Abstract
Deleted in Azoospermia Associated Protein 1 (DAZAP1) is a ubiquitous RNA-binding protein highly expressed in the human and the mouse testes. It shows a dynamic subcellular localization during spermatogenesis, present predominantly in the nuclei of late-stage spermatocytes and round spermatids and translocated to the cytoplasm during spermatid elongation. To test the hypothesis that DAZAP1 shuttles between the nucleus and the cytoplasm, we studied the nuclear transport of DAZAP1 in somatic cells using immunostaining, heterokaryon formation, and mutagenesis. DAZAP1 is detected exclusively in the nucleus and has the ability to shuttle between the nucleus and the cytoplasm using a highly conserved 25 amino acid segment, designated ZNS, at its C terminus. ZNS shares no sequence homology with other known nuclear localization or export signals. Attachment of ZNS to a red fluorescent protein DsRed2 confers the nucleocytoplasmic shuttling ability to that protein. The nuclear localization of DAZAP1 depends on active transcription. In the presence of an RNA polymerase II inhibitor, DAZAP1 is retained in the cytoplasm. DAZAP1 colocalizes with hnRNP A1 and hnRNP C1 in the nucleus and is a component of the heterogeneous nuclear ribonucleoprotein particles. Our results suggest that DAZAP1 plays a key role in mRNA transport during spermatogenesis.
Keywords: RNA-binding protein, nucleocytoplasmic shuttling, DAZAP1
INTRODUCTION
Deleted in Azoospermia Associated Protein 1 (DAZAP1, also known as proline-rich RNA-binding protein [PRRP]) was initially identified through its interaction with the protein products of the Deleted in Azoospermia (DAZ) gene family on the Y chromosome that is frequently deleted in infertile men (Tsui et al. 2000). DAZAP1 orthologs were subsequently isolated from mice and Xenopus and showed a high degree of evolutionary conservation with the human protein (Dai et al. 2001; Zhao et al. 2001; Kurihara et al. 2004). DAZAP1 has a structure similar to those of the 2xRBD members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family, with two RNP-type RNA-binding domains (RBDs) at the N terminus (Dreyfuss et al. 1993; Akindahunsi et al. 2005). However, its C-terminal portion is rich in proline instead of the glycine found in the hnRNP A/B proteins. DAZAP1 binds strongly in vitro to RNA homopolymers and RNA molecules containing two conserved elements, AAUAG and GU1–3AG (Tsui et al. 2000; Hori et al. 2005). It is expressed in multiple human and mouse tissues, with the highest expression level found in the testis (Tsui et al. 2000; Dai et al. 2001). In mouse testes Dazap1 transcripts are detectable in the germ cells only, mainly in type-B spermatogonia and preleptotene spermatocytes (Vera et al. 2002). However, the majority of DAZAP1 protein does not appear until the mid-pachytene stage and shows a dynamic distribution during spermatogenesis. In pachytene spermatocytes, DAZAP1 is present predominatly in the nucleus. Its exclusion from the sex body that contains the transcriptionally inactive X and Y chromosomes suggests its involvement in active transcription. DAZAP1 remains in the nuclei of round spermatids, and relocates to the cytoplasm in elongating spermatids and stays there throughout spermiogenesis. The dynamic distribution of DAZAP1 in the germ cells raised the possibility that it shuttles between the nucleus and the cytoplasm.
Many proteins move between the nucleus and the cytoplasm to carry out their functions. They contain nuclear localization signals (NLSs) and nuclear export signals (NESs) that are recognized by the nuclear transport receptors to guide them through the nuclear pore complex (for review, see Nakielny and Dreyfuss 1999). These transport signals could be short peptide sequences, such as the classical mono- and bipartite basic NLSs and the leucine-rich NESs, or large protein domains or even multicomponents. In addition to the one-way signals, a number of RNA-binding proteins, such as hnRNP A1, hnRNP K, and HuR, carry bidirectional shuttling signals that serve for both nuclear localization and export (Michael et al. 1995, 1997; Fan and Steitz 1998). These shuttling signals are 30–80 amino acid residues in length and bear little sequence homology. The nuclear localization signal activities of most, if not all, of these shuttling signals are transcription sensitive. Thus when the activity of RNA polymerase II is inhibited, the proteins are retained in the cytoplasm. These shuttling RNA-binding proteins usually are involved in mRNA export or mRNA stability.
Here we report our study on the nuclear transport of the human DAZAP1. We found that DAZAP1 is associated with the hnRNP particles in the nucleus and shuttles between the nucleus and the cytoplasm. We located a novel nucleocytoplasmic shuttling signal at the C terminus of DAZAP1 and identified several amino acid residues that are essential for its activity. Our results support a role of DAZAP1 in mRNA transport during spermatogenesis.
