Abstract
The domestication of dogs caused a dramatic change in their way of life compared with that of their ancestor, the gray wolf. We hypothesize that this new life style changed the selective forces that acted upon the species, which in turn had an effect on the dog’s genome. We sequenced the complete mitochondrial DNA genome in 14 dogs, six wolves, and three coyotes. Here we show that dogs have accumulated nonsynonymous changes in mitochondrial genes at a faster rate than wolves, leading to elevated levels of variation in their proteins. This suggests that a major consequence of domestication in dogs was a general relaxation of selective constraint on their mitochondrial genome. If this change also affected other parts of the dog genome, it could have facilitated the generation of novel functional genetic diversity. This diversity could thus have contributed raw material upon which artificial selection has shaped modern breeds and may therefore be an important source of the extreme phenotypic variation present in modern-day dogs.
In The Origin of Species, Darwin (1859) suggested that “several wild species of Canidae have been tamed and that their blood, in some cases mingled together, flows in the veins of our domestic [dog] breeds”. We now know that dogs (Canis familiaris) are entirely derived from the domestication of wolves (Canis lupus) (Vilà et al. 1997); however, the origin of the huge morphological diversity that led Darwin to his speculation remains largely unknown (Sutter and Ostrander 2004). The domestic dog is the most phenotypically diverse mammal on earth. The large differences in size, conformation, behavior, and physiology between dog breeds exceed the differences among species in the dog family, Canidae (Coppinger and Coppinger 2001; Wayne 2001). Recent studies show that the origin of most dog breeds may derive from very recent selective breeding practices and are probably <200 yr old (Parker et al. 2004). However, selection acts upon existing variability. It is remarkable that the potential for such large diversification existed in the ancestral wolf population from where the domestication process was initiated. Furthermore, the time since domestication (at least 14,000 yr; Vilà et al. 1997; Sablin and Khlopachev 2002; Savolainen et al. 2002) seems insufficient to generate substantial additional genetic diversity. What is the origin of this diversity? We hypothesize that changes in the living conditions of dogs as a result of domestication resulted in the release of selective constraint allowing a faster accumulation of functional (non-silent) genetic diversity in a large array of genes.
Weakly deleterious mutations—those with selective effects close to the reciprocal of the effective population size—represent an important class of genetic variability (Ohta and Kimura 1971). Such mutations are expected to accumulate faster in populations with small effective sizes or in populations in which selection has been relaxed, resulting in a decline in fitness. Advantageous mutations, conversely, contribute little to patterns of genetic variation and are enriched in fixed differences between species. To examine whether the accumulation of deleterious mutations is increased in dogs compared with their wild ancestors, we have focused on mitochondrial DNA (mtDNA). The mitochondrial genome represents only a small part of the canine genome and has a unique mode of inheritance. However, while dog and wolf lineages are difficult to separate for nuclear genes (Parker et al. 2004; Vilà et al. 2005), mitochondrial lineages are clearly distinguishable for the two species (Vilà et al. 1997; Savolainen et al. 2002). This offers a good opportunity to evaluate the consequences of life with humans on a portion of the dog genome.
As is often seen in data sets where recombination is rare or absent, mtDNA commonly exhibits an excess of replacement (nonsynonymous) to silent (synonymous) changes in intra-species polymorphism compared with inter-species divergence (Hasegawa et al. 1998). These findings likely reflect an increased preponderance of deleterious alleles segregating within populations. Among the dog mitochondrial genes, 13 encode different proteins of oxidative phosphorylation, which are important for the production of energy and heat. Additionally, the mtDNA includes two ribosomal RNA genes and 22 transfer RNA genes. In humans, large numbers of pathological mutations have been described in mtDNA genes (Chinnery et al. 2000). Studies on human mtDNA have shown that selection may have shaped the patterns of variability observed today (Ruiz-Pesini et al. 2004; Kivisild et al. 2006). In the case of the dog, since wolves still survive across the world, it is possible to compare the genome of the domestic species with that of its ancestor and thus identify both functional and silent nucleotide changes that appeared in the domestic lineages. In this study we compare patterns of molecular evolution in the mtDNA of dogs and wolves in order to understand the genetic consequences of the change in lifestyle associated with domestication.
