Abstract
Prokaryotic RNA polymerases are capable of efficient, continuous synthesis of RNA in vivo, yet purified polymerase-DNA model systems for RNA synthesis typically produce only a limited number of catalytic turnovers. Here, we report that the ribosomal protein S1—which plays critical roles in translation initiation and elongation in Escherichia coli and is believed to stabilize mRNA on the ribosome—is a potent activator of transcriptional cycling in vitro. Deletion of the two C-terminal RNA-binding modules—out of a total of six loosely homologous RNA-binding modules present in S1—resulted in a near-loss of the ability of S1 to enhance transcription, whereas disruption of the very last C-terminal RNA-binding module had only a mild effect. We propose that, in vivo, cooperative interaction of multiple RNA-binding modules in S1 may enhance the transcript release from RNA polymerase, alleviating its inhibitory effect and enabling the core enzyme for continuous reinitiation of transcription.
Keywords: S1, RNA polymerase, transcription, translation
INTRODUCTION
Prokaryotic RNA polymerases are capable of continuous synthesis of RNA in vivo, yet purified polymerase-DNA model systems for RNA synthesis typically produce only a limited number of catalytic turnovers. At near equimolar Escherichia coli RNA polymerase/template DNA ratios—even with relatively strong E. coli promoters such as tac and rrnBP1—in vitro transcription reactions typically produce an average of one to two catalytic turnovers under most experimental conditions (for example, see Sukhodolets etal. 2001). Inhibitory RNA polymerase–transcript interactions present a potential obstacle for continuous transcriptional cycling, with each individual cycle including transcript initiation, elongation, and termination. A number of studies have indicated that elongating E. coli RNA polymerase–transcript complexes are very stable but are destabilized at specific transcription terminators (Berlin and Yanofsky 1983; Wilson and von Hippel 1995; Mooney et al. 1998; Gusarov and Nudler 1999; Yarnell and Roberts 1999). The hexameric ATP-dependent factor Rho has been shown to promote transcript release from the polymerase atcertain terminators (Richardson and Conaway 1980; Shigesada and Wu 1980; for review, see Richardson 2002, 2003). The inability of RNA polymerase to disengage from DNA (in different reaction contexts) may also hinder transcriptional cycling. The transcription-repair coupling factor Mfd and the major E. coli sigma factor (σ70) have been shown to destabilize core RNA polymerase–DNA complexes (Arndt and Chamberlin 1988; Selby and Sancar 1993, 1994). Furthermore, it has recently been proposed that RapA, a bacterial homolog of eukaryotic SWI/SNF proteins, may alter the configuration of DNA to allow release of the polymerase, resulting in enhanced transcriptional cycling (Sukhodolets et al. 2001, 2003). Inhibitory DNA–RNA interactions during transcription may hinder elongation and/or reinitiation of transcription as well. There is ample evidence for the formation of DNA–RNA duplexes during transcription (Zaychikov et al. 1995; Komissarova and Kashlev 1998; Sidorenkov et al. 1998; Korzheva et al. 2000). It has also been suggested that DNA–RNA complexes in which the RNA is wrapped around the DNA are formed during transcription (Drolet et al. 2003). The formation of noncanonical DNA–RNA complexes such as DNA–RNA triplexes (Roberts and Crothers 1992)—which can be promoted by relatively high salt concentrations (Morvan et al. 1997) and influenced by the composition of the DNA and RNA backbone (Roberts and Crothers 1992; Ivanov et al. 2003)—also cannot be ruled out. In an in vitro system, these structures could hinder reinitiation of transcription, as well as transcript elongation.
Here we report that the ribosomal protein S1—which plays critical roles in translation initiation (Tzareva et al. 1994) and elongation (Potapov and Subramanian 1992) in E. coli and is believed to stabilize mRNA on the ribosome through interaction with a pyrimidine-rich region upstream of the Shine-Dalgarno sequence of mRNA (Boni et al. 1991, 2001; Sengupta et al. 2001; Komarova et al. 2002)—is a potent activator of transcriptional cycling in vitro. Deletion of the two C-terminal RNA-binding/S1-modules—out of a total of six loosely homologous RNA-binding modules present in S1 (Bycroft et al. 1997)—resulted in a near-loss of the ability of S1 to enhance transcription. However, the mutant protein retained the ability to bind transcript RNA, suggesting that the transcription-stimulatory effect is more than a mere sequestering of RNA in the in vitro transcription system. Taken together, our results demonstrate significant stimulation of in vitro transcription by S1, an abundant cellular protein belonging to the translational apparatus.
