Abstract
RNA triphosphatase catalyzes the first step in mRNA capping. The RNA triphosphatases of fungi and protozoa are structurally and mechanistically unrelated to the analogous mammalian enzyme, a situation that recommends RNA triphosphatase as an anti-infective target. Fungal and protozoan RNA triphosphatases belong to a family of metal-dependent phosphohydrolases exemplified by yeast Cet1. The Cet1 active site is unusually complex and located within a topologically closed hydrophilic β-barrel (the triphosphate tunnel). Here we probe the active site of Plasmodium falciparum RNA triphosphatase by targeted mutagenesis and thereby identify eight residues essential for catalysis. The functional data engender an improved structural alignment in which the Plasmodium counterparts of the Cet1 tunnel strands and active-site functional groups are located with confidence. We gain insight into the evolution of the Cet1-like triphosphatase family by noting that the heretofore unique tertiary structure and active site of Cet1 are recapitulated in recently deposited structures of proteins from Pyrococcus (PBD 1YEM) and Vibrio (PDB 2ACA). The latter proteins exemplify a CYTH domain found in CyaB-like adenylate cyclases and mammalian thiamine triphosphatase. We conclude that the tunnel fold first described for Cet1 is the prototype of a larger enzyme superfamily that includes the CYTH branch. This superfamily, which we name “triphosphate tunnel metalloenzyme,” is distributed widely among bacterial, archaeal, and eukaryal taxa. It is now clear that Cet1-like RNA triphosphatases did not arise de novo in unicellular eukarya in tandem with the emergence of caps as the defining feature of eukaryotic mRNA. They likely evolved by incremental changes in an ancestral tunnel enzyme that conferred specificity for RNA 5′-end processing.
Keywords: RNA capping, phosphohydrolase, malaria, enzyme evolution
INTRODUCTION
The m7GpppN cap of eukaryotic mRNA is formed via three enzymatic reactions: (1) the 5′ triphosphate end of the pre-mRNA is hydrolyzed to a diphosphate by RNA triphosphatase; (2) the diphosphate RNA end is capped with GMP by RNA guanylyltransferase; and (3) the GpppN cap is methylated by RNA (guanine-N7) methyltransferase (Shuman 2001). We have suggested that the capping enzymes are a good focal point for considering eukaryotic evolution, because the cap structure is ubiquitous in eukaryotic organisms, but absent from the bacterial and archaeal domains of life, signifying that any differences in the capping apparatus between taxa would reflect events that post-date the emergence of ancestral nucleated cells. Previously, we proposed a scheme of eukaryotic phylogeny based on drastic differences in the structure and mechanism of the triphosphatase component (Shuman 2002).
Metazoan and plant RNA triphosphatases belong to the cysteine-phosphatase enzyme superfamily (Takagi et al. 1997; Changela et al. 2001). These RNA triphosphatases catalyze a two-step phosphoryl transfer reaction in which the conserved cysteine of the phosphate-binding loop HCxxxxxR(S/T) attacks the γ phosphorus of triphosphate-terminated RNA to form a covalent protein-cysteinyl-S-phosphate intermediate and expel the diphosphate RNA product. The covalent phosphoenzyme intermediate is hydrolyzed to liberate inorganic phosphate. The reaction does not require a divalent cation cofactor.
The RNA triphosphatases of fungi, protozoa, and several DNA viruses belong to a family of metal-dependent phosphohydrolases (Ho et al. 1998). Members of this family that have been characterized biochemically include the triphosphatase components of the cellular capping systems of Saccharomyces cerevisiae (Bisaillon and Shuman 2001), Schizosaccharomyces pombe (Pei et al. 2001), Candida albicans (Pei et al. 2000), Plasmodium falciparum (Ho and Shuman 2001a; Takagi and Buratowski 2001), Trypanosoma brucei (Ho and Shuman 2001b; Gong et al. 2003), Encephalitozoon cuniculi (Hausmann et al. 2002), and Giardia lamblia (Hausmann et al. 2005), plus the triphosphatases of the Chlorella virus, poxvirus, and baculovirus mRNA capping systems (Myette and Niles 1996; Jin et al. 1998; Martins and Shuman 2001, 2003; Gong and Shuman 2002, 2003). The signature biochemical property of this enzyme family is the ability to hydrolyze NTPs to NDPs and Pi in the presence of manganese or cobalt. The defining primary structure features of the metal-dependent RNA triphosphatases are two glutamate-containing motifs that are required for catalysis by every family member and that comprise the metal-binding site.