Results and Discussion
Previous studies have shown that domestic dog mtDNA sequences cluster in four main clades when compared with wolves, indicating different origination events (Vilà et al. 1997; Savolainen et al. 2002). In order to select samples of dogs representing several mtDNA lineages for the analysis, we sequenced the mitochondrial control region I for 88 dogs from 53 breeds. Among those individuals we selected 14 dogs, which included six from clade I (the clade that encompasses about 71% of today’s dogs; Savolainen et al. 2002) and two or three from each one of the clades II, III, and IV (Vilà et al. 1997). Because we wished to characterize mutations that occurred on dog lineages since the emergence of each clade, the dogs in this study were selected to be representative of the full genetic diversity observed in each clade (Supplemental Fig. S1). Complete mtDNA sequences, excluding the tandem repeat located inside the control region (Hoelzel et al. 1994), were obtained through polymerase chain reaction (PCR) amplification and sequencing. The complete mitochondrial sequence was also obtained for six wolves from throughout the world trying to represent as much of the previously described wolf diversity (Vilà et al. 1999) as possible: Spain, Russia, Saudi Arabia (two individuals), North America, and Sweden. Three coyotes (Canis latrans) from Nebraska and Colorado (two individuals), USA, were also sequenced and used as outgroups.
To construct a gene tree from the 23 complete mtDNA sequences, we first excluded the control region because of the high incidence of homoplasy (Ingman et al. 2000), resulting in a sequence length of 15,547–15,549 base pairs (bp). The average uncorrected pairwise sequence divergence between wolves and dogs was 0.47% (SE = 0.02), whereas average sequence divergence between coyotes and dogs plus wolves was 4.28% (SE = 0.11). A gene tree constructed with these sequences shows that all four clades of dogs are very well supported with bootstrap support values of 100% and Bayesian posterior probabilities of 1.00 (Fig. 1).
Figure 1.
Phylogenetic tree of wolf (W), dog (D), and coyote (C) mtDNA sequences. The tree was constructed using a Bayesian approach. The same topology was obtained with a neighbor-joining approach. Support is indicated at the nodes as percent bootstrap support for 1000 neighbor-joining replicates and Bayesian posterior probabilities. Four clades of dog sequences (I to IV) are indicated as in Vilà et al. (1997). Internal dog branches are marked in orange, and internal wolf branches are marked in light blue. The branch leading to wolf haplotype W1 was basal to the rest of the tree and it was also considered internal. Internal branches that could not be conclusively associated to dogs or to wolves are indicated in discontinuous green.
We used a maximum-likelihood (ML) approach to estimate the rates of synonymous (dS) and nonsynonymous (dN) evolution in mtDNA genes along each individual branch of the gene tree in Figure 1. Branches leading only to one or more dog sequences were considered to be dog branches (in red and orange in Fig. 1) except for the four branches preceding the four clades (dotted lines). These branches were excluded from our analyses because they could not be uniquely assigned to dogs or to wolves. All other branches within the wolf/dog tree (in blue) were considered as wolf branches as they either represent evolution before dog domestication or recent wolf evolution. We first examined the tree for differences in the rate of silent nucleotide substitution between wolves and dogs. We used the ML estimates of dS to perform relative rate tests at the four nodes of the tree associated with the origin of the four dog clades (see Methods). None of the tests showed significant differences between dog and wolf branches (P > 0.05 in all cases, data not shown), although dS was on average 10.9% higher on lineages leading to dogs compared with wolves. We therefore have no evidence to suggest that the mutation rate in mtDNA differs between dogs and wolves. A similar test revealed that dN was on average 40.3% higher on the dog lineages, although none of the individual tests were statistically significant.
