated with Mendelian diseases. In addition, we list complex traits, mutations found in primary tumors/cell lines, and translocations in humans involving PDZ/ligand genes. Those PDZ/ligands targeted by naturally occurring mutations or those derived by ENU mutagenesis of mouse or zebrafish are also furnished.
Microarray filtering and annotation mapping
The public version of the GNF human expression atlas version2 (Su et al. 2004) was obtained from Novartis (http://wombat. gnf.org/), including the primary .cel files, which used the U133a Affymetrix chip and a custom chip (GNF1H). The data set contains the expression values of 33,690 probes reflecting normalization of each array to a set of 100 housekeeping genes common to both the U133a and custom GNF1H array. Subsequently, global median scaling across the arrays was performed, resulting in the expression values across samples for each probe set. The absent/present calls were analyzed, and probe sets with 100% absent calls across all 79 human tissues were not included in the analysis. The data set was further filtered by requiring that a probe set have a threshold value >20 in at least one sample and a maximum–minimum expression value >100. The resultant filtering left 28,852 reliable probe sets for further analysis. For each U133a probe set meeting the above criteria, its corresponding UniGene ID and LocusLink ID were identified based on the combined annotation tables provided at the http://wombat.gnf.org/ and NetAffyx Web site (http://www.affymetrix.com). For the custom GNF1H chip, the mRNA/EST used to design the probe set was blasted against the exemplar sequences of the UniGene database (Build 116). Of the 28,852 probe sets, 26,789 mapped to 16,811 UniGene IDs.
Neighborhood and statistical analysis
Neighborhood analysis was performed using a stand-alone Perl script that was previously described with some modifications and describes the level of enrichment in co-expression between two gene sets (Mootha et al. 2003). In brief, for each query PDZ gene in the atlas, we rank order all other genes in the atlas on the basis of Euclidean distance of gene expression from the query gene after a Z-score transformation of each probe set across the 79 different sample types. For example, the PDZ neighborhood index NR=250(G), for an individual gene G and a radius of 250, is defined as the number of ligands (known literature or potential in our case) in the top 250 ranking genes from each PDZ gene. We subsequently plot the frequency distribution of N250, for example. In order to test the statistical significance of co-expression ligands with PDZ domain-encoding genes, we randomly generated 1000 sets of random ligands equal in size to the gold standard “literature PDZ ligands” (n = 192) or the sum of conserved potential PDZ ligands of Class I, II, and III (i.e., the PDZCBM, n = 899) and calculated for each set the weighted frequency distribution of N(R = 100,250,500). The empirical P-value was defined by permutation testing as the fraction of weighted frequency distributions of random ligand sets that exceeded the observed frequency distribution for literature ligands, and similarly for the composite conserved potential PDZ ligands of Class I, II, and III (both P < 0.01 at radius R = 250 and R = 500). We allowed many to one mappings of probe sets to genes. We adopted this strategy since in 1000 random samplings of gene sets equivalent in size to the PDZCBM (n = 899), we observed that on average 33.6% of genes were represented by multiple probes (minimum 27.5% and maximum 42.0%), with 35.4% and 43.1% of the actual PDZCBM and PDZ, respectively, mapping to multiple probes. These data support our use of the many to one mapping strategy, since the background random gene sets used to calculate empirical P-values had similar numbers of multiple probe sets as compared with the PDZ or PDZCBM. In addition, if we chose one to one mapping, there would be an insufficient number of ligands and PDZ genes to perform the analyses described in this paper. To arrive at a reasonable threshold of correlation between any pair of PDZ and potential ligand that may suggest a functional or physical interaction, we performed permutation testing. The distributions of pair-wise Pearson’s correlation coefficients (PCCs) were computed comparing PDZ genes and known interactors with a random set of ligands. We generated 1000 random ligand gene sets equivalent in size to the PDZCBM genes (n = 899) and plotted the average of frequency distribution of co-efficient for the permutations in Figure 4B. We chose a significant pairwise correlation value for potential PDZ–ligand interactions based on the fact that <5% of the random distribution had a Pearson’s correlation value <ρ = 0.40 in this data set. The matrix of pairwise correlations between PDZ and PDZCBM genes for a single probe per gene was hierarchically clustered and visualized in MATLAB (see Supplemental Table 6 for all correlation values).
Hierarchical clustering
To represent the expression profiles of PDZ genes and the PDZCBM Class I, II, and III ligands, hierarchical clustering with the centroid linkage method was performed using DCHIP (Schadt et al. 2001), using 1 − r as the distance metric, where r is the Pearson correlation coefficient, and the relative expression levels are displayed. For the identified 136 PDZ domain-encoding genes, there were 195 probe sets representing 107 out of the 136 PDZ genes on either the U133a or custom GNF1H arrays, which met our own (described above) as well as dCHIP’s default filtering criteria. For the PDZCBM genes, 86%(433/505), 84% (139/165), and 89% (204/229) of Class I, Class II, and Class III genes, respectively, were represented by at least one probe on the combined U133a and custom GNF1H microarrays. Heatmaps of hierarchical clustering of tissues expression and correlation values are based on a single probe set per gene chosen at random so as to not bias the visual presentation. The probe IDs for all PDZ and PDZCBM genes are included in Supplemental Table 6. We also performed hierarchical clustering using dCHIP on the co-citation expression vectors for PDZ and PDZCBM described above.
