Abstract
Autosomal recessive diseases are those that require mutations in both alleles to exhibit the disorder. Although most recessive conditions are rare, heterozygous carriers of recessive mutations are quite common. In this study, we show that carriers of Nijmegen Breakage Syndrome (NBS) have a distinct gene expression phenotype that differs from that of noncarriers and also from that of carriers of a similar syndrome, Ataxia Telangiectasia (AT). We found 520 genes whose expression levels differ significantly (P ≤ 0.001) between NBS carriers and controls. By linear discriminant analysis, we found a combination of 16 genes that allows 100% correct classification of individuals as either NBS carriers or noncarriers in a training set with 25 individuals, and in a test set with 52 individuals. When applied to AT carriers, the discriminant function misclassified only one out of 18 AT carriers as an NBS carrier. Our result shows that NBS carriers have a specific gene expression phenotype. It suggests that heterozygous mutations can contribute significantly to natural variation in gene expression. This has implications for the role that heterozygosity for recessive diseases plays in the overall genetic architecture of complex human traits and diseases.
Nijmegen Breakage Syndrome (NBS; MIM 251260) is an autosomal recessive disease caused by mutations in the NBS gene (nibrin) on chromosome 8q21 (Saar et al. 1997; Carney et al. 1998; Varon et al. 1998). It is characterized by microcephaly, growth retardation, immunodeficiency, and predisposition to cancer. NBS belongs to a group of chromosomal instability disorders that also includes Ataxia Telangiectasia (AT), Bloom’s syndrome, and Fanconi’s anemia. The cellular hallmarks of these disorders are chromosomal rearrangements, sensitivity to ionizing radiation, and defects in cell cycle checkpoint. Nibrin is a component of the hMre/hRad50 complex that plays a key role in cellular DNA damage response (Carney et al. 1998).
NBS is a rare disease. However, the frequency of heterozygous carriers is estimated to be as high as 1/150 in the Slavic population (Varon et al. 2000; Drabek et al. 2002). A founder mutation, 657del5, was identified in many patients of Slavic descent; nevertheless, there are patients with other mutations in the nibrin gene (Varon et al. 1998). Studies have shown that although NBS is an autosomal recessive disorder, heterozygous carriers have an increased risk of cancer and sensitivity to radiation (Dumon-Jones et al. 2003; Resnick et al. 2003; Cybulski et al. 2004). While polymerase chain reaction with sequence-specific primers can reliably detect carriers of the common 657del5 mutations (Drabek et al. 2002), it is not practical for identifying carriers of other nibrin mutations. Studies have attempted to classify carriers of NBS by radiosensitivity assays, but it was found that the assays lack sensitivity (Tanzanella et al. 2003). Previously, we showed that AT carriers have a phenotype at the gene expression level that differs from that of control individuals (Watts et al. 2002). In this study, we extend this analysis and show that NBS carriers also have a specific gene expression phenotype that differs from that of normal controls and AT carriers.
For many years, geneticists have been interested in phenotypic characteristics of carriers of autosomal recessive diseases (Neel 1947). It has been estimated that most individuals are heterozygous carriers for three or four very deleterious genes (Morton et al. 1956). Even though most recessive disorders are rare, carriers are not. Therefore, even seemingly minor phenotypic characteristics of carriers could be responsible for a substantial part of human biological variation.
Results
Microarray analysis of cells from NBS carriers and normal controls
First, we obtained gene expression profiles of lymphoblastoid cells from nine obligate NBS carriers and 16 normal controls using the Affymetrix Genome Focus arrays that contain ∼8500 genes. From these, we analyzed 3928 genes that are expressed in lymphoblastoid cells. Using the t-statistic, we identified genes whose expression levels differ significantly between controls and NBS carriers. Among the 3928 genes examined, we found 218 that differ at nominal P ≤ 0.0001. At a less stringent threshold of P ≤ 0.001, there are 520 genes that differ significantly.
To correct for the 3928 tests performed and the unknown correlation among the tests, we used a permutation analysis to determine the number of significant genes one would expect by chance alone. In the permutation data, the average number of significant genes (P ≤ 0.001) was 3.9, as expected (SD = 1.95). The number of significantly different genes (520) observed is far greater (P ≪ 0.0001) than would be expected by chance alone, suggesting that NBS carriers have a different gene expression profile compared to controls.