We next examined ML estimates of dN/dS ratios in all branches of the gene tree. The average dN/dS ratio of divergence between a randomly chosen wolf (W5) and a randomly chosen coyote (C2) sequence (0.034; 95% confidence interval CI: 0.023–0.043) is significantly lower than the average ratio along wolf branches (0.091; 95% CI: 0.056–0.127; P < 0.001; confidence intervals and significance testing were calculated by bootstrap), which reflects intraspecific variation. Weakly deleterious mutations are expected to be more common in intra-specific variation than in divergence between species because purifying selection has had less time to act (Akashi 1999; Piganeau and Eyre-Walker 2003; Kivisild et al. 2006). This suggests that many weakly deleterious mutations are segregating within the wolf population. However, the average dN/dS ratio in dog branches (0.183; 95% CI: 0.096–0.279) is significantly higher than for wolves (P = 0.040). This result is especially surprising considering that selection is more effective in growing populations, increasing the probability that deleterious alleles are lost (Otto and Whitlock 1997), and that the domestic population has likely increased by more than six orders of magnitude since the time of domestication (Coppinger and Coppinger 2001). We believe that this could indicate a relaxation of constraint on dog mtDNA genes compared with wolves.
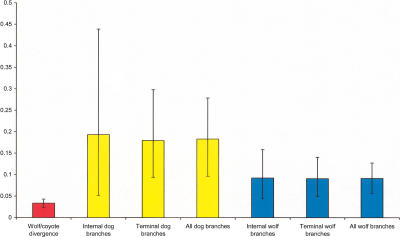
Within populations, the relative number of deleterious compared with neutral changes is expected to decline as a function of allele frequency as a result of purifying selection (Fay et al. 2001). This is because deleterious alleles tend to be removed from the population by selection and hence are unlikely to reach high frequencies. This implies that dN/dS is expected to be higher on terminal branches than internal branches of the wolf/dog portion of the gene tree, which all represent intra-specific variability. As a higher proportion of dog branches are terminal in the gene tree, this could potentially explain the higher dN/dS ratios in dog branches. In order to investigate this possibility, we divided dog and wolf branches into internal (orange for dogs and light blue for wolves, in Fig. 1) and terminal (red and dark blue) branches. The dN/dS ratios are surprisingly consistent within dog and within wolf branches (Fig. 2). This indicates that the effect of allele frequency on the proportion of nonsynonymous mutations has not been that great and that the differences between dN/dS ratios in wolves and dogs represent true differences in selective regimes, rather than simply being an effect of the accumulation of weakly deleterious mutations on terminal branches of the gene tree. It should however be noted that dog branches tend to be more distal in the tree compared with wolf branches, which could potentially contribute to the larger dN/dS in dogs.
Figure 2.
Maximum-likelihood estimates of dN/dS. Values for wolves are indicated in blue, for dogs in yellow, and for the divergence between a random wolf and coyote sequence in red. Ninety-five percent confidence intervals were bootstrap-derived.
We compared the dN/dS ratio along branches representing the divergence between coyotes and wolves, dog diversity, and wolf diversity for individual mitochondrial genes (Supplemental Fig. S2). We also estimated dN/dS ratios for three gene classes: ATPase genes, NADH dehydrogenase (complex I) genes, and cytochrome c oxidase (complex IV) genes (Supplemental Fig. S2). We did not note any trend for the observed differences in dN/dS ratios between branches to be biased toward any particular gene or gene class although the number of changes in individual genes is small and the confidence intervals are very large. Hence our data are compatible with a model whereby dogs are gradually accumulating weakly deleterious changes across all mtDNA genes.