Immune enrichment analysis
In order to ascertain if any individual PDZ gene or PDZCBM gene set member is globally enriched in tissues of the immune system, we used the Wilcoxon rank sum test. To calculate the Wilcoxon statistic, we divided the 79 tissue/cell types into those of immune system origin (n = 22) and those that are not part of the immune system (57), making two sample classes. The list of tissues/cells considered of immune system origin can be found in Supplemental Table 1. The P-values from the Wilcoxon test for each probe representing a PDZ or PDZCBM gene were then calculated. Significant thresholds after multiple hypothesis testing were established by dividing the P-value 0.05 by the number of PDZ probes tested.
Identification of D. melanogaster orthologs and phenotypic characterization
We employed a reciprocal best-hit strategy using an E-cutoff value of 10−10 to identify potential Drosophila melanogaster (http://flybase.bio.indiana.edu/blast/) orthologs for each of the 136 human PDZ and 899 PDZCBM-encoding genes based on the reference protein sequences of the human genes. For the identified ortholog, each FlyBase record was reviewed individually for the gene being a known fly mutant, GO annotation, links to primary literature, and whether it had been tested in an RNAi screen for cell morphology (Kiger et al. 2003). We noted which of the orthologs has been linked to processes that PDZ complexes participate in, defined as actin cytoskeleton rearrangements, polarity (manifested by abnormal bristle morphology, for instance), adhesion (related phenotypes include wound closure), synaptic growth and transmission, and ion channel function (known or predicted by homology with known channels). Supplemental Table 4 contains detailed phenotypic information on fly orthologs.
Immunofluorescence and BRET assays
The bioluminescence resonance energy transfer (BRET) was performed as described previously (Lopez-Ilasaca et al. 2005). 5 × 105 U2OS cells were plated on coverslips. After 24 h, cells were transiently transfected with Transfectin (Biorad) following the manufacturer’s recommendations. Twenty-four hours post-transfection, coverslips were washed in PBS, then fixed and permeabilized in 3.5% PFA/0.1% Tween for 10 min at RT. Cells were incubated with primary antibody (anti-HA [Covance]; anti-FLAG [Sigma, MO]) for 1 h at RT. Coverslips were washed in PBS and incubated with secondary antibody (AlexaFluor 568 anti-mouse IgG [Molecular Probes]; AlexaFluor 350 conjugated-phalloidin [Molecular Probes]) for 1 h at RT before being mounted with Aqua Poly/Mount (Polysciences). Slides were visualized on an Olympus AX70 microscope for BCR and DLG1 and a confocal microscope. For CARD11/SH2D3C immunofluorescence, 7.5 × 106 Jurkat cells were electroporated with 15 μg of GFP- SH2D3C wild-type or GFP vector DNA. Twenty hours post-electroporation, dead cells were removed by centrifugation with Ficoll Paque. Cells were washed twice in media, and allowed to rest for 1 h at 37°C in serum-containing media. After 1 h, cells were resuspended in serum-containing media and loaded on poly-L-lysine (PLL)-coated slides previously incubated with 10 μg/mL each of CD3 and CD28 antibody for 2 h at 37°C. After the indicated time points, the cell suspension was removed, and slides were fixed and permeabilized in 3.5% PFA/0.1% Tween for 10 min at RT. Slides were washed in PBS and blocked for 30 min in 1% BSA/PBS before incubation with primary polyclonal antibody CARD11 Ab (Apotech ALX-210–903). Coverslips were washed in PBS and incubated with secondary antibody AlexaFluor 568 anti-rabbit IgG (Molecular Probes) and AlexaFluor 350 conjugated-phalloidin (Molecular Probes) diluted in blocking solution for 1 h at RT, before being mounted, sealed, and visualized as above.
Plasmids and yeast two-hybrid screen
An IGSF4C plasmid was generously provided by Dr. Y. Murakami (Fukuhara et al. 2001). The carboxy-terminal mutation IGSF4C-AAA was made by PCR-mediated mutagenesis by replacing the most carboxy-terminal three residues with alanine (A) residues. The Myc-CNKSR1 constructs were made by PCR of subregions of the 729-amino-acid wild-type-encoding gene corresponding to the following amino acids: SAM 1–210; CRIC 210–279; PDZ 279–363; PRO/PH-CT 363–279. The yeast two-hybrid screen used full-length CNKSR1 as bait employing a brain cDNA library. The IL-2 promoter luciferase construct was a kind gift of J.O. Liu (Sun et al. 1998). The BCR cDNA was a gift of Dr. J. Groffen (Children’s Hospital of Los Angeles, CA). A 3×HA-tagged BCR gene was cloned into the pKH3 vector at 5′ BamHI and 3′ EcoRI sites by releasing the BCR gene from a GST-tagged BCR gene in pLEF vector (Rudert et al. 1996) by using BamHI and EcoRI partial digestion. Mutant BCR (mutBCR) was constructed by replacing the last four residues LTKL with AAAA by PCR. A Flag-tagged SH2D3C gene was constructed by subcloning the coding sequencing of SH2D3C (Open Biosystems, accession no. BC032365) into the pCMV-FLAG5 vector at 5′ BamHI and 3′ SalI sites after being amplified by PCR. GFP-tagged SH2D3C was inserted into the pEGFP-C1 vector (Clontech) at 5′ BglII and 3′ SalI sites by releasing the full-length gene from the Flag-tagged SH2D3C construct by BamHI and SalI double digestion. GST-tagged SH2D3C was inserted into the modified pEBG-GST vector (pEBG-GST2) at 5′ BamHI to 3′ SalI sites by releasing the full-length gene from the Flag-tagged SH2D3C construct by BamHI and SalI double digestion. All GFP-tagged DLG1 constructs were from Dr. Craig Garner (Wu et al. 1998). All constructs were confirmed by DNA sequencing.