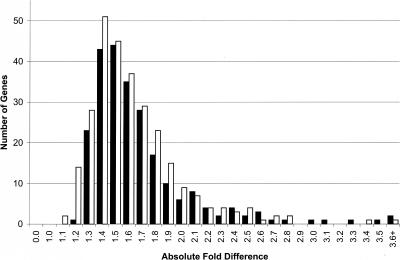
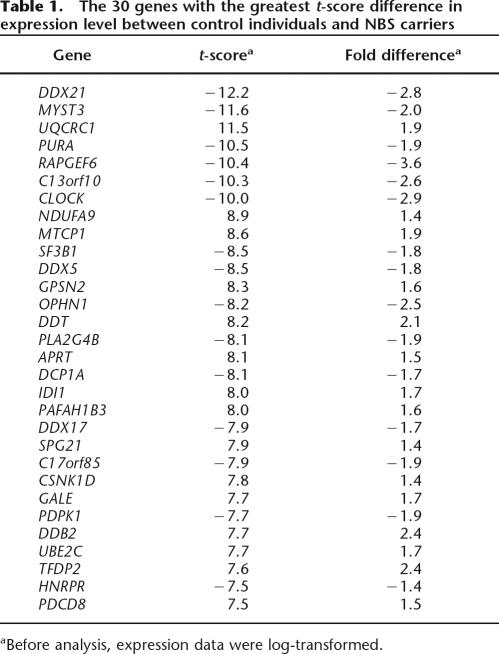
Among these 520 genes, there are 282 genes that are expressed at higher level in NBS carriers than in controls, and 238 genes that are expressed at lower level in NBS carriers than in controls. The distribution of fold differences for the 520 genes is shown in Figure 1. Sixty-nine genes have greater than twofold difference between controls and NBS carriers. The expression level of nibrin, the mutated gene in NBS, was not significantly different between the NBS carriers and controls. Since our goal is to identify genes that are significantly different between the two groups, we ranked the genes by t-scores. Table 1 lists the genes with the highest t-scores and their average fold difference in expression level between controls and NBS carriers.
Figure 1.
Distribution of fold differences for 520 differentially expressed genes (P ≤ 0.001) between normal controls and NBS carriers. Genes whose expression levels are higher among NBS carriers are shown by the white bars, and those whose expression levels are lower among NBS carriers compared to normal controls are shown in black.
Table 1.
The 30 genes with the greatest t-score difference in expression level between control individuals and NBS carriers
aBefore analysis, expression data were log-transformed.
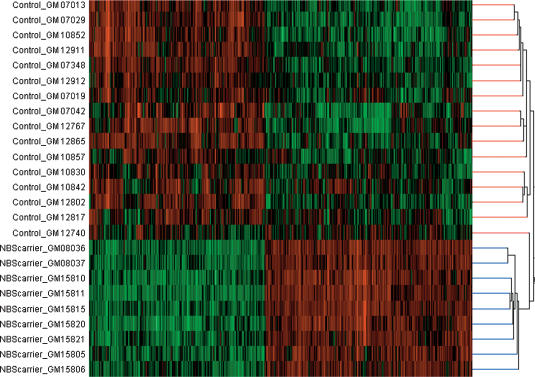
Next, we performed cluster analysis to determine how the 25 subjects (16 controls and nine NBS carriers) can be grouped by the expression levels of the 520 differentially expressed genes (Fig. 2). All but one of the controls was clustered in one branch of the dendrogram, and the NBS carriers were grouped in the other major branch. The cluster analysis, therefore, supports the above findings that based on these 520 genes, the normal controls are more similar to each other than to the NBS carriers, and likewise, the NBS carriers are more similar to each other than to the control individuals.
Figure 2.
Dendrogram showing results of hierarchical cluster analysis to classify nine NBS carriers and 16 normal controls.
Functional classification of differentially expressed genes
We explored the function of the differentially expressed genes by using Gene Ontology annotations (Ashburner et al. 2000; Hosack et al. 2003). Among the 520 differentially expressed genes, there is significant overrepresentation of genes that belong to the categories of RNA/nucleic acid binding and pre-RNA splicing factor activity (P ≤ 0.0001). Differentially expressed genes that belong to RNA/nucleic acid binding include XBP1, EIF4A2, USF2, RFX5, and XRCC1. Those with pre-RNA splicing factor activity include SFRS9, SFPQ, SNRPB2, LSM2, and CUGBP1.
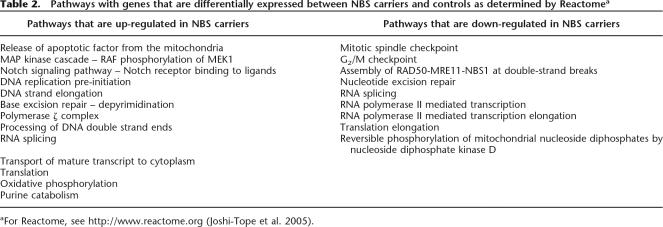
For a more comprehensive analysis of biological pathways that differ between NBS carriers and controls, we combined expression data from the microarray analysis with information in Reactome, a knowledge-base of biological pathways (Joshi-Tope et al. 2005). The results revealed more than 20 different biological pathways that differ in NBS carriers, including nine that are down-regulated and 13 that are up-regulated compared to noncarriers (Table 2; Supplemental Fig. 1).