In order to assign nucleotide changes at each gene to specific branches and investigate their biochemical properties, we compared the maximum-likelihood reconstructed ancestral sequences at each node with those at neighboring nodes. Table 1 shows the number of changes estimated for wolf and dog branches in the tree in Figure 1. These values corroborate the findings of the ML estimation of dN/dS ratios. There is a significant excess of nonsynonymous (NS) changes compared with synonymous (S) along both wolf and dog branches compared with divergence between wolves and coyotes (P = 0.002 and P < 0.001, respectively, G-test of independence). Additionally, a significant excess of nonsynonymous changes is also observed in dog compared with wolf lineages (P = 0.033). In line with our previous findings there is no difference between the ratio of synonymous and nonsynonymous changes between internal and terminal branches within either dogs or wolves (P > 0.05 in both cases). We used three different methods to examine the potential phenotypic effects of mutations: We divided changes into conservative or radical by charge and by polarity (Zhang 2000) and into benign, potentially damaging, or probably damaging using the PolyPhen database (see Methods; Table 1). The relative numbers of conservative to radical (or benign to damaging) changes are not significantly different between dog branches, wolf branches, or in coyote/wolf divergence (G-tests, P > 0.05 in all cases). Hence there appears to be no tendency for dogs to accumulate more radical or damaging changes than wolves, or for such changes to have been preferentially eliminated from the wolf or dog population. This is probably because the majority of radical or damaging changes are strongly deleterious and never reach detectable frequencies in any population. This is consistent with our hypothesis that a relaxation of selective constraint in dogs has resulted in an elevated accumulation of weakly deleterious variants.
Table 1.
Number of nucleotide changes inferred from maximum-likelihood reconstructed ancestral nodes
Although the domestication process was likely initiated by just a few individuals (Vilà et al. 1997, 2005; Savolainen et al. 2002), the total world population of dogs is today estimated to be around 400 million (Coppinger and Coppinger 2001). As the initial dog population was small and was subsequently subdivided, deleterious mutations may have accumulated by genetic drift. In addition, as soon as the first dogs started to live with humans, it is likely that they were strongly selected for behavioral traits like tameness (Saetre et al. 2004). As the dogs’ breeding program was controlled by humans, the intensity of stabilizing selection for other morphologic, behavioral, or physiological traits likely decreased. Individuals with lower metabolic efficiency were more likely to survive and reproduce than they were before. This relaxation of constraint may have allowed the accumulation of additional nonsynonymous mutations on the mitochondrial genome. It is therefore possible that this process led to an increase in functional genetic diversity throughout the entire dog genome, including both genes and elements affecting gene expression. For example, it has been suggested that variation at tandem repeats (Fondon III and Garner 2004) or the presence of short interspersed elements (SINEs; Wang and Kirkness 2005) closely associated with genes could have contributed to the phenotypic diversity in dogs, although there is no direct evidence of these elements being more frequent in dogs than in gray wolves. A relaxation of selective constraint could have contributed not only to the huge phenotypic diversity that exists in today’s dogs but also to the appearance of the large variety of diseases that affect many dog breeds (Ostrander and Kruglyak 2000).
Methods
Sampling strategy and mtDNA sequencing
Blood samples were obtained from the Swedish University of Agricultural Sciences at Uppsala, Sweden. Samples from 88 dogs corresponding to 53 different breeds were initially screened to identify dogs with mitochondrial DNA sequences pertaining to the four clades described by Vilà et al. (1997). Fourteen of these dogs from 13 different breeds were selected to represent all four clades (Supplemental Table S1). Additionally, tissue samples were obtained from six wolves and three coyotes. The wolves originated from Spain, Russia, Saudi Arabia, North America, and Sweden. Two samples were obtained from wolves from Saudi Arabia due to the large mtDNA diversity revealed by a previous study (Vilà et al. 1999). The coyote samples came from Nebraska and Colorado (two individuals), USA. DNA extraction, amplification, and sequencing are described in the Supplemental material.
Tree construction
We constructed a phylogenetic tree of all complete mtDNA sequences except the control region using a Bayesian approach as implemented in MrBayes 3 (Huelsenbeck and Ronquist 2001). The same tree topology was obtained using a neighbor-joining approach in PAUP 4.0 (Swofford 2002) using the model of sequence evolution that best fitted the data according to the program Modeltest 3.6 (Posada and Crandall 1998). Support of nodes was calculated using posterior probabilities for the Bayesian tree and with 1000 bootstrap pseudoreplicates for the neighbor-joining tree.