The crystal structure of the S. cerevisiae RNA triphosphatase Cet1 bound to manganese and sulfate (a mimetic of the product complex with phosphate) revealed a novel tertiary structure in which the active site is situated within a topologically closed hydrophilic tunnel composed of eight antiparallel β-strands (Fig. 1; Lima et al. 1999). Extensive mutational analysis of yeast Cet1 and other family members identified a constellation of 15 side chains within the β-strands that are essential for triphosphatase activity in vitro and in vivo, 10 of which make direct or water-mediated contacts with the divalent cation or the sulfate anion (Bisaillon and Shuman 2001).
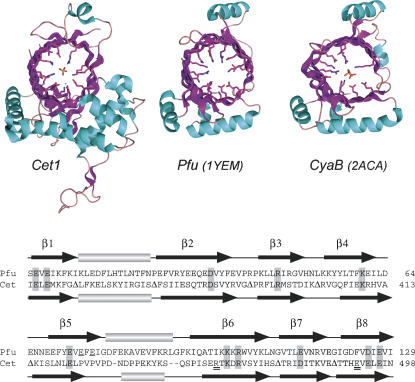
FIGURE 1.
The triphosphate tunnel fold of yeast Cet1 is recapitulated in archaeal and bacterial homologs. (Top panel) The tertiary structures of S. cerevisiae Cet1 (Lima et al. 1999; PDB 1D8H), a P. furiosus protein of unknown function (Yang et al. 2005; PBD 1YEM), and a putative V. parahemolyticus CyaB homolog (Kuzin et al. 2005; PBD 2ACA) are depicted as ribbon diagrams with β-strands colored magenta and helices colored cyan. The amino acids of Cet1 that emanate from the tunnel walls to coordinate the manganese (nonbonded cyan sphere) and sulfate ions in the active site of Cet1 are shown. Conserved hydrophilic residues in tunnels of the Pfu and CyaB structures are also shown. Two phosphate ions are present in the CyaB tunnel. (Bottom panel) The amino acid sequences of Cet1 and Pfu 1YEM were aligned based on the structural superposition above. Residues at the active site of Cet1 that are conserved in the Pfu tunnel are shaded. The two glutamates (Glu74 and Glu76) unique to the β5 strand of 1YEM are underlined. The Arg454–Glu492 pair that is unique to Cet1 and forms a salt bridge within the tunnel are indicated by double underlines. Peptide segments between the secondary structure elements of Cet1 that were omitted from the alignment are denoted by Δ.
The stark differences in the structure and mechanism of the RNA triphosphatases of unicellular eukarya and humans prompted the suggestion that RNA triphosphatase is a potential target for the discovery of antifungal and antiprotozoal drugs (Shuman 2001). The need for new therapeutics is most acute for protozoal illnesses such as malaria, where the increase in resistance of Plasmodium to previously effective drugs such as chloroquine has eroded disease control.
We had initially characterized the biochemical properties of the P. falciparum RNA triphosphatase Prt1 (PFC0985c) by expressing in bacteria an active fragment lacking the C-terminal 140 amino acids (Ho and Shuman 2001a). Purified recombinant Prt1 catalyzed manganese-dependent hydrolysis of the γ-phosphate of ATP and magnesium-dependent hydrolysis of the γ-phosphate of triphosphate-terminated RNA. A manual alignment of the Prt1 primary structure with other Cet1-like RNA triphosphatases hinted at the presence of putative counterparts of the eight strands that comprise the Cet1 triphosphate tunnel. The glutamate-containing motifs that likely form two of the tunnel strands are readily identified (and are underlined in Fig. 2). However, further structure prediction was confounded by the insertion of peptide segments, enriched in poly-asparagine tracts and acidic residues, that have no counterparts in other Cet1-like proteins (highlighted in gray in Fig. 2).
FIGURE 2.