RESULTS
Nuclear localization of DAZAP1
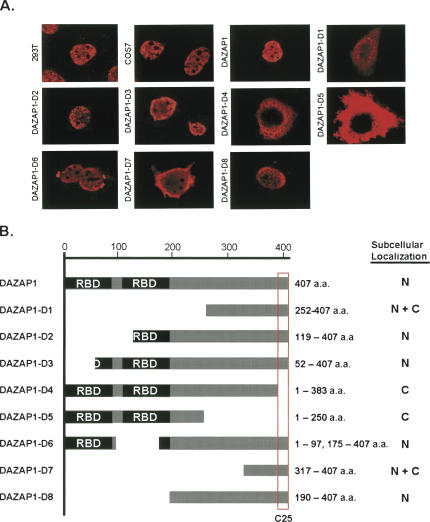
Our previous study showed that in mouse testes the 45-kDa DAZAP1 was present predominantly in the nuclei of pachytene spermatocytes and round spermatids, and relocated to the cytoplasm of elongating and elongated spermatids (Vera et al. 2002). To determine the subcellular localization of DAZAP1 in somatic cells, we performed immunofluorescence staining of the endogenous DAZAP1 in both human kidney epithelial 293T cells and monkey COS7 cells using a DAZAP1-specific antibody. In both cell types, the staining was restricted to the nuclei (Fig. 1A). The signals were excluded from the nucleoli and appeared as speckles, suggesting the association of DAZAP1 with specific nuclear structures. We next mapped the region required for DAZAP1 nuclear localization by constructing expression vectors encoding either intact or truncated DAZAP1 linked at the N terminus to an Xpress epitope (Fig. 1B). Immunostaining of COS7 and 293T cells transfected with the vectors yielded similar results. The nuclear staining pattern of the Xpress-tagged DAZAP1 (Fig. 1A) was similar to that of the endogenous protein, indicating that addition of the epitope did not affect the subcellular localization of the protein. The expression patterns of DAZAP1 derivatives, summarized in Figure 1B, indicated that the segment containing the last 25 amino acid residues at the C-terminal end, designated C25, was required for nuclear localization. C25 was present in derivatives D2, D3, D6, and D8, which were restricted to the nuclei, and absent from D4 and D5, which were found only in the cytoplasm. The presence of D1 and D7 in both the cytoplasm and the nucleus is probably due to their small size (<18 kDa), which allows passive diffusion through the nuclear pore complexes (Fenaroli et al. 2004).
FIGURE 1.
Nuclear localization of DAZAP1. (A) Immunostaining of DAZAP1 and its derivatives. The two top left panels show the endogenous DAZAP1 in 293T and COS7 cells, respectively, detected with an anti-DAZAP1 antibody. The remaining panels show the expression of Xpress-tagged DAZAP1 and its derivatives in COS7 cells transfected with the respective expression vectors as indicated at the left side of each panel. The structures of DAZAP1 and its derivatives are shown in B. The expressed proteins were detected using an anti-Xpress antibody, and the locations of the signals are summarized in B. (B) Structures of DAZAP1 and its derivatives and their subcellular localization. DAZAP1 contains two RNA-binding domains (RBDs) at the N-terminal portion and a proline-rich C-terminal portion. The C25 segment that is required for the nuclear localization of DAZAP1 is boxed. (N) nuclear, (C) cytoplasmic.
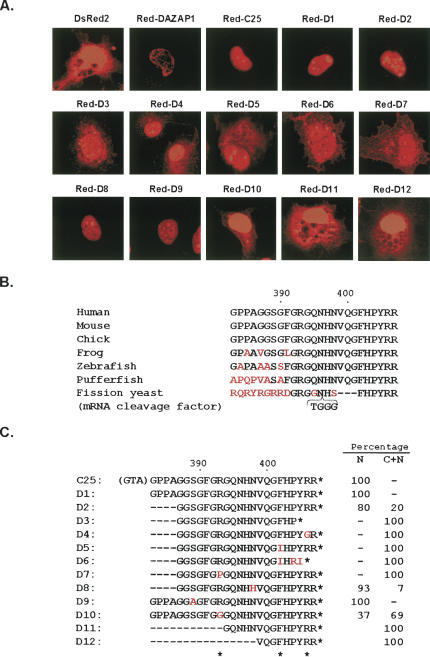
To investigate whether C25 contained a nuclear localization signal, we used a derivative of the Discosoma sp. red fluorescent protein, DsRed2, as a reporter (Matz et al. 1999). Cells transfected with the empty pDsRed2-C1 vector expressed the 27-kDa DsRed2 in both the nucleus and the cytoplasm, with the contour of the nucleus barely visible (Fig. 2A). A fusion between DsRed2 and the intact DAZAP1 localized the protein to the nucleus with a distribution pattern similar to that of the endogenous DAZAP1. Attachment of C25 to the C terminus of DsRed2 also conferred nuclear localization to the protein, indicating that C25 was sufficient for nuclear localization. Red-C25 had a more homogeneous nuclear distribution, suggesting that it was present in the nucleoplasm and no longer associated with specific nuclear entities. The results indicate that C25 contains a NLS that is capable of targeting a heterologous protein to the nucleus.