In order to distinguish between mutations along different branches of the gene tree, we classified wolf/dog branches as wolf internal, wolf terminal, dog internal, and dog terminal. The internal branches leading to each of the four dog clades were excluded from the analysis (Fig. 1) because they could not be conclusively assigned to dogs or to wolves. Internal wolf branches were defined as internal branches leading to only wolves or both wolves and dogs. Since the branch leading to wolf haplotype W1 was basal to the rest of the tree, it was also considered internal. Internal dog branches were defined as internal branches leading only to dog sequences. Terminal wolf and dog branches lead directly to a wolf or dog sequence, respectively.
Maximum-likelihood estimation of evolutionary rates
We used the PAML package (Yang 1997) to reconstruct the changes that occurred along each branch of the gene tree at all of the site classes within the mtDNA molecule. For sites outside of protein coding genes, we used the BaseML program to provide ML estimates of ancestral nucleotide states at each node of the tree and the number of changes along each branch using the Hasegawa et al. (1985) model of base substitution. These sites were classified as RNA genes, control region (excluding tandem repeat), and other noncoding regions. We used CodeML, implementing the codon substitution model of Goldman and Yang (1994) to provide ML estimates of the ancestral codons at each node in the tree and the nonsynonymous (dN) and synonymous (dS) substitution rates along each branch. The free-ratio model, where dN/dS is allowed to vary among branches, was used. Codon frequencies were calculated from the average nucleotide frequencies at the three codon positions.
We tested whether the rate of molecular evolution differs between dogs and wolves using an extension of the relative rate test (Sarich and Wilson 1973) based on lineage-specific ML estimates of dN and dS. This was done by first choosing the four nodes in the gene tree leading to the branches before each of the four dog clades (discontinuous green branches in Fig. 1). For each node, we then compared the average dN and dS values from the node to the tip of a dog branch in the clade in question with the average dN and dS values from the node to the tip of a wolf branch. We also estimated the average dN/dS values of the internal and external branch categories described in the previous section using ML estimates of dN/dS from each individual branch of the tree. For both procedures, generation of confidence intervals and estimation of the significance of differences between statistics on different branch categories were performed by nonparametric bootstrapping. We recalculated each statistic from data sets produced by resampling each codon with replacement with 1000 replicates (Felsenstein 1985). When comparing statistics for each individual gene, we resampled each gene separately.
We assigned each inferred amino acid change to a specific branch in the gene tree by comparison of ML inferred sequences at each node. Nonsynonymous changes on each branch in the tree were categorized as radical or conservative by the criteria of both polarity and charge presented by Zhang (2000) and by using the PolyPhen application, which incorporates sequence, phylogenetic, and structural information to characterize the potential severity of the phenotypic effect produced by each mutation.
Acknowledgments
Åke Hedhammar, from the Department of Small Animal Clinical Sciences, Swedish University of Agricultural Sciences in Uppsala, Sweden, helped us to obtain samples from dogs. Ron Ruthstrom, from the Kansas Department of Wildlife and Parks, USA, and Jennifer Leonard, from the Department of Evolutionary Biology, Uppsala University, provided the coyote samples. Frank Hailer and Jennifer Leonard provided valuable comments and discussion. Financial support was provided by a grant of the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning to CV.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. DQ480489–DQ480511.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5117706