RESULTS
S1 promotes transcriptional cycling
S1 was originally identified as one of the proteins loosely associated with the ribosome, and ribosome preparations often have been reported to contain less-than-stoichiometric amounts of S1 (Subramanian and van Duin 1977). Free S1 has also been identified as a part of the bacteriophage Qβ replication complex (Inouye et al. 1974; Wahba etal. 1974). Earlier we reported copurification with E. coli RNA polymerase ofstoichiometric amounts of S1 (Sukhodolets and Garges 2003). These studies suggested a role of S1 beyond that in the ribosome. Here, we report the results of a detailed study of the effect of purified S1 on E. coli RNA polymerase transcriptional activity on DNA templates incorporating the extensively characterized bacteriophage T7A1 promoter (Dayton et al. 1984; Reynolds et al. 1992; Daube et al. 1994; Susa et al. 2002).
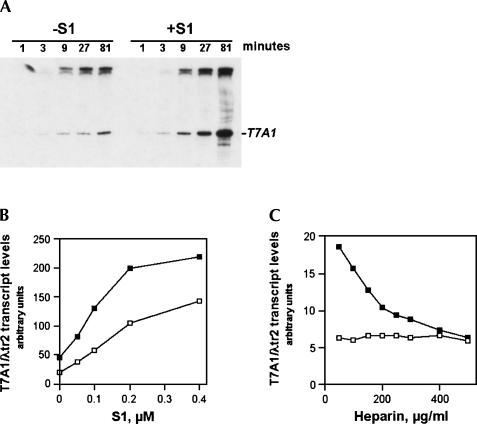
Figure 1A shows a typical transcription-stimulatory effect resulting from the addition of purified native S1 to invitro transcription reactions. With native S1, half-maximal transcription stimulatory effect was observed in the 100–200 nM protein concentration range (Fig. 1B). Although S1 showed a mild transcription-stimulatory effect in the presence of low to moderate concentrations of the DNA competitor heparin—a polyanion and DNA analog that is known to disrupt the relatively weak nonspecific RNA polymerase–DNA interaction and to inhibit DNA binding to the polymerase (Walter et al. 1967; Kadesch etal. 1980; Schlax et al. 1995)—a further increase in heparin concentration virtually abolished the effect (Fig. 1C). Because excess heparin is known to inhibit reinitiation of transcription (resulting in a “single-round” transcription reaction), this result indicates that the transcription-stimulatory effect of S1 resulted primarily from an increase in the number of completed transcriptional rounds, referred to here as enhanced transcriptional cycling.
FIGURE 1.
The ribosomal protein S1 enhances transcriptional cycling. In vitro transcription reactions with supercoiled DNA template pCPGλtr2 (Reynolds et al. 1992) were carried out as described in Materials and Methods. (A) Kinetics of the transcription-stimulatory activity of S1. Reactions were carried out in the presence or absence of 0.2 μM purified native S1. The position of the T7A1 promoter-generated transcript RNA terminated at a specific transcription terminator (λtr2) is indicated. (B) The yields of T7A1/λtr2 transcript plotted as a function of native S1 concentration. In vitro transcription reactions were carried out in the presence of 50mM NaCl (open symbols) and 100 mM NaCl (closed symbols). (C) The transcription-stimulatory effect of S1 is abolished in the presence of high concentrations of the DNA competitor heparin. The reaction conditions were as described in Materials and Methods, with the exception that heparin was added to some of the reactions along with the rNTP premix. In vitro transcription reactions were performed in the absence (open boxes) or in the presence (solid boxes) of 0.2 μM purified native S1.
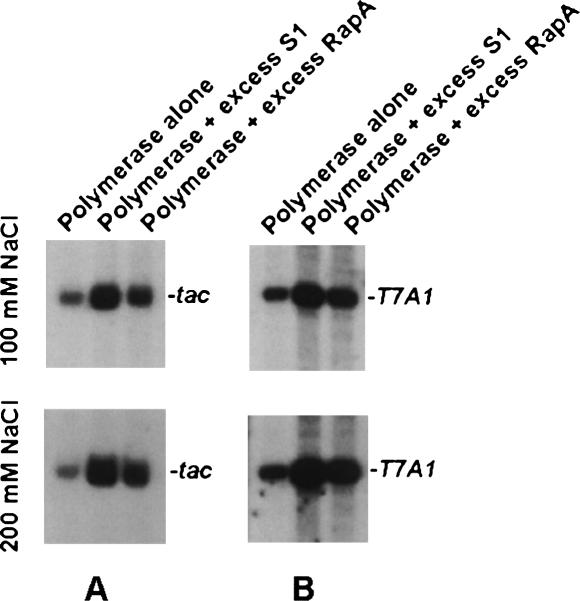
Back-to-back comparison of the transcription-stimulatory activities of S1 with that of RapA, an RNA polymerase-associated ATPase, and a member of the SWI/SNF superfamily (an excess of which was shown to promote transcriptional cycling; see Sukhodolets et al. 2001), revealed that the magnitude of the transcription-stimulatory effect ofS1 generally superseded that of RapA (Fig. 2).