FIGURE 2.
Mutagenesis analyses of the nuclear localization signal of DAZAP1. (A) Subcellular localization of the red fluorescent protein DsRed2 and its fusions with C25 and its derivatives in COS7 cells transfected with the respective expression vectors. (B) Evolutionary conservation of the C25 sequence. Sequences at the C termini of putative DAZAP1 orthologs in the various animals as well as that at the C terminus of the S. pombe mRNA cleavage factor protein are shown, with the positions in the human sequence indicated at the top. Chimpanzee, bovine, and rat DAZAP1 have the same C25 sequence as that of the humans and the mice. Different amino acids are shown in red. (C) Sequences of C25 and its derivatives in the DsRed2 fusion proteins. The first three residues shown in parentheses in front of C25 are encoded by the polylinker of the vector. These residues are removed from all the derivatives. The amino acid residues essential for nuclear localization are indicated by asterisks at the bottom. The subcellular localization of the fusion proteins are indicated at the right. The percentages of cells with nuclear restricted expression (N) and both cytoplasmic and nuclear expression (C + N) are indicated. For fusion proteins D2, D8, and D10, which have two different expression patterns, the patterns present in the majority of cells are shown in A.
We next used mutagenesis to identify the amino acid residues within C25 that are essential for nuclear localization. C25 bears no sequence similarity to other NLS or shuttling signals. Its sequence is highly conserved, and DAZAP1 orthologs in all mammals as well as chicken have the same C25 sequence at their C termini (Fig. 2B). Even the mRNA cleavage factor protein in the fission yeast Schizosaccharomyces pombe contains the same six amino acids at the end, suggesting the importance of these terminal residues. We generated a series of deletions and substitutions within the C25 segment of Red-C25 and studied their effects on nuclear localization (Fig. 2A,C). Most derivatives of Red-C25 were either confined to the nucleus or distributed in both the nucleus and the cytoplasm. However, in cells expressing Red-D2, Red-D8, and Red-D10, both patterns of expression were observed. The cells with both nuclear and cytoplasmic staining in general had much weaker signals in the cytoplasm, and some of them could represent cells that have just finished mitotic division, with the Red fusion proteins not completely transported into the daughter cell nuclei (Pinol-Roma and Dreyfuss 1991). The mutagenesis results are summarized in Figure 2C. In Red-C25 the polylinker of the vector added a tripeptide sequence G-T-A in front of C25 that is similar to the G-S-G sequence right in front of C25 in DAZAP1. Deletion of this tripeptide (in D1) or four additional residues from the N terminus of C25 (in D2) did not significantly affect its activity. However, deletion of 11 or more residues from the N terminus (in D11 and D12) or three residues from the C terminus (in D3) caused the fusion protein to appear in the cytoplasm. Single amino acid changes at R393 (in D7 and D10), F402 (in D5), and R406 (in D4) also impaired nuclear localization of the fusion proteins, whereas changes at S389 (D9) and N398 (D8) had few effects. Our analyses thus identified three residues, R393, F402, and R406, that are essential for the NLS activity.