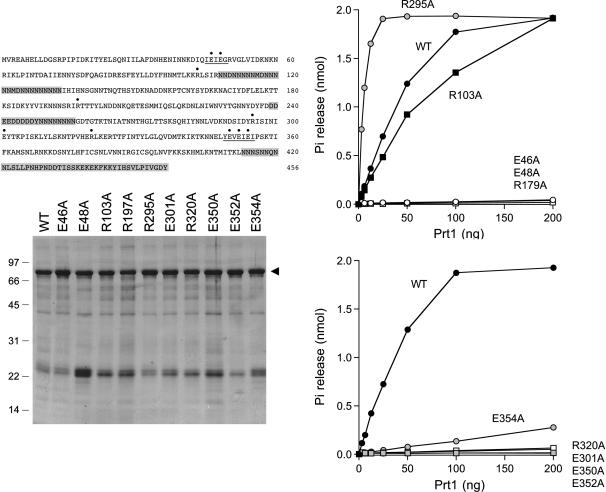
Triphosphatase activities of wild-type and mutant Prt1 proteins. (Top left panel) The sequence of the Prt1 polypeptide component of the His10/Smt3-Prt1 fusion is shown. The residues subjected to alanine substitution in the present study are indicated by •. The metal-binding motifs 45IEIEG49 and 349YEVEIEI355 are underlined. Segments of degenerate primary structure are shaded gray. (Bottom left panel) Aliquots (4 μg) of the Ni-agarose preparations of wild-type (WT) His10/Smt3-Prt1 and the Prt1-Ala mutant proteins were analyzed by electrophoresis through a 15% polyacrylamide gel containing 0.1% SDS. Polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (in kilodaltons) of marker proteins are indicated on the left. His10/Smt3-Prt1 is denoted by the arrowhead on the right. (Right panels) Reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 2 mM MnCl2, 0.2 mM [γ-32P]ATP, and either wild-type or mutant proteins as specified were incubated for 15 min at 30°C. 32Pi release is plotted as a function of input Prt1.
Here, we probed the active site of Prt1 by targeted mutagenesis and thereby identified eight residues essential for catalysis. The functional data were used to revise the secondary structure model so that the Prt1 counterparts of the Cet1 tunnel strands are now defined with a high degree of confidence. Also, we shed light on the evolution of the Cet1 family by noting that its heretofore unique tertiary structure and active site are recapitulated in a P. furiosus protein (PBD 1YEM) of unknown biochemical function. The Cet1-like archaeal protein exemplifies a so-called CYTH domain found in CyaB-like adenylate cyclases and mammalian thiamine triphosphatase (Iyer and Aravind 2002). Thus, Cet1 is the founder of a large and widely distributed enzyme superfamily, defined by the triphosphate tunnel, with functions not limited to RNA 5′ processing.
RESULTS AND DISCUSSION
Mutational mapping of the Prt1 active site
Ten amino acids of Prt1 were chosen for alanine-scanning: Glu46, Glu48, Arg103, Arg197, Arg295, Glu301, Arg320, Glu350, Glu352, and Glu354 (indicated by • over the sequence in Fig. 2). Wild-type Prt1 and the Prt1-Ala mutants were produced in Escherichia coli as His10/Smt3 fusions and isolated from soluble bacterial lysates by Ni-agarose chromatography (Fig. 2). Triphosphatase activity was gauged by the release of 32Pi from 0.2 mM [γ-32P]ATP in the presence of manganese. Enzyme titrations were performed for each of the Prt1 preparations (Fig. 3). The specific activities calculated from the slopes of the titration curves were normalized to that of wild-type Prt1 (defined as 100%). The values for the mutant proteins were as follows: E46A (0.6%); E48A (0.3%); R103A (66%); R197A (0.6%); R295A (660%); E301A (≤0.1%); R320A (1%); E350A (0.8%); E352A (0.1%); E354A (5%).
FIGURE 3.