Cell culture and transient transfection
Human embryonic kidney 293T cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal calf serum, 100 units/mL penicillin G, and 100 μg/mL streptomycin sulfate at 37°C and 5% CO2. One day before transfection, 1.5 × 106 293T cells were plated in 2 mL of DMEM medium per well in a six-well plate. The cells were transiently transfected with the indicated amount of plasmids by using lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Twenty-four hours later, cells were rinsed in ice-cold PBS and lysed with rotation for 30 min at 4°C in modified RIPA buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% Triton-X-100, 0.1% SDS, 5 mM EDTA, 0.5% sodium deoxycholate, 10 mM sodium fluoride, 1 mM sodium vanadate, 0.4 mM PMSF, and protease cocktail [1 tablet for 10 mL buffer, Roche]), and insoluble materials were removed by centrifugation at 14,000 rpm for 15 min. Jurkat cells (1 107) were electroporated with 15 μg of GST vector or GST-SH2D3C. Twenty-four hours post-electroporation, cells were lysed in 1% Triton lysis buffer, and GST pull-downs were probed for endogenous CARD11. For immunoprecipitation, cell lysates were incubated with appropriate specific antibodies for 3 h at 4°C and subsequently mixed with antibody affinity gel (goat affinity-purified antibody to mouse IgG [ICN Pharmaceuticals]) for an additional 90 min at 4°C. The immunoprecipitates were washed three times with modified RIPA buffer. The immunoprecipitated proteins and total cell lysates were resolved by SDS-PAGE, transferred to immobilon-P transfer membranes (Millipore), and immounoblotted with the indicated antibodies. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (DakoCytomation California) were used as secondary reagents. Detection was performed by enhanced chemiluminescence with the Western Lightning Chemiluminescence Regent (PerkinElmer Life Sciences).
Acknowledgments
We thank Dan Podolsky, Joe Avruch, Vamsi Mootha, Brian Seed, and Frederick Alt for their support and helpful review of the manuscript. We thank Julio Bernabe-Ortiz for his work on the protein–protein interaction assays. We thank A.I. Su for his helpful discussions. C.G. is supported by a Crohn’s and Colitis Foundation Research Fellowship Grant. R.X. is supported by the Faculty Development Fund (GI unit) and the CCFA.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5285206
References
- Balda M.S., Matter K., Matter K. Transmembrane proteins of tight junctions. Semin. Cell Dev. Biol. 2000;11:281–289. doi: 10.1006/scdb.2000.0177. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Knoblich J.A., Knoblich J.A. Dare to be different: Asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A., Mechtler K., Knoblich J.A., Knoblich J.A. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- Beuming T., Skrabanek L., Niv M.Y., Mukherjee P., Weinstein H., Skrabanek L., Niv M.Y., Mukherjee P., Weinstein H., Niv M.Y., Mukherjee P., Weinstein H., Mukherjee P., Weinstein H., Weinstein H. PDZBase: A protein–protein interaction database for PDZ-domains. Bioinformatics. 2005;21:827–828. doi: 10.1093/bioinformatics/bti098. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: Links from the Drosophila neoplastic tumor suppressors. Genes & Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Black D.S., Marie-Cardine A., Schraven B., Bliska J.B., Marie-Cardine A., Schraven B., Bliska J.B., Schraven B., Bliska J.B., Bliska J.B. The Yersinia tyrosine phosphatase YopH targets a novel adhesion-regulated signalling complex in macrophages. Cell. Microbiol. 2000;2:401–414. doi: 10.1046/j.1462-5822.2000.00061.x. [DOI] [PubMed] [Google Scholar]
- Bork P., Jensen L.J., von Mering C., Ramani A.K., Lee I., Marcotte E.M., Jensen L.J., von Mering C., Ramani A.K., Lee I., Marcotte E.M., von Mering C., Ramani A.K., Lee I., Marcotte E.M., Ramani A.K., Lee I., Marcotte E.M., Lee I., Marcotte E.M., Marcotte E.M. Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Butz S., Okamoto M., Sudhof T.C., Okamoto M., Sudhof T.C., Sudhof T.C. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Gao W., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Egawa T., Albrecht B., Favier B., Sunshine M.J., Mirchandani K., O’Brien W., Thome M., Littman D.R., Albrecht B., Favier B., Sunshine M.J., Mirchandani K., O’Brien W., Thome M., Littman D.R., Favier B., Sunshine M.J., Mirchandani K., O’Brien W., Thome M., Littman D.R., Sunshine M.J., Mirchandani K., O’Brien W., Thome M., Littman D.R., Mirchandani K., O’Brien W., Thome M., Littman D.R., O’Brien W., Thome M., Littman D.R., Thome M., Littman D.R., Littman D.R. Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr. Biol. 2003;13:1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- Fuja T.J., Lin F., Osann K.E., Bryant P.J., Lin F., Osann K.E., Bryant P.J., Osann K.E., Bryant P.J., Bryant P.J. Somatic mutations and altered expression of the candidate tumor suppressors CSNK1 ε, DLG1, and EDD/hHYD in mammary ductal carcinoma. Cancer Res. 2004;64:942–951. doi: 10.1158/0008-5472.can-03-2100. [DOI] [PubMed] [Google Scholar]