Nucleocytoplasmic shuttling of DAZAP1
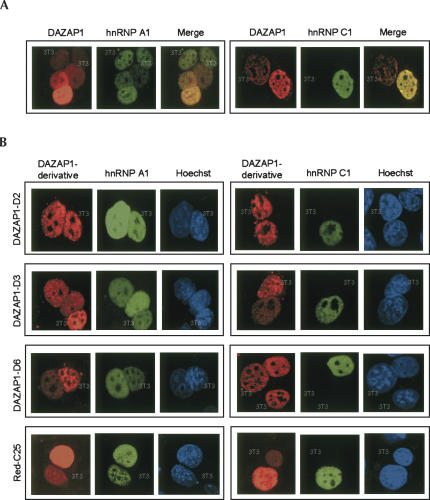
We used the well-established interspecies heterokaryon assay to investigate whether DAZAP1 is able to shuttle between the nucleus and the cytoplasm (Pinol-Roma and Dreyfuss 1992). We fused mouse 3T3 cells with human HeLa cells expressing Xpress-tagged DAZAP1 and GFP-hnRNP A1 or GFP-hnRNP C1 in the absence of new protein synthesis, and detected the presence of DAZAP1 in the mouse nuclei by immunostaining. HnRNP A1 and hnRNP C1, which possess and lack nucleocytoplasmic shuttling ability, respectively, served as controls. As shown in Figure 3A, DAZAP1 and hnRNP A1, but not hnRNP C1, were detected in the mouse nuclei that could be easily distinguished from the HeLa nuclei by their characteristic Hoechst staining, indicating that DAZAP1 could shuttle between the nucleus and the cytoplasm. DAZAP1 derivatives with exclusive nuclear localization, such as D2, D3, and D6, as well as Red-C25, were also found in the mouse nuclei and thus could shuttle too (Fig. 3B). Our results show that C25 contains both nuclear localization and nuclear export signal. We designated this nucleocytoplasmic shuttling signal “ZNS.”
FIGURE 3.
Nucleocytoplasmic shuttling of DAZAP1. Heterokaryons between mouse 3T3 cells and HeLa cells expressing Xpress-tagged DAZAP1 (A) or its derivatives (B), and either GFP-hnRNP A1 or GFP-hnRNP C1 are shown. Pictures within a frame show the same set of nuclei stained for the Xpress epitope (red), GFP (green), and DNA (blue). The nuclei of 3T3 cells are indicated. The presence of DAZAP1 and HnRNP A1 but not hnRNP C1 in the mouse nuclei indicates that the former two proteins can shuttle between the nucleus and the cytoplasm.
Transcription-dependent nuclear localization of DAZAP1
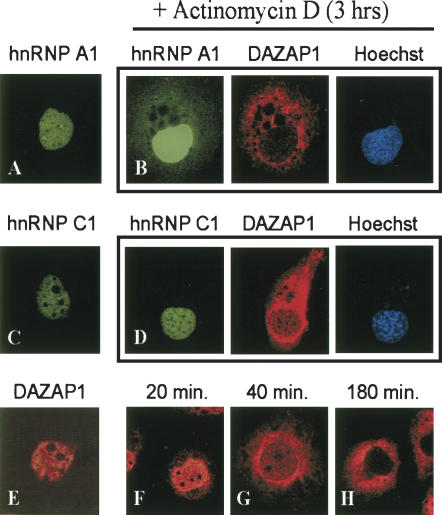
During spermatogenesis, DAZAP1 is in the nuclei of round spermatids and relocates to the cytoplasm in elongating spermatids when the nuclear transcription is shutting down. It is also reported that the nuclear localization of hnRNP A1 and HuR, but not hnRNP C1, depends on active transcription in the nucleus (Pinol-Roma and Dreyfuss 1991; Fan and Steitz 1998). We therefore studied the effect of actinomycin D (5 μg/mL), an inhibitor of RNA polymerase II, on the nuclear localization of DAZAP1. COS7 cells transfected with expression vectors of either GFP-hnRNP A1 (Fig. 4A) or GFP-hnRNP C1 (Fig. 4C) expressed the proteins exclusively in the nucleus together with the endogenous DAZAP1 (Fig. 4E). After treatment with actinomycin D for 3 h, both hnRNP A1 and DAZAP1, but not hnRNP C1, appeared in the cytoplasm (Fig. 4B,D). Similar results were obtained when the cells were simultaneously treated with cycloheximide to inhibit protein synthesis (data not shown). The cells being examined were well separated from others and unlike newly divided cells. Thus the cytoplasmic hnRNP A1 and DAZAP1 observed were probably those exported from the nucleus and accumulated in the cytoplasm during the course of cycloheximide treatment. A comparison of the signals in the nucleus with that in the cytoplasm indicates that DAZAP1 has a much faster export rate than hnRNP A1 (Fig. 4B). At the end of the 3-h treatment, the vast majority of hnRNP A1 remained in the nucleus whereas most DAZAP1 was present in the cytoplasm. An additional time-course study showed that at 40 min after the addition of cycloheximide, more than half of nuclear DAZAP1 had already exited to the cytoplasm (Fig. 4G).
FIGURE 4.
Transcription-dependent nuclear localization of DAZAP1. COS7 cells transfected with pEGFP-hnRNP A1 (A,B) or pEGFP-hnRNP C1 (C,D) were treated with (B,D) or without (A,C) actinomycin D, and the expression of GFP fusion proteins and endogenous DAZAP1 was examined. Pictures in a frame show the same cells. E–H show the accumulation of DAZAP1 in the cytoplasm at various time points after actinomycin D treatment.