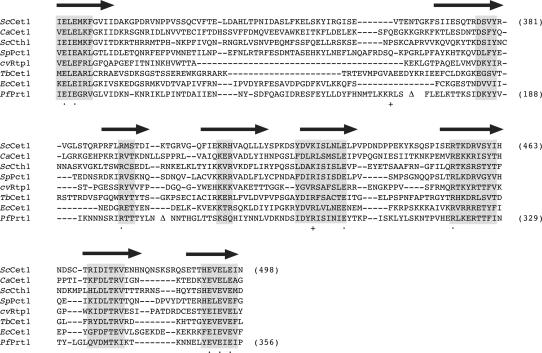
Revised primary structure alignment of Prt1 to other RNA triphosphatases. The amino acid sequence of P. falciparum Prt1 is aligned to the sequences of the catalytic domains of RNA triphosphatases S. cerevisiae Cet1, C. albicans CaCet1, S. cerevisiae Cth1, S. pombe Pct1, Chlorella virus cvRtp1, T. brucei Cet1, and E. cuniculi Cet1. Gaps in the alignment are indicated by dashes. Poly-asparagine inserts in Prt1 are omitted from the alignment and are denoted by Δ. The β-strands that form the triphosphate tunnel of Cet1 are denoted above the sequence. Peptide segments with the highest degree of conservation in all proteins are highlighted in shaded boxes. The eight side chains of Prt1 presently identified as essential by alanine scanning are denoted by • below the sequence. The nonessential Prt1 side chains Arg103 and Arg295 are indicated by + below the sequence.
The eight Prt1 side chains defined as essential by the alanine scan—Glu46, Glu48, Arg197, Glu301, Arg320, Glu350, Glu352, and Glu354—are the putative equivalents of essential Cet1 residues Glu305, Glu307, Arg393, Glu433, Arg454, Glu492, Glu494, and Glu496, respectively. These positions are denoted by • under the aligned sequences of fungal, viral, and protozoan tunnel-family RNA triphosphatases shown in Figure 3. The alignment of Prt1 is modified compared to that reported initially (Ho and Shuman 2001a) in light of the present finding that Arg103 (formerly predicted to define the third β-strand of the tunnel and to correspond to Cet1 Arg393) is clearly not essential for catalysis. In the revised structural alignment, Arg103 is located upstream of the newly assigned equivalents of the second and third β-strands of the Cet1 triphosphate tunnel (Fig. 3). Arg103 is separated from the second β-strand by a 68-amino-acid insert (denoted by Δ in Fig. 3) that includes a long poly-asparagine tract (Fig. 2). The assignments of the Prt1 counterparts of the other six strands of the tunnel are unchanged from the earlier model and are consistent with the present mutational data.
The roles of the eight essential Prt1 side chains can be inferred by reference to the Cet1 structure and available mutational data for Cet1 and other tunnel family members (Lima et al. 1999; Bisaillon and Shuman 2001; Gong and Shuman 2002). Four of the essential Prt1 glutamates—Glu46, Glu48, Glu352, Glu354—are predicted to bind the divalent cation, with Glu46, Glu48, and Glu354 coordinating the metal directly and Glu352 making a water-mediated metal contact. Our finding that alanine mutations of each of the four metal-binding glutamates abolish (Glu46, Glu48, Glu352) or severely reduce (Glu364) specific activity in manganese-dependent ATPase hydrolysis is consistent with the deleterious effects of conservative mutations at three of these positions (E48Q, E352D, and E354D) on Prt1 ATPase function noted by Takagi and Buratowski (2001). Glu301 is predicted to coordinate the water nucleophile for its attack on the γ phosphorus, potentially serving as a general base catalyst. Arg197 makes a bidentate contact to the γ phosphate oxygens and stabilizes the predicted pentacoordinate transition state. Arg320 and Glu350, located in the sixth and eighth β-strands, respectively, are predicted to form a salt bridge that stabilizes the tunnel architecture.
One of the mutations studied here, R295A, resulted in a sixfold increase in the ATPase activity of Prt1. Arg295 is predicted to reside within the fifth β-strand of the triphosphate tunnel and is the counterpart of Lys427 in Cet1. Cet1 Lys427 does not contact the metal or the sulfate ion within the triphosphate tunnel. The K427A mutation of Cet1 had no effect on triphosphatase activity in vitro or Cet1 function in vivo (Bisaillon and Shuman 2001), notwithstanding that this position is conserved as a lysine or arginine in all of the tunnel family members shown in Figure 3. Thus, the basis for the gain of function by the Prt1 R295A mutant is not obvious at present.