- Fukuhara H., Kuramochi M., Nobukuni T., Fukami T., Saino M., Maruyama T., Nomura S., Sekiya T., Murakami Y., Kuramochi M., Nobukuni T., Fukami T., Saino M., Maruyama T., Nomura S., Sekiya T., Murakami Y., Nobukuni T., Fukami T., Saino M., Maruyama T., Nomura S., Sekiya T., Murakami Y., Fukami T., Saino M., Maruyama T., Nomura S., Sekiya T., Murakami Y., Saino M., Maruyama T., Nomura S., Sekiya T., Murakami Y., Maruyama T., Nomura S., Sekiya T., Murakami Y., Nomura S., Sekiya T., Murakami Y., Sekiya T., Murakami Y., Murakami Y. Isolation of the TSLL1 and TSLL2 genes, members of the tumor suppressor TSLC1 gene family encoding transmembrane proteins. Oncogene. 2001;20:5401–5407. doi: 10.1038/sj.onc.1204696. [DOI] [PubMed] [Google Scholar]
- Garcia E.P., Mehta S., Blair L.A., Wells D.G., Shang J., Fukushima T., Fallon J.R., Garner C.C., Marshall J., Mehta S., Blair L.A., Wells D.G., Shang J., Fukushima T., Fallon J.R., Garner C.C., Marshall J., Blair L.A., Wells D.G., Shang J., Fukushima T., Fallon J.R., Garner C.C., Marshall J., Wells D.G., Shang J., Fukushima T., Fallon J.R., Garner C.C., Marshall J., Shang J., Fukushima T., Fallon J.R., Garner C.C., Marshall J., Fukushima T., Fallon J.R., Garner C.C., Marshall J., Fallon J.R., Garner C.C., Marshall J., Garner C.C., Marshall J., Marshall J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron. 1998;21:727–739. doi: 10.1016/s0896-6273(00)80590-5. [DOI] [PubMed] [Google Scholar]
- Gaudet S., Branton D., Lue R.A., Branton D., Lue R.A., Lue R.A. Characterization of PDZ-binding kinase, a mitotic kinase. Proc. Natl. Acad. Sci. 2000;97:5167–5172. doi: 10.1073/pnas.090102397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Ooi C.E., Godwin B., Vitols E., Godwin B., Vitols E., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Hanada T., Lin L., Tibaldi E.V., Reinherz E.L., Chishti A.H., Lin L., Tibaldi E.V., Reinherz E.L., Chishti A.H., Tibaldi E.V., Reinherz E.L., Chishti A.H., Reinherz E.L., Chishti A.H., Chishti A.H. GAKIN, a novel kinesin-like protein associates with the human homologue of the Drosophila discs large tumor suppressor in T lymphocytes. J. Biol. Chem. 2000;275:28774–28784. doi: 10.1074/jbc.M000715200. [DOI] [PubMed] [Google Scholar]
- Hara H., Wada T., Bakal C., Kozieradzki I., Suzuki S., Suzuki N., Nghiem M., Griffiths E.K., Krawczyk C., Bauer B., Wada T., Bakal C., Kozieradzki I., Suzuki S., Suzuki N., Nghiem M., Griffiths E.K., Krawczyk C., Bauer B., Bakal C., Kozieradzki I., Suzuki S., Suzuki N., Nghiem M., Griffiths E.K., Krawczyk C., Bauer B., Kozieradzki I., Suzuki S., Suzuki N., Nghiem M., Griffiths E.K., Krawczyk C., Bauer B., Suzuki S., Suzuki N., Nghiem M., Griffiths E.K., Krawczyk C., Bauer B., Suzuki N., Nghiem M., Griffiths E.K., Krawczyk C., Bauer B., Nghiem M., Griffiths E.K., Krawczyk C., Bauer B., Griffiths E.K., Krawczyk C., Bauer B., Krawczyk C., Bauer B., Bauer B., et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- Hara H., Bakal C., Wada T., Bouchard D., Rottapel R., Saito T., Penninger J.M., Bakal C., Wada T., Bouchard D., Rottapel R., Saito T., Penninger J.M., Wada T., Bouchard D., Rottapel R., Saito T., Penninger J.M., Bouchard D., Rottapel R., Saito T., Penninger J.M., Rottapel R., Saito T., Penninger J.M., Saito T., Penninger J.M., Penninger J.M. The molecular adapter Carma1 controls entry of IκB kinase into the central immune synapse. J. Exp. Med. 2004;200:1167–1177. doi: 10.1084/jem.20032246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M., Chelly J., Carter N., Danek A., Crocker P., Monaco A.P., Chelly J., Carter N., Danek A., Crocker P., Monaco A.P., Carter N., Danek A., Crocker P., Monaco A.P., Danek A., Crocker P., Monaco A.P., Crocker P., Monaco A.P., Monaco A.P. Isolation of the gene for McLeod syndrome that encodes a novel membrane transport protein. Cell. 1994;77:869–880. doi: 10.1016/0092-8674(94)90136-8. [DOI] [PubMed] [Google Scholar]
- Hoover K.B., Bryant P.J., Bryant P.J. Drosophila Yurt is a new protein-4.1-like protein required for epithelial morphogenesis. Dev. Genes Evol. 2002;212:230–238. doi: 10.1007/s00427-002-0231-6. [DOI] [PubMed] [Google Scholar]