Association of DAZAP1 with the hnRNP particles
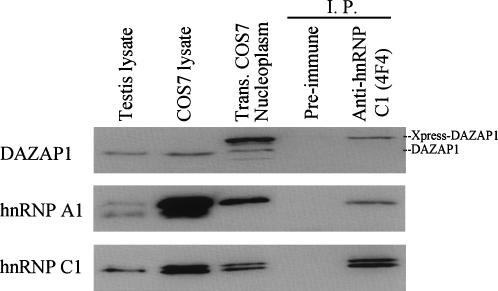
DAZAP1 has a nuclear staining pattern partially overlapping those of hnRNP A1 and hnRNP C1, raising the possibility that it is associated with the hnRNP particles (Fig. 3A). We tested the possibility by isolating hnRNP particles from the nucleoplasm of COS7 cells expressing Xpress-tagged DAZAP1 using the anti-hnRNP C1 antibody 4F4 (Pinol-Roma et al. 1988). Western blot analyses showed the presence of Xpress-DAZAP1 as well as hnRNP A1 and hnRNP C1 in the isolated hnRNP particles (Fig. 5). These proteins could not be precipitated with a preimmune antibody. A reciprocal experiment using the anti-DAZAP1 antibody failed to bring down hnRNP A1 or hnRNP C1 from the nucleoplasmic fraction (data not shown). It is possible that the C terminus of DAZAP1 is buried inside the hnRNP particles, unavailable to the binding of the antibody.
FIGURE 5.
Association of DAZAP1 with hnRNP particles. HnRNP particles were immunoprecipitated from the nucleoplasm of COS7 cells transfected with an expression vector of Xpress-tagged DAZAP1 and Western blotted with antibodies against the various proteins shown at the left. Preimmune serum was used as a control for the specificity of immunoprecipitation.
DISCUSSION
DAZAP1 has a dynamic subcellular distribution during spermatogenesis (Tsui et al. 2000; Dai et al. 2001; Vera et al. 2002). Because it was difficult to study nuclear trafficking in germ cells, we studied it in somatic cells that express a significant amount of DAZAP1. Our results show that DAZAP1 is present in the nucleus of somatic cells as a component of the hnRNP particles and has the ability to shuttle between the nucleus and the cytoplasm using a 25–amino acid ZNS at its C terminus. Our conclusion contradicts a previous report stating that the entire C-terminal half of DAZAP1 was required for nuclear localization (Kurihara et al. 2004). In that report, the investigators monitored the expression of fusion proteins between DAZAP1 fragments and a single copy of GFP; such an approach has been found recently to be unreliable (Beetz et al. 2004). However, our approach using tagged proteins overexpressed in cultured cells may not truthfully mimic the in vivo situation either. The final proof awaits the generation and characterization of mice with a knock-in Dazap1 gene containing mutations in the ZNS sequence. ZNS represents a novel nucleocytoplasmic shuttling signal that shares little sequence similarity with other characterized shuttling signals such as the M9 domain of hnRNP A1 (Siomi and Dreyfuss 1995), KNS of hnRNP K (Michael et al. 1997), or HNS of HuR (Fan and Steitz 1998), though ZNS is rich in glycine similar to M9. ZNS contains three arginine residues at two clusters separated by 12 residues, and two of the residues have been shown to be important for the nuclear localization activity. This structure of ZNS is similar to that of the typical bipartite basic NLS that contains two essential clusters of basic amino acids (Robbins et al. 1991). However, the spacer in the latter usually can be mutated without affecting its activity, whereas substitution of a phenylalanine residue within the “spacer” of ZNS abolishes its activity. DAZAP1 relies on ZNS for its nuclear transport, and the process is likely regulated by its binding to RNA since in the absence of new nuclear RNA synthesis DAZAP1 is retained in the cytoplasm. Additional work is required to identify the nuclear transport receptors and the pathways by which the DAZAP1 passes through the nuclear pores. The mechanisms that regulate DAZAP1 nuclear transport also await elucidation, and they could involve post-translational modification of DAZAP1 such as phosphorylation, arginine methylation, or sumoylation (Wood et al. 2003; Xu and Henry 2004; Poon and Jans 2005).