In summary, our mutational data for Prt1 engender an improved secondary structure model for this enzyme and build a compelling case for membership of Prt1 in the Cet1-like triphosphate tunnel family of RNA capping enzymes. The similarity of the active sites of fungal and Plasmodium RNA triphosphatases lends support to the idea that a mechanism-based inhibitor of one of the tunnel family members might display broad spectrum activity across the family, in which case it might be reasonable to direct efforts at identifying inhibitors of Plasmodium RNA triphosphatase, given that malaria is the most consequential human health problem caused by protozoan or fungal pathogens.
New clues to the evolution and distribution of Cet1-like proteins
The case for the tunnel family RNA triphosphatases as anti-infective targets rests primarily on the complete divergence in structures and mechanisms of the RNA triphosphatases of the unicellular pathogen and the mammalian host. In addition, the fact that psi-BLAST searches fail to identify a metazoan protein with primary structure similarity to the tunnel family of RNA triphosphatases suggests that (1) metazoans either have no homolog of Cet1-like enzymes, for example, because the tunnel-family RNA triphosphatase gene was lost after its replacement by a cysteine-phosphatase; or (2) homologs do exist in metazoa, but have diverged from the Cet1-like RNA triphosphatases to the point that they are no longer recognizable as such at the primary structure level. Either scenario might augur well for achieving selective inhibition of the pathogen-encoded metal-dependent RNA triphosphatases with minimal impact on host metabolism.
Yet, from the perspective of enzyme evolution, it is of clear interest to discern whether the signature Cet1-like enzyme fold arose uniquely in unicellular eukarya for the purpose of RNA capping, or if nature has exploited the triphosphate tunnel for other purposes in other taxa. Structural genomics projects are generating large numbers of crystal structures of proteins from diverse species in an effort to illuminate novel folds and occult evolutionary connections between protein families. Thus, it was remarkable that a DALI search (Holm and Sander 1993) of the Protein Data Bank against the 288-amino-acid protomer of the Cet1 crystal structure yielded Cet1 itself (Z score 44.9) and a single significant “hit” to the crystal structure of a 171-amino-acid P. furiosus protein of unknown function (Z score 9.3) deposited in PDB (1YEM) by Yang et al. (2005) from the Southeast Collaboratory for Structural Genomics. Although the primary structure conservation was low (14% identity in the DALI alignment), the folds were remarkably similar. A manual alignment of the structures shows that 1YEM recapitulates almost perfectly the eight-stranded β-barrel of the triphosphate tunnel of Cet1 (Fig. 1). The tunnels of Cet1 and 1YEM superimpose with an RMSD of 1.33 Å at 72 Cα positions. 1YEM also includes counterparts of two of the helices that comprise the globular pedestal on which the Cet1 tunnels rests and the single helix situated over the “roof” of the tunnel. It appears that the 1YEM protein is a minimized version of the triphosphate tunnel enzyme fold.
Although its biochemical function is not known, we predict that the Pfu protein is a metal-dependent phosphohydrolase, insofar as the hydrophilic interior of its tunnel resembles that of Cet1 (Fig. 1). Indeed, a structure-guided alignment of the Pfu 1YEM and yeast Cet1 amino acid sequences reveals side-chain identity or similarity at 11 of the residues that comprise the Cet1 active site (highlighted in Fig. 1). The conserved functional groups include all four acidic residues in strands β1 and β8 that bind the divalent cation, plus the putative glutamate general base in β5, and the basic side chains in strands β3 and β6 that coordinate the sulfate in the Cet1 structure. There are two notable differences between the tunnels of Cet1 and 1YEM. First, the Pfu protein has no counterparts of Cet1 residues Arg454 and Glu492, which are essential for the activity of the yeast triphosphatase (and the Plasmodium triphosphatase, as shown herein). Rather, the equivalent positions in 1YEM are occupied by isoleucine and phenylalanine, respectively. This difference testifies to the coevolution of the Arg–Glu pair that forms a salt bridge within the tunnel of the Cet1-like RNA triphosphatases. Second, the β5 strand of 1YEM extends further in the C-terminal direction than does the equivalent strand of Cet1. The 1YEM strand includes within its extra segment two glutamates (Glu74 and Glu76) not found in Cet1. The alternating sequence of glutamates and hydrophobic residues in the 71YEVEFEI77 β5-strand orients all three of the glutamates on the same face of the strand. Glu72 and Glu74 project into the tunnel. Although Glu72 is a candidate general base, as mentioned above, we speculate that Glu72 and Glu74 might also comprise a second metal-binding site within the tunnel of 1YEM.