- Hosack D.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Sherman B.T., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Yoshida M., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S., Morita K., Kubota K., Saitou M., Tsukita S., Kubota K., Saitou M., Tsukita S., Saitou M., Tsukita S., Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Sakakibara M., Yamasaki S., Takeuchi A., Arase H., Miyazaki M., Nakajima N., Okada M., Saito T., Sakakibara M., Yamasaki S., Takeuchi A., Arase H., Miyazaki M., Nakajima N., Okada M., Saito T., Yamasaki S., Takeuchi A., Arase H., Miyazaki M., Nakajima N., Okada M., Saito T., Takeuchi A., Arase H., Miyazaki M., Nakajima N., Okada M., Saito T., Arase H., Miyazaki M., Nakajima N., Okada M., Saito T., Miyazaki M., Nakajima N., Okada M., Saito T., Nakajima N., Okada M., Saito T., Okada M., Saito T., Saito T. Cutting edge: Negative regulation of immune synapse formation by anchoring lipid raft to cytoskeleton through Cbp-EBP50-ERM assembly. J. Immunol. 2002;168:541–544. doi: 10.4049/jimmunol.168.2.541. [DOI] [PubMed] [Google Scholar]
- Jansen R., Greenbaum D., Gerstein M., Greenbaum D., Gerstein M., Gerstein M. Relating whole-genome expression data with protein–protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelen F., Oleksy A., Smietana K., Otlewski J., Oleksy A., Smietana K., Otlewski J., Smietana K., Otlewski J., Otlewski J. PDZ domains—Common players in the cell signaling. Acta Biochim. Pol. 2003;50:985–1017. [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Aravin A., Tuschl T., Sander C., Marks D.S., Tuschl T., Sander C., Marks D.S., Sander C., Marks D.S., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J.E., Wilson L.E., Vinuesa C.G., Lesage S., Blery M., Miosge L.A., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., Wilson L.E., Vinuesa C.G., Lesage S., Blery M., Miosge L.A., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., Vinuesa C.G., Lesage S., Blery M., Miosge L.A., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., Lesage S., Blery M., Miosge L.A., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., Blery M., Miosge L.A., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., Miosge L.A., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., Kucharska E.M., Hara H., Penninger J.M., Hara H., Penninger J.M., Penninger J.M., et al. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18:751–762. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- Kennedy M.B. Origin of PDZ (DHR, GLGF) domains. Trends Biochem. Sci. 1995;20:350. doi: 10.1016/s0968-0004(00)89074-x. [DOI] [PubMed] [Google Scholar]
- Kiger A.A., Baum B., Jones S., Jones M.R., Coulson A., Echeverri C., Perrimon N., Baum B., Jones S., Jones M.R., Coulson A., Echeverri C., Perrimon N., Jones S., Jones M.R., Coulson A., Echeverri C., Perrimon N., Jones M.R., Coulson A., Echeverri C., Perrimon N., Coulson A., Echeverri C., Perrimon N., Echeverri C., Perrimon N., Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Sheng M., Sheng M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Knetsch M.L., Schafers N., Horstmann H., Manstein D.J., Schafers N., Horstmann H., Manstein D.J., Horstmann H., Manstein D.J., Manstein D.J. The Dictyostelium Bcr/Abr-related protein DRG regulates both Rac- and Rab-dependent pathways. EMBO J. 2001;20:1620–1629. doi: 10.1093/emboj/20.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L., Larsson B., von Heijne G., Sonnhammer E.L.L., von Heijne G., Sonnhammer E.L.L., Sonnhammer E.L.L. Predicting transmembrane protein topology with a Hidden Markov Model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Labouesse M. Epithelium-mesenchyme: A balancing act of RhoGAP and RhoGEF. Curr. Biol. 2004;14:R508–R510. doi: 10.1016/j.cub.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Lee I., Date S.V., Adai A.T., Marcotte E.M., Date S.V., Adai A.T., Marcotte E.M., Adai A.T., Marcotte E.M., Marcotte E.M. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Vidalain P.O., Han J.D., Chesneau A., Hao T., Han J.D., Chesneau A., Hao T., Chesneau A., Hao T., Hao T., et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Kang H., Raab M., da Silva A.J., Kraeft S.K., Rudd C.E., Kang H., Raab M., da Silva A.J., Kraeft S.K., Rudd C.E., Raab M., da Silva A.J., Kraeft S.K., Rudd C.E., da Silva A.J., Kraeft S.K., Rudd C.E., Kraeft S.K., Rudd C.E., Rudd C.E. FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl. Acad. Sci. 1998;95:8779–8784. doi: 10.1073/pnas.95.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ilasaca M.A., Bernabe-Ortiz J.C., Na S.Y., Dzau V.J., Xavier R.J., Bernabe-Ortiz J.C., Na S.Y., Dzau V.J., Xavier R.J., Na S.Y., Dzau V.J., Xavier R.J., Dzau V.J., Xavier R.J., Xavier R.J. Bioluminescence resonance energy transfer identify scaffold protein CNK1 interactions in intact cells. FEBS Lett. 2005;579:648–654. doi: 10.1016/j.febslet.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Ludford-Menting M.J., Oliaro J., Sacirbegovic F., Cheah E.T., Pederson N., Thomas S.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., Oliaro J., Sacirbegovic F., Cheah E.T., Pederson N., Thomas S.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., Sacirbegovic F., Cheah E.T., Pederson N., Thomas S.