DAZAP1 has a 2xRBD structure similar to that of hnRNP A1 and is present in the nucleus. It is therefore no surprise that DAZAP1 is a component of the hnRNP particles, and its escape from identification in previous studies could be due to its low abundance in HeLa cells (Pinol-Roma et al. 1988). The observation that the nuclear staining of DAZAP1 only partially overlapped those of hnRNP A1 and hnRNP C1 is not unexpected, since the various hnRNP proteins do not exist in a fixed stoichiometry in different tissues (Kamma et al. 1995). Several hnRNP proteins shuttle between the nucleus and the cytoplasm and participate in mRNA transport (Dreyfuss et al. 1993). Our results also support a role for DAZAP1 in mRNA export. DAZAP1 was originally identified through its interaction with DAZ and DAZL, two closely related male germ-cell-specific RNA-binding proteins involved in the regulation of protein synthesis (Tsui et al. 2000; Collier et al. 2005; Pan et al. 2005; Reynolds et al. 2005). The association between DAZAP1 and DAZ/DAZL appears to be transient, since DAZ and DAZL are present most abundantly in the cytoplasm of pachytene spermatocytes when the majority of DAZAP1 is present in the nucleus (Ruggiu et al. 1997; Reijo et al. 2000). It is possible that DAZAP1 transports its mRNA cargos from the nuclei to the cytoplasm and hands them over to DAZ and DAZL for protein synthesis. Numerous putative RNA substrates/downstream genes of DAZ and DAZL have been identified (Venables et al. 2001; Jiao et al. 2002; Fox et al. 2005; Reynolds et al. 2005). Whether DAZAP1 mediated the transport of these RNA substrates remained to be determined.
It is interesting that the nuclear localization of DAZAP1 is transcription-dependent. This is consistent with the observation that DAZAP1 is found in the cytoplasm of elongating spermatids when the nuclear transcription is turning off (Vera et al. 2002). Additional proteins that are involved in the regulation of transcription, such as ACT (activator of CREM in the testis) and the spermatid-expressed kinesin KIF17b, also relocate from the nucleus to the cytoplasm in elongating spermatids (Kimmins et al. 2004; Hogarth et al. 2005). The mechanisms underlying the cytoplasmic accumulation of DAZAP1 during transcription inhibition in somatic cells and at spermatid elongation in the testis could be different, however. In somatic cells, the shuttling DAZAP1 is likely prevented from re-entering into the nucleus when there are no new mRNA cargos to be transported. On the other hand, the cytoplasmic accumulation of DAZAP1 and other nuclear proteins in elongating spermatids may be for a different reason. This is at a time when the nucleus is unloading unnecessary proteins in preparation for DNA packaging and dramatic reduction in size to transform into the sperm head. There is a need for active transport of most proteins out of the nucleus since the nucleus apparently does not have the capacity to degrade these proteins. The exported nuclear proteins together with most cytoplasmic components are eventually disposed as residue bodies that are phagocytized by the Sertoli cells (Kerr and de Kretser 1974). Very little is known about the regulation of the nuclear export in elongating and elongated spermatocytes and the consequences of its deregulation. Future studies on the regulation of nuclear transport of DAZAP1 may provide a tool to investigate this process during spermiogenesis.
MATERIALS AND METHODS
Antibodies
The anti-DAZAP1 antibody was generated by immunizing a goat with the last 19 amino acid residues of the human DAZAP1 (Dai et al. 2001). Other antibodies used in the study were purchased, including the mouse monocloncal anti-Xpress antibody (Invitrogen), rhodamine conjugated donkey antibodies against mouse and goat IgG (Santa Cruz Biotechnology), mouse monoclonal [4B10] to hnRNP A1 (Novus Biologicals), and mouse monoclonal [4F4] to hnRNP C1 (Abcam).
Expression vectors
pDAZAP1-Xpress was constructed by PCR amplification of the coding region of a human DAZAP1 cDNA clone (Tsui et al. 2000) followed by insertion of the fragment in-frame into the EcoRV and XbaI sites of pcDNA4/His A (Invitrogen), which added an Xpress epitope at the N terminus of the protein. To construct expression vectors for truncated DAZAP1, pDAZAP1-Xpress was cut open with various restriction enzymes and religated with or without treatment with the mung bean nuclease (MBN) or the Klenow fragment of Escherichia coli DNA polymerase I (KF) as for D1: BstXI + PstI, MBN; D2: EcoRI; D3: BstXI + Xho I, MBN; D4: ApaI; D5: SbfI + ApaI, MBN; and D7: SmaI + EcoRI, KF. For D6 and D8, the coding region of DAZAP1 was first cloned into pUC19 to make deletions (D6: EagI, KF, NsiI, MBN; D8: NsiI + XbaI, MBN) before recloning back into pcDNA4/His A. Plasmid pRed-DAZAP1 was constructed by ligating a KF-treated BamHI + XbaI fragment of pDAZAP1-Xpress into the BamHI site of pDsRed2-C1 (Clontech) after filling up the ends of the vector with KF. The construct was digested with ApaI and religated to generate pRed-C25. All the constructs were verified by DNA sequencing across the junctions and by in vitro synthesis (using the TNT coupled reticulocyte lysate system, Promega) of proteins of the expected size that could be immunoprecipitated by the antibodies. Plasmids pEGFP-hnRNP A1 and pEGFP-hnRNP C1 were gifts from Dr. Joan Steitz at Yale University (Lai et al. 2003).