Table 2.
Pathways with genes that are differentially expressed between NBS carriers and controls as determined by Reactomea
aFor Reactome, see http://www.reactome.org (Joshi-Tope et al. 2005).
Nibrin is known to play a role in DNA repair. Thus it is not surprising that among the down-regulated pathways is homologous recombination repair, in which NBS forms a complex with RAD50 and MRE11 at double-strand breaks. In addition, two other repair pathways, the MAD1- and MAD2-mediated mitotic spindle checkpoint, and the XPG- and ERCC1-dependent nucleotide excision repair, are also down-regulated. These findings suggest that although NBS is a recessive disorder, carriers have detectable differences in their DNA repair pathways compared to noncarriers. Besides DNA damage repair, other pathways such as transcription, translation elongation, and nucleotide metabolism are also down-regulated.
While there are down-regulated pathways, we also found ones that are up-regulated in NBS carriers. These include signaling pathways such as MAP-kinase cascade and NOTCH signaling. For example, the NOTCH ligand JAG2 is significantly up-regulated. Base excision repair and polymerase ζ complex-mediated DNA damage bypass are also up-regulated. Polymerase ζ allows DNA replication in the presence of DNA lesions. To cope with DNA damage, cells either stop replication in order to repair the damage or bypass the lesion. In the heterozygous NBS cells, polymerase ζ is overexpressed to bypass the DNA lesions. Such translesion synthesis allows damaged DNA to serve as a template, thus promoting the accumulation of mutations. In addition, components of several general housekeeping pathways such as DNA replication, RNA polymerase II- and III-mediated transcription, and translation are also up-regulated in NBS carriers.
Discriminant analysis: NBS carriers and normal controls
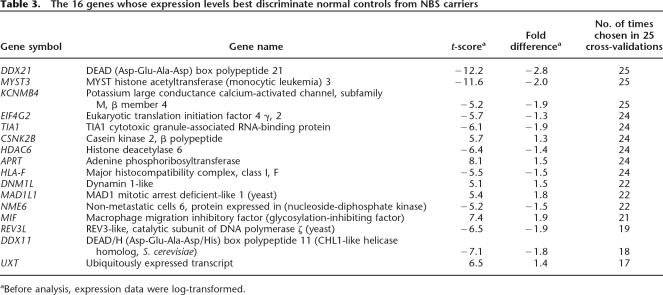
Identifying the genes that differ between NBS carriers and controls will allow a better understanding of the biological differences between these individuals. For this purpose, an extensive list is needed since it is not clear which genes contribute most to biologically relevant differences. However, for classification purposes, we want to have the smallest set of genes that when combined provide a highly accurate method of distinguishing between the two groups of individuals. To find this set of genes, we performed linear discriminant analysis with the 218 genes that nominally differ at P ≤ 0.0001 between the two groups. By stepwise discriminant analysis, the best linear discriminant function was formed with 16 genes. Those genes and their average fold differences between NBS carriers and controls are shown in Table 3. The average discriminant score for the 16 controls is −109 and for the nine carriers is +194, and the distributions of scores for carriers and controls do not overlap.
Table 3.
The 16 genes whose expression levels best discriminate normal controls from NBS carriers
aBefore analysis, expression data were log-transformed.
Three of the 16 genes (DDX21, MYST3, and APRT) are among the 30 genes with highest t-statistic (Tables 1 and 3). The remaining 27 genes with high t-scores were not selected in the discriminant analysis, presumably because their expression levels are highly correlated with one or several of the genes that formed the discriminant functions. For example, the correlation coefficients between UQCRC1 and DDX21 and between PURA and DDX21 are 0.45 and 0.65, respectively. The inclusion of correlated genes would add little to the ability to predict whether an individual is a NBS carrier or not.