Sequence-based searches for homologs of 1YEM immediately revealed it to be an example of a so-called CYTH domain, recently described by Iyer and Aravind (2002). The founders of this family are the bacterial adenylate cyclase enzyme CyaB (Sismeiro et al. 1998) and the mammalian enzyme thiamine triphosphatase (Lakaye et al. 2002). Whereas CYTH domains were predicted to exist in many organisms in all three domains of the universal phylogenetic tree, there was no relationship noted at the time between the CYTH domain and the Cet1-like RNA triphosphatases. Indeed, it was stated that CYTH proteins are not present in yeast (Iyer and Aravind 2002). Recently, a crystal structure of a Vibrio parahemolyticus homolog of CyaB was deposited in the PDB (2ACA) by the Northeast Structural Genomics Consortium (Kuzin et al. 2005). The CyaB structure adheres to the Cet1-like triphosphate tunnel fold and includes two phosphate ions in the active site (Fig. 1). The tunnels of Cet1 and CyaB superimpose with an RMSD of 1.36 Å at 59 Cα positions.
In light of the now clear similarities between yeast Cet1, Pfu 1YEM, and CyaB 2ACA (the first CYTH domains for which structures are available), and the fact that Cet1, CyaB, and thiamine triphosphatase are all metal-dependent enzymes that act on triphosphate-containing substrates, we surmise that the triphosphate tunnel fold first described for yeast Cet1 (Lima et al. 1999) is the prototype of a larger enzyme superfamily that includes the CYTH branch. This superfamily, which we name “triphosphate tunnel metalloenzyme” to reflect the defining structural features, is apparently of deep evolutionary origin. Thus, the Cet1 fold did not arise de novo in unicellular eukarya with the emergence of caps as the defining feature of eukaryotic mRNA. Instead, the Cet1-like RNA triphosphatases would have evolved by incremental changes to a precursor triphosphate tunnel metalloenzyme protein (perhaps of archaeal origin), which eventually imparted biological specificity for RNA 5′-end processing.
MATERIALS AND METHODS
Expression and purification of recombinant Prt1
The bacterial expression plasmid pET-His10/Smt3-Prt1 encodes a catalytically active N-terminal domain of P. falciparum RNA triphosphatase (amino acids 1–456) fused in-frame to an N-terminal His10/Smt3 domain, consisting of a His10 leader peptide (MGHHHHHHHHHHSSGHIEGRH) followed by the 98-amino-acid S. cerevisiae Smt3 protein and a single serine. (Smt3 is the yeast ortholog of the small ubiquitin-like modifier SUMO.) pET-His10/Smt3-Prt1 was transformed into E. coli BL21(DE3). A 500-mL bacterial culture was grown at 37°C in Luria-Bertani medium containing 60 μg/mL kanamycin until the A 600 reached 0.5. The culture was placed on ice for 10 min, adjusted to 2% (v/v) ethanol and 0.4 mM isopropyl-1-thio-β-D-galactopyranoside, and then incubated at 17°C for 18 h with constant shaking. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All of the subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 20 mL of buffer A (50 mM Tris-HCl at pH 7.5, 0.25 M NaCl, 10% sucrose). Cell lysis was achieved by the addition of lysozyme and Triton X-100 to final concentrations of 50 μg/mL and 0.1%, respectively. The lysate was sonicated to reduce viscosity, and insoluble material was removed by centrifugation. The soluble extract was applied to a 1-mL column of nickel-nitrilotriacetic acid-agarose resin (QIAGEN) that had been equilibrated with buffer A containing 0.1% Triton X-100. The column was washed with 10 mL of the same buffer and then eluted stepwise with 3-mL aliquots of buffer B (50 mM Tris-HCl at pH 8.0, 0.25 M NaCl, 10% glycerol, 0.1% Triton X-100) containing 0, 50, 100, 200, and 500 mM imidazole. The polypeptide compositions of the column fractions were monitored by SDS-PAGE. The 68-kDa recombinant His10/Smt3-Prt1 polypeptide was recovered predominantly in the 200 mM imidazole fraction (containing ∼0.5 mg of protein).