References
- Akashi H. Inferring the fitness effects of DNA mutations from polymorphism and divergence data: Statistical power to detect directional selection under stationarity and free recombination. Genetics. 1999;151:221–238. doi: 10.1093/genetics/151.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P.F., Johnson M.A., Wardell T.M., Singh-Kler R., Hayes C., Brown D.T., Taylor R.W., Bindoff L.A., Turnbull D.M., Johnson M.A., Wardell T.M., Singh-Kler R., Hayes C., Brown D.T., Taylor R.W., Bindoff L.A., Turnbull D.M., Wardell T.M., Singh-Kler R., Hayes C., Brown D.T., Taylor R.W., Bindoff L.A., Turnbull D.M., Singh-Kler R., Hayes C., Brown D.T., Taylor R.W., Bindoff L.A., Turnbull D.M., Hayes C., Brown D.T., Taylor R.W., Bindoff L.A., Turnbull D.M., Brown D.T., Taylor R.W., Bindoff L.A., Turnbull D.M., Taylor R.W., Bindoff L.A., Turnbull D.M., Bindoff L.A., Turnbull D.M., Turnbull D.M. The epidemiology of pathogenic mitochondrial DNA mutations. Ann. Neurol. 2000;48:188–193. [PubMed] [Google Scholar]
- Coppinger R., Coppinger L., Coppinger L. Dogs. The University of Chicago Press; Chicago: 2001. [Google Scholar]
- Darwin C. On the origin of species by means of natural selection. Murray; London: 1859. [Google Scholar]
- Fay J.C., Wyckoff G.J., Wu C.I., Wyckoff G.J., Wu C.I., Wu C.I. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fondon J.W., III, Garner H.R., Garner H.R. Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N., Yang Z., Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T., Kishino H., Yano T., Yano T. Dating of the human–ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Cao Y., Yang Z., Cao Y., Yang Z., Yang Z. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: Nonsynonymous/synonymous rate ratio is much higher within species than between species. Mol. Biol. Evol. 1998;15:1499–1505. doi: 10.1093/oxfordjournals.molbev.a025877. [DOI] [PubMed] [Google Scholar]
- Hoelzel A.R., Lopez J.V., Dover G.A., O’Brien S.J., Lopez J.V., Dover G.A., O’Brien S.J., Dover G.A., O’Brien S.J., O’Brien S.J. Rapid evolution of a heteroplasmic repetitive sequence in the mitochondrial DNA control region of carnivores. J. Mol. Evol. 1994;39:191–199. doi: 10.1007/BF00163807. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F., Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ingman M., Kaessmann H., Pääbo S., Gyllensten U., Kaessmann H., Pääbo S., Gyllensten U., Pääbo S., Gyllensten U., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Kivisild T., Shen P., Wall D.P., Do B., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A., Shen P., Wall D.P., Do B., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A., Wall D.P., Do B., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A., Do B., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A., Sung R., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A., Davis K., Passarino G., Underhill P.A., Scharfe C., Torroni A., Passarino G., Underhill P.A., Scharfe C., Torroni A., Underhill P.A., Scharfe C., Torroni A., Scharfe C., Torroni A., Torroni A., et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Kimura M., Kimura M. On the constancy of the evolutionary rate of cistrons. J. Mol. Evol. 1971;1:18–25. doi: 10.1007/BF01659391. [DOI] [PubMed] [Google Scholar]
- Ostrander E.A., Kruglyak L., Kruglyak L. Unleashing the canine genome. Genome Res. 2000;10:1271–1274. doi: 10.1101/gr.155900. [DOI] [PubMed] [Google Scholar]
- Otto S.P., Whitlock M.C., Whitlock M.C. The probability of fixation in populations of changing size. Genetics. 1997;146:723–733. doi: 10.1093/genetics/146.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H.G., Kim L.V., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Kim L.V., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., DeFrance H.B., Ostrander E.A., Kruglyak L., Ostrander E.A., Kruglyak L., Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1166. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Piganeau G., Eyre-Walker A., Eyre-Walker A. Estimating the distribution of fitness effects from DNA sequence data: Implications for the molecular clock. Proc. Natl. Acad. Sci. 2003;100:10335–10340. doi: 10.1073/pnas.1833064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E., Mishmar D., Brandon M., Procaccio V., Wallace D.C., Mishmar D., Brandon M., Procaccio V., Wallace D.C., Brandon M., Procaccio V., Wallace D.C., Procaccio V., Wallace D.C., Wallace D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- Sablin M.V., Khlopachev G.A., Khlopachev G.A. The earliest Ice Age dogs: Evidence from Eliseevichi. Curr. Anthropol. 2002;43:795–799. [Google Scholar]