Mutagenesis
Single-base substitutions were introduced into pRed2-C25 using the QuikChange site-directed mutagenesis kit from Stratagene. The method is based on the sensitivity of the template plasmid DNA, but not the PCR products of primers containing the desired mutations, to the digestion of the methylation-sensitive restriction enzyme DpnI. Deletions within C25 were generated by first introducing new restriction sites within the sequence followed by restriction digestion and religation.
Cell transfection and expression detection
Human 293T cells and monkey COS7 cells were grown on glass slides in 24-well plates and transfected with the various expression vectors using lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Forty-eight hours after transfection, the cells were fixed in 3% formaldehyde and permeabilized with 0.5% Triton X-100. The expression of Xpress-tagged DAZAP1 and its derivatives was detected by incubating the fixed cells with a mouse monocloncal anti-Xpress antibody (Invitrogen) followed by a secondary rhodamine-conjugated donkey anti-mouse IgG antibody (Santa Cruz Biotechnology). The cells were counterstained with Hoechst 33,258 (Sigma) before being mounted and photographed using a Bio-Rad Radiance 2100 laser scanning confocal microscope. The expression of GFP and DsRed2 fusion proteins was examined directly using the proper fluorescent light. Expression of the endogenous DAZAP1 was detected using a goat anti-DAZAP1 antibody followed by a secondary rhodamine-conjugated donkey anti-goat IgG antibody.
Heterokaryon assays
The heterokaryon assays were carried out according to a published procedure (Lai et al. 2003). Briefly, HeLa cells were transfected with a DAZAP1 expression vector together with either pEGFP-hnRNP A1 or pEGFP-hnRNP C1. Forty-eight hours after transfection, NIH 3T3 cells were added to the culture. The cells were treated with cycloheximide at 50 μg/mL for 3 h and at 100 μg/mL for 0.5 h to inhibit new protein synthesis before they were induced to fuse by the treatment of 50% polyethylene glycol. After another 3 h of incubation in the presence of cycloheximide (100 μg/mL), the cells were fixed and stained with antibodies as described in the previous section.
Inhibition of transcription
Cells were treated with actinomycin D to inhibit transcription by RNA polymerase II according to a published protocol (Michael et al. 1997). Briefly, COS7 cells were transfected with pEGFP-hnRNP A1 or pEGFP-hnRNP C1. After 2 d of growth, the cells were treated with actinomycin D (5 μg/mL) alone or together with cycloheximide (100 μg/mL) for 3 h at 37°C before being fixed for immunostaining of the endogenous DAZAP1 and detection of the GFP fusion proteins.
Isolation of the hnRNP particles
Immunopurification of the hnRNP particles was carried out according to Pinol-Roma et al. (1988). Briefly, COS7 cells were transfected with pDAZAP1-Xpress and harvested 48 h after transfection. The cells were lysed by passing through a 25-gauge needle, and the nuclei were collected by centrifugation and lysed by sonication. The nuclear lysate was cleared of chromatin and nucleoli by centrifugation on top of a 30% sucrose cushion. The resulting nucleoplasm was used in the immunopurification of hnRNP particles using a mouse monoclonal antibody [4F4] against hnRNP C1.
ACKNOWLEDGMENTS
We thank Joan Steitz for pEGFP-hnRNP A1 and pEGFP-hnRNP C1, Woan-Yuh Tarn for helpful advice and critical reading of the manuscript, and Kuan-Yu Chou for assistance with the confocal microscope. The work was supported by the Academia Sinica in Taiwan.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.42206.