Alanine mutagenesis of Prt1
Silent diagnostic restriction sites and single alanine substitutions were introduced into the Prt1 gene by two-stage overlap extension PCR. BamHI restriction fragments of the PCR-amplified mutated DNAs were inserted into the T7-based expression plasmid pET28-His10/Smt3 that had been digested with BamHI. The presence of the desired mutations was confirmed in each case by sequencing the entire insert. The occurrence of PCR-generated mutations outside the targeted region was thereby excluded. The His10/Smt3-tagged Prt1-Ala proteins were produced in E. coli and purified by nickel-agarose affinity chromatography as described above.
Determination of Prt1 protein concentration
The total protein concentrations of the Ni-agarose preparations of the recombinant Prt1 proteins were initially determined by using Bio-Rad dye reagent with bovine serum albumin as a standard. Aliquots (∼4 μg) of the protein preparations were then analyzed by SDS-PAGE in parallel with 1.27, 2.54, and 3.81 μg of BSA. The gel was fixed and stained with Coomassie blue dye. The concentrations of the His10/Smt3-Prt1 polypeptide concentrations were then determined by quantifying the staining intensities of the His10/Smt3-Prt1 and BSA polypeptides using a Digital Imaging and Analysis System from Alpha Innotech Corp. and interpolating the intensity of the His10/Smt3-Prt1 staining intensity to the BSA standard curve.
Nucleoside triphosphate phosphohydrolase assay
Reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 7.5), 5 mM DTT, 1 mM MnCl2, 0.2 mM [γ-32P]ATP, and serial twofold dilutions of either wild-type or mutant Prt1 proteins (0, 6.25, 12.5, 25, 50, 100, and 200 ng) were incubated for 15 min at 37°C. The reactions were quenched by adding 2.5 μL of 5 M formic acid. Aliquots of the mixtures were applied to polyethyleneimine-cellulose TLC plates, which were developed with 1 M formic acid, 0.5 M LiCl. 32Pi release was quantified by scanning the chromatogram with a Fujix imaging device. The specific activities were calculated from the slopes of the titration curves.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM52470 and a New Initiatives in Malaria Award from the Burroughs Wellcome Fund. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.119806.
REFERENCES
- Bisaillon M., Shuman S. Structure-function analysis of the active site tunnel of yeast RNA triphosphatase. J. Biol. Chem. 2001;276:17261–17266. doi: 10.1074/jbc.M100980200. [DOI] [PubMed] [Google Scholar]
- Changela A., Ho C.K., Martins A., Shuman S., Mondragon A. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 2001;20:2575–2586. doi: 10.1093/emboj/20.10.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Shuman S. Chlorella virus RNA triphosphatase: Mutational analysis and mechanism of inhibition by tripolyphosphate. J. Biol. Chem. 2002;277:15317–15324. doi: 10.1074/jbc.M200532200. [DOI] [PubMed] [Google Scholar]
- Gong C., Shuman S. Mapping the active site of vaccinia virus RNA triphosphatase. Virology. 2003;309:125–134. doi: 10.1016/s0042-6822(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Gong C., Martins A., Shuman S. Structure-function analysis of Trypanosoma brucei RNA triphosphatase and evidence for a two-metal mechanism. J. Biol. Chem. 2003;278:50843–50852. doi: 10.1074/jbc.M309188200. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Vivarès C.P., Shuman S. Characterization of the mRNA capping apparatus of the microsporidian parasite Encephalitozoon cuniculi . J. Biol. Chem. 2002;277:96–103. doi: 10.1074/jbc.M109649200. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Altura M.A., Witmer M., Singer S.M., Elmendorf H.G., Shuman S. Yeast-like mRNA capping apparatus in Giardia lamblia . J. Biol. Chem. 2005;280:12077–12086. doi: 10.1074/jbc.M412063200. [DOI] [PubMed] [Google Scholar]