FIGURE 2.
Back-to-back comparison of the transcription-stimulatory activity of S1 and RapA. Reactions with ptac1617 (Sukhodolets et al. 2001) (A) and pCPGλtr2 (B) supercoiled DNA templates were carried out in the presence of 2.1 μM S1 and 2.4 μM RapA. RNA polymerase/DNA template ratio and concentrations and conditions for in vitro transcription reactions were as described earlier (Sukhodolets et al. 2001). Promoter-specific transcripts resulting from in vitro transcription reactions carried out in the presence of 100 mM NaCl (top) and 200 mM NaCl (bottom) are shown.
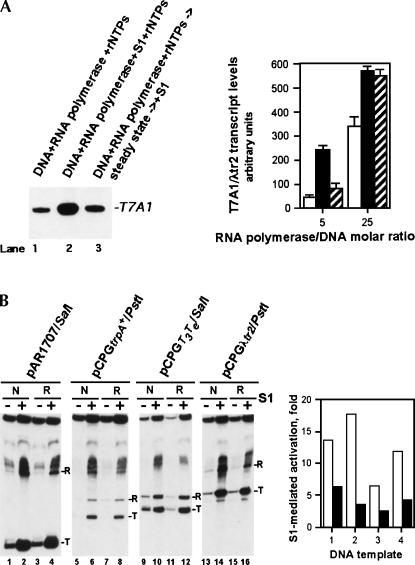
Order-of-addition experiments revealed that once the product RNA level reached the initial plateau, the addition of S1 failed to stimulate transcription significantly further (Fig. 3A). Increasing the RNA polymerase/DNA molar ratio reduced the magnitude of the effect (Fig. 3A). Because in our previous study with RapA the transcription termination and/or post-termination events were identified as likely rate-limiting steps for continuous transcriptional cycling (see Sukhodolets et al. 2001), the apparent similarity of the transcription-stimulatory activity of S1 and RapA prompted us to explore the effect of S1 on the efficiency of transcription termination. These studies suggested onlya marginal (two- to threefold) (Fig. 3B, graph) enhancement of termination efficiency in the presence of S1: The levels of runoff RNA and RNA transcripts terminated at specific terminators were up-regulated to a comparable extent by S1 (Fig. 3B), with native and recombinant S1 showing similar transcription-stimulatory properties (Fig. 3B). Linear DNA templates showed S1-stimulatory effects similar to those seen with supercoiled DNA templates (Figs. 1, 3; Sukhodolets and Garges 2003).
FIGURE 3.
Effect of delayed addition of S1 to in vitro transcription reactions and the effect of S1 on the efficiency of transcription termination. (A) The delayed addition of S1 to in vitro transcription reactions results in greatly reduced transcription-stimulatory activity. The quantitated results of two independent, parallel experiments are shown. Reactions in lanes 1 (open columns) and 2 (black columns) are similar to those in lanes 5 and 10 of Figure 1A; in lane 3 (hatched columns), purified native S1 was added 30 min after the initiation of the in vitro transcription reaction, and the reactions were incubated at 37°C for an additional 60 min. The reactions were performed with an approximately fivefold (see Materials and Methods) or 25-fold molar excess of RNA polymerase over the supercoiled pCPGλtr2 DNA template. The RNA polymerase/DNA template molar ratios are indicated. Incubation of RNA polymerase alone at 37°C up to 90 min produced no significant reduction in the enzyme's activity. (B) Mild stimulation of the transcription termination efficiency in the presence of 0.3 μM purified native S1 (lanes “N”) or 0.6 μM recombinant His-tagged S1 (lanes “R”). RNA transcripts terminated at specific terminators (T) and runoff RNA transcripts (R) are indicated. The quantitated results of the experiment are shown on the right; open and black columns show the magnitudes of the S1-mediated transcriptional activation for RNA transcripts terminated at specific terminators and runoff RNA transcripts, respectively. The linearized plasmid DNA templates used in this experiment were pAR1707 (1), pCPGtrpA + (2), pCPGT3Te (3), and pCPGλtr2 (4). DNA templates carried the T7A1 promoter followed by a defined transcription terminator downstream (the type of terminator is apparent from the plasmids' names); all fourtemplates are described in Reynolds et al. (1992).