REFERENCES
- Akindahunsi A.A., Bandiera A., Manzini G. Vertebrate 2xRBD hnRNP proteins: A comparative analysis of genome, mRNA and protein sequences. Comput. Biol. Chem. 2005;29:13–23. doi: 10.1016/j.compbiolchem.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Beetz C., Brodhun M., Moutzouris K., Kiehntopf M., Berndt A., Lehnert D., Deufel T., Bastmeyer M., Schickel J. Identification of nuclear localisation sequences in spastin (SPG4) using a novel Tetra-GFP reporter system. Biochem. Biophys. Res. Commun. 2004;318:1079–1084. doi: 10.1016/j.bbrc.2004.03.195. [DOI] [PubMed] [Google Scholar]
- Collier B., Gorgoni B., Loveridge C., Cooke H.J., Gray N.K. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai T., Vera Y., Salido E.C., Yen P.H. Characterization of the mouse Dazap1 gene encoding an RNA-binding protein that interacts with infertility factors DAZ and DAZL. BMC Genomics. 2001;2 doi: 10.1186/1471-2164-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Fan X.C., Steitz J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaroli A., Vujanac M., De Cesare D., Zimarino V. A small-scale survey identifies selective and quantitative nucleo-cytoplasmic shuttling of a subset of CREM transcription factors. Exp. Cell Res. 2004;299:209–226. doi: 10.1016/j.yexcr.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Fox M., Urano J., Reijo Pera R.A. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics. 2005;85:92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Hogarth C., Itman C., Jans D.A., Loveland K.L. Regulated nucleocytoplasmic transport in spermatogenesis: A driver of cellular differentiation? Bioessays. 2005;27:1011–1025. doi: 10.1002/bies.20289. [DOI] [PubMed] [Google Scholar]
- Hori T., Taguchi Y., Uesugi S., Kurihara Y. The RNA ligands for mouse proline-rich RNA-binding protein (mouse Prrp) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 2005;33:190–200. doi: 10.1093/nar/gki153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Trifillis P., Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol. Reprod. 2002;66:475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- Kamma H., Portman D.S., Dreyfuss G. Cell type-specific expression of hnRNP proteins. Exp. Cell Res. 1995;221:187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]
- Kerr J.B., de Kretser D.M. Proceedings: The role of the Sertoli cell in phagocytosis of the residual bodies of spermatids. J. Reprod. Fertil. 1974;36:439–440. doi: 10.1530/jrf.0.0360439. [DOI] [PubMed] [Google Scholar]
- Kimmins S., Kotaja N., Davidson I., Sassone-Corsi P. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction. 2004;128:5–12. doi: 10.1530/rep.1.00170. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe H., Kawaguchi A., Hori T., Mishiro K., Ono M., Sawada H., Uesugi S. Dynamic changes in intranuclear and subcellular localizations of mouse Prrp/DAZAP1 during spermatogenesis: The necessity of the C-terminal proline-rich region for nuclear import and localization. Arch. Histol. Cytol. 2004;67:325–333. doi: 10.1679/aohc.67.325. [DOI] [PubMed] [Google Scholar]
- Lai M.C., Kuo H.W., Chang W.C., Tarn W.Y. A novel splicing regulator shares a nuclear import pathway with SR proteins. EMBO J. 2003;22:1359–1369. doi: 10.1093/emboj/cdg126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., Lukyanov S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Choi M., Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Eder P.S., Dreyfuss G. The K nuclear shuttling domain: A novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Pan H.A., Lin Y.S., Lee K.H., Huang J.R., Lin Y.H., Kuo P.L. Expression patterns of the DAZ-associated protein DAZAP1 in rat and human ovaries. Fertil. Steril. 2005;84(Suppl. 2):1089–1094. doi: 10.1016/j.fertnstert.2005.03.075. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Choi Y.D., Matunis M.J., Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes & Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Poon I.K., Jans D.A. Regulation of nuclear transport: Central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Reijo R.A., Dorfman D.M., Slee R., Renshaw A.A., Loughlin K.R., Cooke H., Page D.C. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol. Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- Reynolds N., Collier B., Maratou K., Bingham V., Speed R.M., Taggart M., Semple C.A., Gray N.K., Cooke H.J. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum. Mol. Genet. 2005;14:3899–3909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- Robbins J., Dilworth S.M., Laskey R.A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Ruggiu M., Speed R., Taggart M., McKay S.J., Kilanowski F., Saunders P., Dorin J., Cooke H.J. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- Siomi H., Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui S., Dai T., Roettger S., Schempp W., Salido E.C., Yen P.H. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- Venables J.P., Ruggiu M., Cooke H.J. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 2001;29:2479–2483. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera Y., Dai T., Hikim A.P., Lue Y., Salido E.C., Swerdloff R.S., Yen P.H. Deleted in azoospermia associated protein 1 shuttles between nucleus and cytoplasm during normal germ cell maturation. J. Androl. 2002;23:622–628. [PubMed] [Google Scholar]
- Wood L.D., Irvin B.J., Nucifora G., Luce K.S., Hiebert S.W. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Natl. Acad. Sci. 2003;100:3257–3262. doi: 10.1073/pnas.0637114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Henry M.F. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell. Biol. 2004;24:10742–10756. doi: 10.1128/MCB.24.24.10742-10756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.M., Jiang C., Kroll T.T., Huber P.W. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 2001;20:2315–2325. doi: 10.1093/emboj/20.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]