Validation of discriminant genes
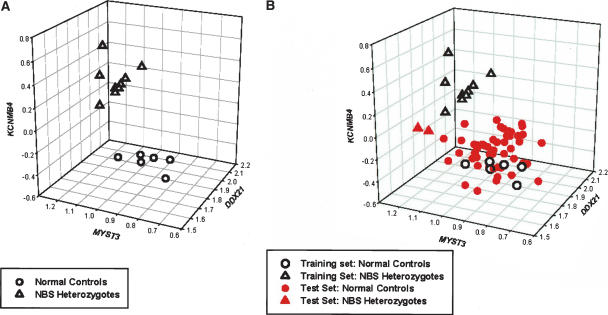
To determine how well we could classify individuals who are not part of the “training set,” we performed cross-validation and also tested a new set of individuals. In the cross-validation, the individual to be classified was left out of the selection of genes, and the discriminant function was calculated on the basis of the remaining 24 individuals. All 25 individuals were correctly classified under this more stringent criterion. In the 25 cross-validation analyses, 17 resulted in exactly the same 16 genes as classifiers. Three genes—DDX21, MYST3, and KCNMB4—were selected as classifiers in all 25 analyses. The number of analyses in which each gene was chosen as classifier is shown in Table 3. Expression levels of DDX21, MYST3, and KCNMB4 for the NBS carriers and controls are shown in Figure 3A. Even based on expression levels of just these three genes, NBS carriers and controls are well classified. Thus, it is not too surprising that there is a large difference in average discriminant scores between controls (−109) and NBS carriers (194) when all 16 genes were used as in the discriminant function.
Figure 3.
Expression level of three genes (DDX21, MYST3, KCNMB4) for (A) 16 normal controls and nine NBS carriers, and (B) same individuals as in A and 50 additional normal controls and two additional NBS carriers. Some data points overlap each other because some of the individuals have very similar expression levels of these genes.
Next, to examine how well this scheme will work when applied to an independent sample, we considered the preceding 25 samples as a training set and used the discriminant function formed by them to classify two other NBS carriers and 50 additional unrelated individuals who are part of the Center d’Etude du Polymorphisme Humain (CEPH) collections. All 50 CEPH individuals were correctly classified as normal individuals, and the two NBS carriers were classified as NBS carriers. The frequency of NBS carriers is 0.7% in the Slavic population and likely to be lower in other populations; so it is very possible that none of the 50 CEPH individuals tested are NBS carriers. In Figure 3B, expression levels for DDX21, MYST3, and KCNMB4 for the new samples are shown along with those for individuals in the training set. Similar to the samples in the training set, the additional NBS carriers and normal controls have different expression levels of DDX21, MYST3, and KCNMB4.
One of the characteristics of NBS is radiosensitivity, thus we compared results from the discriminant analysis with those for another radiosensitivity syndrome, Ataxia Telangiectasia (AT). At the cellular level, AT and NBS are quite similar (van der Burgt et al. 1996). To determine if the discriminant function can be applied to distinguish AT carriers from NBS carriers, we used it to classify 18 AT carriers. We applied the linear function with the 16 predictor genes to classify the 18 AT carriers as controls or NBS carriers. Among them, 17 were classified with the control group and one was classified with the NBS carriers. From this result, it appears that the discriminant function determined in our study is quite specific for NBS carriers. It allows highly accurate discrimination of NBS carriers from controls, and yet in almost all cases it does not assign AT carriers as NBS carriers.
Discussion
Diseases that are transmitted in recessive fashion are by definition those that have no marked heterozygous phenotypes. However, studies from the early days of human genetics had already suggested that there are subtle manifestations in carriers that are different from the norm (Neel 1947; Hsia 1957, 1966). For many recessive mutations, heterozygous carriers are quite common. Therefore, they could contribute significantly to phenotypic diversity, if they have unique phenotypes. In order to quantify this contribution, an accurate method to identify and phenotype carriers is needed. Efforts to phenotype carriers have included physical examination and biochemical assays; however, those findings have been quite inconclusive. Recent development in genomic technologies has extended the tools for phenotyping to the measurement of expression levels of genes. Such studies have identified specific gene expression signatures for various tumors that result from somatic mutations (DeRisi et al. 1996; Golub et al. 1999) and subtle changes that result from germline mutations such as heterozygous mutations in the ATM gene (Watts et al. 2002). Here, we show that carriers of NBS have a gene expression phenotype that differs from that of noncarriers and of carriers of AT. We found 520 genes whose expression levels differ significantly (P ≤ 0.001) between NBS carriers and controls. We show that baseline expression levels of 16 genes allowed discrimination of NBS carriers from noncarriers. This finding is validated on a new sample of 50 normal controls and two carriers. In addition, AT carriers were classified as non-NBS carriers, therefore allowing discrimination between carriers of these two closely related radiosensitivity syndromes.
There are several implications to our findings. First, identification of carriers is important medically. NBS and AT patients are known to be radiosensitive. Studies have suggested that carriers of these diseases are also radiosensitive (Dumon-Jones et al. 2003; Resnick et al. 2003). However, these studies have been hampered by the lack of accurate assays to identify carriers. Accurate diagnosis of carrier status of various radiosensitivity syndromes will facilitate studies to determine the radiosensitivity of these individuals. This is particularly important as diagnostic and therapeutic use of radiation is becoming increasingly common and for future studies to understand the basis of radiosensitivity.