- Saetre P., Lindberg J., Leonard J.A., Olsson K., Pettersson U., Ellegren H., Bergstrom T.F., Vilà C., Jazin E., Lindberg J., Leonard J.A., Olsson K., Pettersson U., Ellegren H., Bergstrom T.F., Vilà C., Jazin E., Leonard J.A., Olsson K., Pettersson U., Ellegren H., Bergstrom T.F., Vilà C., Jazin E., Olsson K., Pettersson U., Ellegren H., Bergstrom T.F., Vilà C., Jazin E., Pettersson U., Ellegren H., Bergstrom T.F., Vilà C., Jazin E., Ellegren H., Bergstrom T.F., Vilà C., Jazin E., Bergstrom T.F., Vilà C., Jazin E., Vilà C., Jazin E., Jazin E. From wolf to domestic dog: Changes in brain gene expression. Brain Res. Mol. Brain Res. 2004;126:198–206. doi: 10.1016/j.molbrainres.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Sarich V.M., Wilson A.C., Wilson A.C. Generation time and genomic evolution in primates. Science. 1973;179:1144–1147. doi: 10.1126/science.179.4078.1144. [DOI] [PubMed] [Google Scholar]
- Savolainen P., Zhang Y.P., Luo J., Lundeberg J., Leitner T., Zhang Y.P., Luo J., Lundeberg J., Leitner T., Luo J., Lundeberg J., Leitner T., Lundeberg J., Leitner T., Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Sutter N.B., Ostrander E.A., Ostrander E.A. Dog star rising: The canine genetic system. Nat. Rev. Genet. 2004;5:900–910. doi: 10.1038/nrg1492. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Version 4. Sinauer Associates; Sunderland, Massachusetts: 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). [Google Scholar]
- Vilà C., Savolainen P., Maldonado J.E., Amorim I.R., Rice J.E., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K., Savolainen P., Maldonado J.E., Amorim I.R., Rice J.E., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K., Maldonado J.E., Amorim I.R., Rice J.E., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K., Amorim I.R., Rice J.E., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K., Rice J.E., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K., Crandall K.A., Lundeberg J., Wayne R.K., Lundeberg J., Wayne R.K., Wayne R.K. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- Vilà C., Amorim I.R., Leonard J.A., Posada D., Castroviejo J., Petrucci-Fonseca F., Crandall K.A., Ellegren H., Wayne R.K., Amorim I.R., Leonard J.A., Posada D., Castroviejo J., Petrucci-Fonseca F., Crandall K.A., Ellegren H., Wayne R.K., Leonard J.A., Posada D., Castroviejo J., Petrucci-Fonseca F., Crandall K.A., Ellegren H., Wayne R.K., Posada D., Castroviejo J., Petrucci-Fonseca F., Crandall K.A., Ellegren H., Wayne R.K., Castroviejo J., Petrucci-Fonseca F., Crandall K.A., Ellegren H., Wayne R.K., Petrucci-Fonseca F., Crandall K.A., Ellegren H., Wayne R.K., Crandall K.A., Ellegren H., Wayne R.K., Ellegren H., Wayne R.K., Wayne R.K. Mitochondrial DNA phylogeography and population history of the Gray Wolf Canis lupus. Mol. Ecol. 1999;8:2089–2103. doi: 10.1046/j.1365-294x.1999.00825.x. [DOI] [PubMed] [Google Scholar]
- Vilà C., Seddon J., Ellegren H., Seddon J., Ellegren H., Ellegren H. Genes of domestic mammals augmented by backcrossing with wild ancestors. Trends Genet. 2005;21:214–218. doi: 10.1016/j.tig.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Wang W., Kirkness E.F., Kirkness E.F. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005;15:1798–1808. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne R.K. Consequences of domestication: Morphological diversity of the dog. In: Ruvinsky A., Sampson J., Sampson J., et al., editors. The genetics of the dog. CABI Publishing; Oxon, UK: 2001. pp. 43–60. [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 2000;50:56–68. doi: 10.1007/s002399910007. [DOI] [PubMed] [Google Scholar]