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., Cheah E.T., Pederson N., Thomas S.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., Pederson N., Thomas S.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., Thomas S.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., Pasam A., Iazzolino R., Dow L.E., Waterhouse N.J., Iazzolino R., Dow L.E., Waterhouse N.J., Dow L.E., Waterhouse N.J., Waterhouse N.J., et al. A network of PDZ-containing proteins regulates T cell polarity and morphology during migration and immunological synapse formation. Immunity. 2005;22:655–656. doi: 10.1016/j.immuni.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Macara I.G. Parsing the polarity code. Nat. Rev. Mol. Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- Mancini A., Koch A., Stefan M., Niemann H., Tamura T., Koch A., Stefan M., Niemann H., Tamura T., Stefan M., Niemann H., Tamura T., Niemann H., Tamura T., Tamura T. The direct association of the multiple PDZ domain containing proteins (MUPP-1) with the human c-Kit C-terminus is regulated by tyrosine kinase activity. FEBS Lett. 2000;482:54–58. doi: 10.1016/s0014-5793(00)02036-6. [DOI] [PubMed] [Google Scholar]
- Marie-Cardine A., Verhagen A.M., Eckerskorn C., Schraven B., Verhagen A.M., Eckerskorn C., Schraven B., Eckerskorn C., Schraven B., Schraven B. SKAP-HOM, a novel adaptor protein homologous to the FYN-associated protein SKAP55. FEBS Lett. 1998;435:55–60. doi: 10.1016/s0014-5793(98)01040-0. [DOI] [PubMed] [Google Scholar]
- Massimi P., Gardiol D., Roberts S., Banks L., Gardiol D., Roberts S., Banks L., Roberts S., Banks L., Banks L. Redistribution of the discs large tumor suppressor protein during mitosis. Exp. Cell Res. 2003;290:265–274. doi: 10.1016/s0014-4827(03)00317-3. [DOI] [PubMed] [Google Scholar]
- Matsumine A., Ogai A., Senda T., Okumura N., Satoh K., Baeg G.H., Kawahara T., Kobayashi S., Okada M., Toyoshima K., Ogai A., Senda T., Okumura N., Satoh K., Baeg G.H., Kawahara T., Kobayashi S., Okada M., Toyoshima K., Senda T., Okumura N., Satoh K., Baeg G.H., Kawahara T., Kobayashi S., Okada M., Toyoshima K., Okumura N., Satoh K., Baeg G.H., Kawahara T., Kobayashi S., Okada M., Toyoshima K., Satoh K., Baeg G.H., Kawahara T., Kobayashi S., Okada M., Toyoshima K., Baeg G.H., Kawahara T., Kobayashi S., Okada M., Toyoshima K., Kawahara T., Kobayashi S., Okada M., Toyoshima K., Kobayashi S., Okada M., Toyoshima K., Okada M., Toyoshima K., Toyoshima K., et al. Binding of APC to the human homolog of the Drosophila discs large tumor suppressor protein. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- Matsumoto R., Wang D., Blonska M., Li H., Kobayashi M., Pappu B., Chen Y., Lin X., Wang D., Blonska M., Li H., Kobayashi M., Pappu B., Chen Y., Lin X., Blonska M., Li H., Kobayashi M., Pappu B., Chen Y., Lin X., Li H., Kobayashi M., Pappu B., Chen Y., Lin X., Kobayashi M., Pappu B., Chen Y., Lin X., Pappu B., Chen Y., Lin X., Chen Y., Lin X., Lin X. Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-κB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Menon S.D., Chia W., Chia W. Drosophila rolling pebbles: A multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev. Cell. 2001;1:691–703. doi: 10.1016/s1534-5807(01)00075-2. [DOI] [PubMed] [Google Scholar]
- Mok H., Shin H., Kim S., Lee J.R., Yoon J., Kim E., Shin H., Kim S., Lee J.R., Yoon J., Kim E., Kim S., Lee J.R., Yoon J., Kim E., Lee J.R., Yoon J., Kim E., Yoon J., Kim E., Kim E. Association of the kinesin superfamily motor protein KIF1Bα with postsynaptic density-95 (PSD-95), synapse-associated protein-97, and synaptic scaffolding molecule PSD-95/discs large/zona occludens-1 proteins. J. Neurosci. 2002;22:5253–5258. doi: 10.1523/JNEUROSCI.22-13-05253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J.M., Zamorano P.L., Garner C.C., Zamorano P.L., Garner C.C., Garner C.C. MAGUKs in synapse assembly and function: An emerging view. Cell. Mol. Life Sci. 2004;61:911–929. doi: 10.1007/s00018-003-3364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Delmonte T., Villeneuve A., Sladek R., Xu F., Villeneuve A., Sladek R., Xu F., Sladek R., Xu F., Xu F., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K., Dixit V.M., Dixit V.M. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr. Biol. 2003;13:1247–1251. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- Nourry C., Grant S.G., Borg J.P., Grant S.G., Borg J.P., Borg J.P. PDZ domain proteins: Plug and play! Sci. STKE. 2003;2003 doi: 10.1126/stke.2003.179.re7. RE7. [DOI] [PubMed] [Google Scholar]