Second, our results contribute to an understanding of phenotypic characteristics of carriers of NBS. The 520 genes whose expression levels define the gene expression phenotype of NBS carriers belong to several major DNA repair pathways. In NBS carriers, homologous recombination repair, nucleotide excision repair, and mitotic checkpoint were down-regulated, while steps in DNA damage bypass, processing of double-strand ends, and apoptosis were up-regulated. These data suggest that cells from NBS carriers are less efficient in DNA repair. This is expected in cells from NBS patients but not in cells from carriers. With time or under stress such as radiation exposure, this deficiency in DNA repair may increase the carriers’ risk of developing malignancies.
Lastly, our results identify heterozygous mutations as a contributor of natural variation in human gene expression. We and others have shown that there is extensive natural variation in human gene expression (Cheung and Spielman 2002; Yan et al. 2002; Cheung et al. 2003; Lo et al. 2003; Pastinen et al. 2004). Linkage (Schadt et al. 2003; Morley et al. 2004) and association studies (Cheung et al. 2005; Stranger et al. 2005) have identified polymorphic genetic determinants of this variation. Results from the present study identify heterozygous mutations as another contributor. We showed that expression levels of hundreds of genes differ between NBS carriers and noncarriers. If expression levels of several hundred genes differ in carriers of one mutation and carriers of recessive mutations are common, heterozygous mutations likely contribute a significant proportion of natural variation in human gene expression.
In this study, we showed that NBS carriers have a distinct gene expression phenotype. Although mutations in the nibrin gene are recessive, they result in phenotypes that differ between carriers and noncarriers. This finding suggests the importance of determining clinically important phenotypes in carriers and the contribution of heterozygous carriers to phenotypic diversity.
Methods
Study subject and expression phenotyping
Lymphoblastoid cell lines (from Coriell Cell Repositories) were grown to a density of 5 × 105 cells/mL in RPMI 1640 with 15% fetal bovine serum (FBS), 100 U penicillin/mL, 100 μg streptomycin/mL sulfate, and 1% L-glutamine. To identify differentially expressed genes between NBS carriers and controls and as a training set for the discriminant analysis, samples from 16 normal controls (GM07013, GM07019, GM07029, GM07042, GM07348, GM10830, GM10842, GM10852, GM10857, GM12740, GM12767, GM12802, GM12817, GM12865, GM12911, GM12912) and nine obligate NBS carriers (GM08036, GM08037, GM15805, GM15806, GM15810, GM15811, GM15815, GM15820, GM15821) were used. Total RNA from each sample was extracted with the RNeasy Mini-Kit (Qiagen) and hybridized onto the Affymetrix Genome Focus arrays per the manufacturer’s protocol. The growing and processing of cell lines were randomized by genotypes to eliminate batch effects that may contribute to genotype-specific gene expression.
To validate results from the discriminant analysis, expression data for 50 unrelated individuals that were part of another study (Morley et al. 2004) and those for two NBS carriers and 16 AT carriers were used. Those expression data were also obtained from lymphoblastoid cells (Coriell Cell Repositories) and grown with the above protocol.
Expression intensity was scaled to 500 using the Affymetrix MAS 5.0 software, log2 transformed, and normalized by Z-transformation. The 3928 genes that were called “Present” by the MAS 5.0 software in 90% or more of the controls or NBS carriers were used for further analysis.
Data analysis
Identify differentially expressed genes between normal controls and NBS carriers
To help identify the genes that were differentially expressed between the 16 controls and nine NBS carriers, the t-statistic was used (Watts et al. 2002). The significance of the t-scores was determined by a permutation test (Manly 1997). In each permutation, for each gene the expression intensities for the 25 subjects were randomly assigned to one of two groups of 16 and nine individuals (corresponding to the real sample size); we then calculated the absolute value of the t-score. This was repeated 10,000 times to obtain a distribution of t-scores for each gene. The distribution was then used to determine the ranking and therefore the P-value of the absolute value of the t-score from the real data. When several t-scores among the 10,000 had the same value as that from the real data, we assigned conservatively the percentile of the real value as the least extreme in the set.
Among the 3928 genes, 218 genes had t-scores that were the highest among the 10,000 replicates, thus those were given a P-value ≤0.0001. There were 520 genes (including the 218 genes) that were ranked in the top 10 among the 10,000 replicates (the highest 0.1%) and were given a P-value ≤0.001.
To correct for the problem of multiple testing and the unknown correlation between the genes, we used the sets of permutation to give us 10,000 estimates of the number of extreme t-scores expected under the null hypothesis that no genes differ in expression level between controls and NBS carriers. Among the replicates, the average number of genes with P ≤ 0.001 was 3.9. This was as expected since irrespective of the correlation structure, we expect ∼3928(0.001) = 3.9 genes with P ≤ 0.001, just by chance. The correlation between genes will, however, affect the variance of this number, so that this cannot be predicted in advance. We found that the variance was 3.80. The observed number (520 genes) is thus ∼260 standard deviations in excess of the mean. This is thus very highly significant, and we therefore rejected the null hypothesis that NBS carriers and noncarriers have the same gene expression phenotypes.