- Ho C.K., Shuman S. A yeast-like mRNA capping apparatus in Plasmodium falciparum . Proc. Natl. Acad. Sci. 2001a;98:3050–3055. doi: 10.1073/pnas.061636198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.K., Shuman S. Trypanosoma brucei RNA triphosphatase. Antiprotozoal drug target and guide to eukaryotic phylogeny. J. Biol. Chem. 2001b;276:46182–46186. doi: 10.1074/jbc.M108706200. [DOI] [PubMed] [Google Scholar]
- Ho C.K., Pei Y., Shuman S. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J. Biol. Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distant matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Iyer L.M., Aravind L. The catalytic domains of thiamine triphosphatase and CyaB-like adenylyl cyclase define a novel superfamily of domains that bind organic phosphates. BMC Genomics. 2002;3 doi: 10.1186/1471-2164-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Dong W., Guarino L.A. The LEF-4 subunit of baculovirus RNA polymerase has RNA 5′-triphosphatase and ATPase activities. J. Virol. 1998;72:10011–10019. doi: 10.1128/jvi.72.12.10011-10019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin A.P., Abashidze M., Vorobiev S.M., Forouhar F., Chen Y., Acton T., Xiao R., Conover K., Ma L.C., Cunningham K.E., et al. Protein Data Bank 2ACA. 2005. X-ray structure of a putative adenylate cyclase Q87NV8 from Vibrio parahemolyticus at the 2.5 Å resolution. [Google Scholar]
- Lakaye B., Makarchikov A.F., Antunes A.F., Zorzi W., Coumans B., DePauw E., Wins P., Grisar T., Bettendorf L. Molecular characterization of a specific thiamine triphosphatase widely expressed in mammalian tissues. J. Biol. Chem. 2002;277:13771–13777. doi: 10.1074/jbc.M111241200. [DOI] [PubMed] [Google Scholar]
- Lima C.D., Wang L.K., Shuman S. Structure and mechanism of yeast RNA triphosphatase: An essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- Martins A., Shuman S. Mutational analysis of baculovirus capping enzyme Lef4 delineates an autonomous triphosphatase domain and structural determinants of divalent cation specificity. J. Biol. Chem. 2001;276:45522–45529. doi: 10.1074/jbc.M107615200. [DOI] [PubMed] [Google Scholar]
- Martins A., Shuman S. Mapping the triphosphatase active site of baculovirus mRNA capping enzyme Lef4 and evidence for a two-metal mechanism. Nucleic Acids Res. 2003;31:1455–1463. doi: 10.1093/nar/gkg244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myette J., Niles E.G. Characterization of the vaccinia virus RNA 5′-triphosphatase and nucleoside triphosphate phosphohydrolase activities: Demonstration that both activities are carried out at the same active site. J. Biol. Chem. 1996;271:11945–11952. doi: 10.1074/jbc.271.20.11945. [DOI] [PubMed] [Google Scholar]
- Pei Y., Lehman K., Tian L., Shuman S. Characterization of Candida albicans RNA triphosphatase and mutational analysis of its active site. Nucleic Acids Res. 2000;28:1885–1892. doi: 10.1093/nar/28.9.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Schwer B., Hausmann S., Shuman S. Characterization of Schizosaccharomyces pombe RNA triphosphatase. Nucleic Acids Res. 2001;29:387–396. doi: 10.1093/nar/29.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harb. Symp. Quant. Biol. 2001;66:301–312. doi: 10.1101/sqb.2001.66.301. [DOI] [PubMed] [Google Scholar]
- Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- Sismeiro O., Trotot P., Biville F., Vivares C., Danchin A. Aeromonas hydrophila adenylyl cyclase 2: A new class of adenylyl cyclases with thermophilic properties and sequence similarities to proteins from hyperthermophilic archaebacteria. J. Bacteriol. 1998;180:3339–3344. doi: 10.1128/jb.180.13.3339-3344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T., Buratowski S. A Plasmodium falciparum protein related to fungal RNA 5′-triphosphatases. Mol. Biochem. Parasitol. 2001;114:239–244. doi: 10.1016/s0166-6851(01)00254-7. [DOI] [PubMed] [Google Scholar]
- Takagi T., Moore C.R., Diehn F., Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- Yang H., Chang J., Shah A., Ng J.D., Liu Z.J., Chen L., Lee D., Tempel W., Praissman J.L., Line D., et al. Protein Data Bank 1YEM. 2005. Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus . [Google Scholar]