- Owen A.B., Stuart J., Mach K., Villeneuve A.M., Kim S., Stuart J., Mach K., Villeneuve A.M., Kim S., Mach K., Villeneuve A.M., Kim S., Villeneuve A.M., Kim S., Kim S. A gene recommender algorithm to identify coexpressed genes in C. elegans. Genome Res. 2003;13:1828–1837. doi: 10.1101/gr.1125403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkert R.R., DiVittorio H.M., Prehoda K.E., DiVittorio H.M., Prehoda K.E., Prehoda K.E. Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nat. Struct. Mol. Biol. 2004;11:1122–1127. doi: 10.1038/nsmb839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N. The maternal effect of lethal(1)discs-large-1: A recessive oncogene of Drosophila melanogaster. Dev. Biol. 1988;127:392–407. doi: 10.1016/0012-1606(88)90326-0. [DOI] [PubMed] [Google Scholar]
- Peterson F.C., Penkert R.R., Volkman B.F., Prehoda K.E., Penkert R.R., Volkman B.F., Prehoda K.E., Volkman B.F., Prehoda K.E., Prehoda K.E. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol. Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- Rahmouni S., Vang T., Alonso A., Williams S., van Stipdonk M., Soncini C., Moutschen M., Schoenberger S.P., Mustelin T., Vang T., Alonso A., Williams S., van Stipdonk M., Soncini C., Moutschen M., Schoenberger S.P., Mustelin T., Alonso A., Williams S., van Stipdonk M., Soncini C., Moutschen M., Schoenberger S.P., Mustelin T., Williams S., van Stipdonk M., Soncini C., Moutschen M., Schoenberger S.P., Mustelin T., van Stipdonk M., Soncini C., Moutschen M., Schoenberger S.P., Mustelin T., Soncini C., Moutschen M., Schoenberger S.P., Mustelin T., Moutschen M., Schoenberger S.P., Mustelin T., Schoenberger S.P., Mustelin T., Mustelin T. Removal of C-terminal SRC kinase from the immune synapse by a new binding protein. Mol. Cell. Biol. 2005;25:2227–2241. doi: 10.1128/MCB.25.6.2227-2241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A.K., Bunescu R.C., Mooney R.J., Marcotte E.M., Bunescu R.C., Mooney R.J., Marcotte E.M., Mooney R.J., Marcotte E.M., Marcotte E.M. Consolidating the set of known human protein–protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau A., Buttgereit D., Holz A., Fetter R., Doberstein S.K., Paululat A., Staudt N., Skeath J., Michelson A.M., Renkawitz-Pohl R., Buttgereit D., Holz A., Fetter R., Doberstein S.K., Paululat A., Staudt N., Skeath J., Michelson A.M., Renkawitz-Pohl R., Holz A., Fetter R., Doberstein S.K., Paululat A., Staudt N., Skeath J., Michelson A.M., Renkawitz-Pohl R., Fetter R., Doberstein S.K., Paululat A., Staudt N., Skeath J., Michelson A.M., Renkawitz-Pohl R., Doberstein S.K., Paululat A., Staudt N., Skeath J., Michelson A.M., Renkawitz-Pohl R., Paululat A., Staudt N., Skeath J., Michelson A.M., Renkawitz-Pohl R., Staudt N., Skeath J., Michelson A.M., Renkawitz-Pohl R., Skeath J., Michelson A.M., Renkawitz-Pohl R., Michelson A.M., Renkawitz-Pohl R., Renkawitz-Pohl R. rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development. 2001;128:5061–5073. doi: 10.1242/dev.128.24.5061. [DOI] [PubMed] [Google Scholar]
- Reczek D., Bretscher A., Bretscher A. Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J. Cell Biol. 2001;153:191–206. doi: 10.1083/jcb.153.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein R., Simon I., Simon I. MILANO–Custom annotation of microarray results using automatic literature searches. BMC Bioinformatics. 2005;6:12. doi: 10.1186/1471-2105-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudert F., Visser E., Gradl G., Grandison P., Shemshedini L., Wang Y., Grierson A., Watson J., Visser E., Gradl G., Grandison P., Shemshedini L., Wang Y., Grierson A., Watson J., Gradl G., Grandison P., Shemshedini L., Wang Y., Grierson A., Watson J., Grandison P., Shemshedini L., Wang Y., Grierson A., Watson J., Shemshedini L., Wang Y., Grierson A., Watson J., Wang Y., Grierson A., Watson J., Grierson A., Watson J., Watson J. pLEF, a novel vector for expression of glutathione S-transferase fusion proteins in mammalian cells. Gene. 1996;169:281–282. doi: 10.1016/0378-1119(95)00820-9. [DOI] [PubMed] [Google Scholar]
- Sakakibara A., Hattori S., Nakamura S., Katagiri T., Hattori S., Nakamura S., Katagiri T., Nakamura S., Katagiri T., Katagiri T. A novel hematopoietic adaptor protein, Chat-H, positively regulates T cell receptor-mediated interleukin-2 production by Jurkat cells. J. Biol. Chem. 2003;278:6012–6017. doi: 10.1074/jbc.M207942200. [DOI] [PubMed] [Google Scholar]