Discriminant analysis
Conventional stepwise discriminant analysis (SPSS) was performed with the expression intensity of the 218 genes (with P = 0.0001) on all 25 individuals. Fisher’s classification function coefficients and within-groups’ correlation matrix for the independent variables were used. At each step, the variable that minimizes the overall Wilk’s λ is entered; the minimal F-value for entering a variable is 3.84 and that for removing a variable is 2.71.
Sixteen genes (Table 3) were identified as the minimum set required to best discriminate between the controls and the NBS carriers. Validation with leave-one-out-classification and additional subjects (CEPH individuals and AT carriers) were also carried out using SPSS with the above parameters. The discriminant score for each individual is the value of the discriminant function for the 16 observations.
Clustering
The similarity of the 25 subjects (16 normal controls and nine NBS carriers) was assessed by Pearson’s correlation with expression levels of 520 genes (P < 0.001), and the phenotypes were grouped by hierarchical clustering using the centroid linkage method (as implemented in GeneSpring, Silicon Genetics).
Gene Ontology classification and pathway analysis
Gene Ontology (GO) categories were determined using EASEOnline (Hosack et al. 2003; http://david.niaid.nih.gov/david/). EASEOnline determines overrepresentation by comparing the differentially expressed genes to all the genes on the array using the one-tailed Fisher exact test. Pathways to which the genes belong were determined using the Reactome Knowledgebase (Joshi-Tope et al. 2005; http://www.reactome.org).
Acknowledgments
We thank Nancy King, Michael Morley, Jason Watts, and Marc Burock for performing the microarray experiments and data analysis; and Richard Spielman for comments. This work is supported by grants from the National Institutes of Health.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data from this study have been submitted to NCBI/GEO under accession no. GSE3894.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5320706
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney J.P., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R., III, Hays L., Morgan W.F., Petrini J.H., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R., III, Hays L., Morgan W.F., Petrini J.H., Olivares H., Davis E.M., Le Beau M., Yates J.R., III, Hays L., Morgan W.F., Petrini J.H., Davis E.M., Le Beau M., Yates J.R., III, Hays L., Morgan W.F., Petrini J.H., Le Beau M., Yates J.R., III, Hays L., Morgan W.F., Petrini J.H., Yates J.R., III, Hays L., Morgan W.F., Petrini J.H., Hays L., Morgan W.F., Petrini J.H., Morgan W.F., Petrini J.H., Petrini J.H. The hMre11/hRad50 protein complex and Nijmegen Breakage Syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Spielman R.S., Spielman R.S. The genetics of variation in gene expression. Nat. Genet. 2002;32:522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.-Y., Morley M., Spielman R.S., Conlin L.K., Weber T.M., Arcaro M., Jen K.-Y., Morley M., Spielman R.S., Weber T.M., Arcaro M., Jen K.-Y., Morley M., Spielman R.S., Arcaro M., Jen K.-Y., Morley M., Spielman R.S., Jen K.-Y., Morley M., Spielman R.S., Morley M., Spielman R.S., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J., Ewens K.G., Weber T.M., Morley M., Burdick J., Weber T.M., Morley M., Burdick J., Morley M., Burdick J., Burdick J. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C., Gorski B., Debniak T., Gliniewicz B., Mierzejewski M., Masojc B., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., Gorski B., Debniak T., Gliniewicz B., Mierzejewski M., Masojc B., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., Debniak T., Gliniewicz B., Mierzejewski M., Masojc B., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., Gliniewicz B., Mierzejewski M., Masojc B., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., Mierzejewski M., Masojc B., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., Masojc B., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., Jakubowska A., Matyjasik J., Zlowocka E., Sikorski A., Matyjasik J., Zlowocka E., Sikorski A., Zlowocka E., Sikorski A., Sikorski A., et al. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64:1215–1219. doi: 10.1158/0008-5472.can-03-2502. [DOI] [PubMed] [Google Scholar]
- DeRisi J., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M., Ray M., Chen Y., Su Y.A., Trent J.M., Chen Y., Su Y.A., Trent J.M., Su Y.A., Trent J.M., Trent J.M. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Drabek J., Hajduch M., Gojova L., Weigl E., Mihal V., Hajduch M., Gojova L., Weigl E., Mihal V., Gojova L., Weigl E., Mihal V., Weigl E., Mihal V., Mihal V. Frequency of 657del(5) mutation of the NBS1 gene in the Czech population by polymerase chain reaction with sequence specific primers. Cancer Genet. Cytogenet. 2002;138:157–159. doi: 10.1016/s0165-4608(02)00594-0. [DOI] [PubMed] [Google Scholar]
- Dumon-Jones V., Frappart P.O., Tong W.M., Sajithlal G., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z.Q., Frappart P.O., Tong W.M., Sajithlal G., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z.Q., Tong W.M., Sajithlal G., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z.Q., Sajithlal G., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z.Q., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z.Q., Schmid G., Herceg Z., Digweed M., Wang Z.Q., Herceg Z., Digweed M., Wang Z.Q., Digweed M., Wang Z.Q., Wang Z.Q. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63:7263–7269. [PubMed] [Google Scholar]
- Golub T.R., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., Loh M.L., Downing J.R., Caligiuri M.A., Downing J.R., Caligiuri M.A., Caligiuri M.A., et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Hosack D.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Sherman B.T., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia D.Y. The laboratory detection of heterozygotes. Am. J. Hum. Genet. 1957;9:98–112. [PMC free article] [PubMed] [Google Scholar]
- Hsia D.Y. The diagnosis of carriers of disease-producing genes. Ann. N.Y. Acad. Sci. 1966;66:946–964. [Google Scholar]
- Joshi-Tope G., Gillespie M., Vastrik I., D’Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., Gillespie M., Vastrik I., D’Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., Vastrik I., D’Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., D’Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., Jassal B., Gopinath G.R., Wu G.R., Matthews L., Gopinath G.R., Wu G.R., Matthews L., Wu G.R., Matthews L., Matthews L., et al. Reactome: A knowledge base of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Yang H.H., Gere S., Buetow K.H., Lee M.P., Gere S., Buetow K.H., Lee M.P., Buetow K.H., Lee M.P., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly B.J.F. 2nd ed. Chapman & Hall; London: 1997. Randomization, bootstrap and Monte Carlo methods in biology. [Google Scholar]
- Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Ewens K.G., Spielman R.S., Cheung V.G., Spielman R.S., Cheung V.G., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton N.E., Crow J.F., Muller H.J., Crow J.F., Muller H.J., Muller H.J. An estimate of the mutational damage in man from data on consanguineous marriages. Proc. Natl. Acad. Sci. 1956;42:855–863. doi: 10.1073/pnas.42.11.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel J.V. The clinical detection of the genetic carriers of inherited disease. Medicine. 1947;26:135–153. doi: 10.1097/00005792-194705000-00001. [DOI] [PubMed] [Google Scholar]
- Pastinen T., Sladek R., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Sladek R., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Villeneuve A., Gaudin T., Brandstrom H., Gaudin T., Brandstrom H., Brandstrom H., et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol. Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- Resnick I.B., Kondratenko I., Pashanov E., Maschan A.A., Karachunsky A., Togoev O., Timakov A., Polyakov A., Tverskaya S., Evgrafov O., Kondratenko I., Pashanov E., Maschan A.A., Karachunsky A., Togoev O., Timakov A., Polyakov A., Tverskaya S., Evgrafov O., Pashanov E., Maschan A.A., Karachunsky A., Togoev O., Timakov A., Polyakov A., Tverskaya S., Evgrafov O., Maschan A.A., Karachunsky A., Togoev O., Timakov A., Polyakov A., Tverskaya S., Evgrafov O., Karachunsky A., Togoev O., Timakov A., Polyakov A., Tverskaya S., Evgrafov O., Togoev O., Timakov A., Polyakov A., Tverskaya S., Evgrafov O., Timakov A., Polyakov A., Tverskaya S., Evgrafov O., Polyakov A., Tverskaya S., Evgrafov O., Tverskaya S., Evgrafov O., Evgrafov O., et al. 657del5 mutation in the gene for Nijmegen Breakage Syndrome (NBS1) in a cohort of Russian children with lymphoid tissue malignancies and controls. Am. J. Med. Genet. A. 2003;120:174–179. doi: 10.1002/ajmg.a.20188. [DOI] [PubMed] [Google Scholar]