- Sakisaka T., Takai Y., Takai Y. Biology and pathology of nectins and nectin-like molecules. Curr. Opin. Cell Biol. 2004;16:513–521. doi: 10.1016/j.ceb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Schadt E.E., Li C., Ellis B., Wong W.H., Li C., Ellis B., Wong W.H., Ellis B., Wong W.H., Wong W.H. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell. Biochem. Suppl. 2001;37:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- Sheng M., Sala C., Sala C. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Skop A.R., Liu H., Yates J., III, Meyer B.J., Heald R., Liu H., Yates J., III, Meyer B.J., Heald R., Yates J., III, Meyer B.J., Heald R., Meyer B.J., Heald R., Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z., Fanning A.S., Fu C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C., Fanning A.S., Fu C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C., Fu C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., Cantley L.C., Crompton A., Chan A.C., Anderson J.M., Cantley L.C., Chan A.C., Anderson J.M., Cantley L.C., Anderson J.M., Cantley L.C., Cantley L.C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Stanfield G.M., Horvitz H.R., Horvitz H.R. The ced-8 gene controls the timing of programmed cell deaths in C. elegans. Mol. Cell. 2000;5:423–433. doi: 10.1016/s1097-2765(00)80437-2. [DOI] [PubMed] [Google Scholar]
- Stanyon C.A., Liu G., Mangiola B.A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Liu G., Mangiola B.A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Mangiola B.A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Zhang H., Zhong J., Finley R.L., Jr., Zhong J., Finley R.L., Jr., Finley R.L., Jr. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P., Edkins S., Davies H., Greenman C., Cox C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., Edkins S., Davies H., Greenman C., Cox C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., Davies H., Greenman C., Cox C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., Greenman C., Cox C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., Cox C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., Hunter C., Bignell G., Teague J., Smith R., Stevens C., Bignell G., Teague J., Smith R., Stevens C., Teague J., Smith R., Stevens C., Smith R., Stevens C., Stevens C., et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat. Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugie K., Jeon M.S., Grey H.M., Jeon M.S., Grey H.M., Grey H.M. Activation of naive CD4 T cells by anti-CD3 reveals an important role for Fyn in Lck-mediated signaling. Proc. Natl. Acad. Sci. 2004;101:14859–14864. doi: 10.1073/pnas.0406168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Youn H.D., Loh C., Stolow M., He W., Liu J.O., Youn H.D., Loh C., Stolow M., He W., Liu J.O., Loh C., Stolow M., He W., Liu J.O., Stolow M., He W., Liu J.O., He W., Liu J.O., Liu J.O. Cabin 1 a negative regulator of Calcineurin signaling in T lymphocytes. Immunity. 1998;8:703–711. doi: 10.1016/s1074-7613(00)80575-0. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O.G., Dolinski K., Owen A.B., Altman R.B., Botstein D., Dolinski K., Owen A.B., Altman R.B., Botstein D., Owen A.B., Altman R.B., Botstein D., Altman R.B., Botstein D., Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae). Proc. Natl. Acad. Sci. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Lockshon D., Narayan V., Srinivasan M., Pochart P., Narayan V., Srinivasan M., Pochart P., Srinivasan M., Pochart P., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Ham M., Hendriks W., Hendriks W. PDZ domains—Glue and guide. Mol. Biol. Rep. 2003;30:69–82. doi: 10.1023/a:1023941703493. [DOI] [PubMed] [Google Scholar]
- Wang G.S., Hong C.J., Yen T.Y., Huang H.Y., Ou Y., Huang T.N., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Hong C.J., Yen T.Y., Huang H.Y., Ou Y., Huang T.N., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Yen T.Y., Huang H.Y., Ou Y., Huang T.N., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Huang H.Y., Ou Y., Huang T.N., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Ou Y., Huang T.N., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Huang T.N., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Jung W.G., Kuo T.Y., Sheng M., Wang T.F., Kuo T.Y., Sheng M., Wang T.F., Sheng M., Wang T.F., Wang T.F., et al. Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron. 2004;42:113–128. doi: 10.1016/s0896-6273(04)00139-4. [DOI] [PubMed] [Google Scholar]
- Wolff T., Rubin G.M., Rubin G.M. Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development. 1998;125:1149–1159. doi: 10.1242/dev.125.6.1149. [DOI] [PubMed] [Google Scholar]
- Woods D.F., Bryant P.J., Bryant P.J. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- Wu H., Reuver S.M., Kuhlendahl S., Chung W.J., Garner C.C., Reuver S.M., Kuhlendahl S., Chung W.J., Garner C.C., Kuhlendahl S., Chung W.J., Garner C.C., Chung W.J., Garner C.C., Garner C.C. Subcellular targeting and cytoskeletal attachment of SAP97 to the epithelial lateral membrane. J. Cell Sci. 1998;111:2365–2376. doi: 10.1242/jcs.111.16.2365. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Leitenberg D., Li B., Flavell R.A., Leitenberg D., Li B., Flavell R.A., Li B., Flavell R.A., Flavell R.A. Deficiency of small GTPase Rac2 affects T cell activation. J. Exp. Med. 2001;194:915–926. doi: 10.1084/jem.194.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]