- Saar K., Chrzanowska K.H., Stumm M., Jung M., Nurnberg G., Wienker T.F., Seemanova E., Wegner R.D., Reis A., Sperling K., Chrzanowska K.H., Stumm M., Jung M., Nurnberg G., Wienker T.F., Seemanova E., Wegner R.D., Reis A., Sperling K., Stumm M., Jung M., Nurnberg G., Wienker T.F., Seemanova E., Wegner R.D., Reis A., Sperling K., Jung M., Nurnberg G., Wienker T.F., Seemanova E., Wegner R.D., Reis A., Sperling K., Nurnberg G., Wienker T.F., Seemanova E., Wegner R.D., Reis A., Sperling K., Wienker T.F., Seemanova E., Wegner R.D., Reis A., Sperling K., Seemanova E., Wegner R.D., Reis A., Sperling K., Wegner R.D., Reis A., Sperling K., Reis A., Sperling K., Sperling K. The gene for the ataxia-telangiectasia variant, Nijmegen Breakage Syndrome, maps to a 1-cM interval on chromosome 8q21. Am. J. Hum. Genet. 1997;60:605–610. [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Milligan S.B., Lamb J.R., Cavet G., Lamb J.R., Cavet G., Cavet G., et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Stranger B.E., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Hunt S., Kahl B., Antonarakis S.E., Tavare S., Kahl B., Antonarakis S.E., Tavare S., Antonarakis S.E., Tavare S., Tavare S., et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzanella C., Antoccia A., Spadoni E., di Masi A., Pecile V., Demori E., Varon R., Marseglia G.L., Tiepolo L., Maraschio P., Antoccia A., Spadoni E., di Masi A., Pecile V., Demori E., Varon R., Marseglia G.L., Tiepolo L., Maraschio P., Spadoni E., di Masi A., Pecile V., Demori E., Varon R., Marseglia G.L., Tiepolo L., Maraschio P., di Masi A., Pecile V., Demori E., Varon R., Marseglia G.L., Tiepolo L., Maraschio P., Pecile V., Demori E., Varon R., Marseglia G.L., Tiepolo L., Maraschio P., Demori E., Varon R., Marseglia G.L., Tiepolo L., Maraschio P., Varon R., Marseglia G.L., Tiepolo L., Maraschio P., Marseglia G.L., Tiepolo L., Maraschio P., Tiepolo L., Maraschio P., Maraschio P. Chromosome instability and nibrin protein variants in NBS heterozygotes. Eur. J. Hum. Genet. 2003;11:297–303. doi: 10.1038/sj.ejhg.5200962. [DOI] [PubMed] [Google Scholar]
- van der Burgt I., Chrzanowska K.H., Smeets D., Weemaes C., Chrzanowska K.H., Smeets D., Weemaes C., Smeets D., Weemaes C., Weemaes C. Nijmegen Breakage Syndrome. J. Med. Genet. 1996;33:153–156. doi: 10.1136/jmg.33.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R., Vissinga C., Platzer M., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., Vissinga C., Platzer M., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., Platzer M., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., Seemanova E., Cooper P.R., Nowak N.J., Cooper P.R., Nowak N.J., Nowak N.J., et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen Breakage Syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Varon R., Seemanova E., Chrzanowska K., Hnateyko O., Piekutowska-Abramczuk D., Krajewska-Walasek M., Sykut-Cegielska J., Sperling K., Reis A., Seemanova E., Chrzanowska K., Hnateyko O., Piekutowska-Abramczuk D., Krajewska-Walasek M., Sykut-Cegielska J., Sperling K., Reis A., Chrzanowska K., Hnateyko O., Piekutowska-Abramczuk D., Krajewska-Walasek M., Sykut-Cegielska J., Sperling K., Reis A., Hnateyko O., Piekutowska-Abramczuk D., Krajewska-Walasek M., Sykut-Cegielska J., Sperling K., Reis A., Piekutowska-Abramczuk D., Krajewska-Walasek M., Sykut-Cegielska J., Sperling K., Reis A., Krajewska-Walasek M., Sykut-Cegielska J., Sperling K., Reis A., Sykut-Cegielska J., Sperling K., Reis A., Sperling K., Reis A., Reis A. Clinical ascertainment of Nijmegen Breakage Syndrome (NBS) and prevalence of the major mutation, 657del5, in three Slav populations. Eur. J. Hum. Genet. 2000;8:900–902. doi: 10.1038/sj.ejhg.5200554. [DOI] [PubMed] [Google Scholar]
- Watts J.A., Morley M., Burdick J.T., Fiori J.L., Ewens W.J., Spielman R.S., Cheung V.G., Morley M., Burdick J.T., Fiori J.L., Ewens W.J., Spielman R.S., Cheung V.G., Burdick J.T., Fiori J.L., Ewens W.J., Spielman R.S., Cheung V.G., Fiori J.L., Ewens W.J., Spielman R.S., Cheung V.G., Ewens W.J., Spielman R.S., Cheung V.G., Spielman R.S., Cheung V.G., Cheung V.G. Gene expression phenotype in heterozygous carriers of Ataxia Telangiectasia. Am. J. Hum. Genet. 2002;71:791–800. doi: 10.1086/342974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Vogelstein B., Kinzler K.W., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]