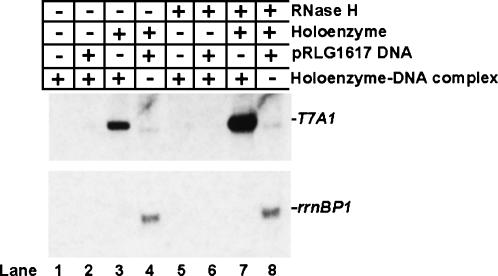
To determine whether nonproductive complexes formed during the course of in vitro transcription reactions resulted from the inability of RNA polymerase to initiate new rounds of transcription or the inability of the DNA template to support the reaction, purified RNA polymerase–DNA complexes obtained after the fractionation of in vitro transcription reaction mixtures carried out to the initial plateau in the transcript production were concentrated and tested again for their transcriptional activity under various conditions. In the presence of rNTPs, purified RNA polymerase–DNA complexes failed to show transcriptional activity even in the presence of challenger DNA templates (Fig. 4, lanes 1,2,5,6). At the same time, the DNA template present in purified RNA polymerase–DNA complexes supported a robust promoter-specific transcriptional activity in the presence of supplemental (trans) RNA polymerase holoenzyme (Fig. 4, lanes 3,7). Moreover, the activity of the DNA template thus reused in the transcription reaction increased significantly after treatment with RNase H, suggesting the formation of potentially inhibitory DNA–RNA complexes (Fig. 4, cf. lanes 3 and 7).
FIGURE 4.
Inactivation of RNA polymerase rather than the DNA template is the principal reason for poor transcriptional cycling in in vitro transcription reactions with supercoiled DNA. Five microliters of 500 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 1 M NaCl were mixed with 25 μL of purified water, 5 μL RNA polymerase holoenzyme (1.2mg/mL), 5 μL pCPGλtr2 supercoiled DNA (0.89 mg/mL) with the T7A1 promoter and λtr2 terminator, 10 μL of 5× rNTP mix containing 1 mM each of ATP, GTP, CTP, and UTP, and 0.01 μCi of [α32P] UTP. Following a 30-min incubation at 37°C, the entire 50-μL reaction mixture was passed through a Superdex 200 HR 10/30 column pre-equilibrated with TGED buffer containing 100 mM NaCl; 0.5-mL fractions were collected. Aliquots of 250 μL from fractions containing the DNA-bound transcription complexes were concentrated to ∼25 μL using Microcon-10 concentrators (Amicon), and 3-μL aliquots of the resulting purified RNA polymerase–DNA complex were used in in vitro transcription reactions, performed as described in Materials and Methods, with the exception that RNA polymerase and the DNA template were omitted. When indicated, RNA polymerase holoenzyme was added to a final concentration of 0.05 mg/mL; pRLG1617 supercoiled DNA template with the ribosomal rrnBP1 promoter and T1T2 terminator (Ross et al. 1990) to 0.08 mg/mL; and RNase H (New England Biolabs), 5 U per 20 μL reaction. A representative result of two independent experiments is shown.
S1 interacts with the transcription complex
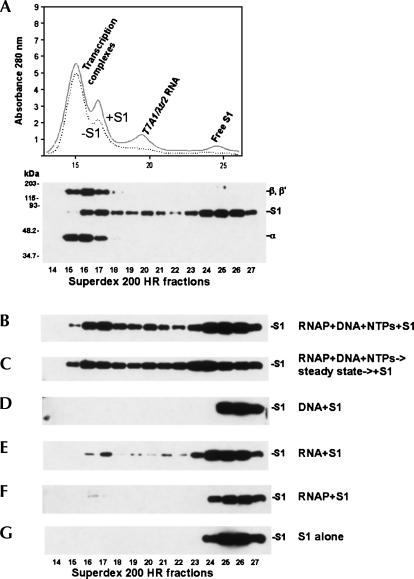
Our next objective was to monitor the binding of S1 to functional RNA polymerase–RNA–DNA complexes in comparison to its binding to isolated components of the transcription complex, such as transcript RNA, DNA, or RNA polymerase. Entire in vitro transcription reactions, carried out to a steady state, or mixtures of the purified enzymes or enzymes and nucleic acids were passed through a Superdex 200 HR column, and the content of the column fractions was analyzed (Fig. 5). This technique allowed for the separation of free S1 from S1 bound to high-molecular weight fractions (Fig. 5A–C). This set of experiments demonstrated that S1 interacted with high affinity with high-molecular weight fractions presumably containing the DNA-bound transcription complexes and their subassembly components (resulting from gel-filtration) (Fig. 5A,B). Greater than 50% of the entire amount of S1 present in the samples showed characteristic mobility shifts, interacting with high-molecular weight fractions (Fig. 5B,C). This extent of recruitment of S1 into protein–nucleic acid complexes could not be achieved by substituting an in vitro transcription premixture of RNA polymerase, DNA template, and rNTPs with various combinations of individual reaction substrates/products (Fig. 5D,E,F, and data not shown). However, RNA or RNA polymerase alone produced detectable S1 mobility shifts suggestive of both S1–RNA and S1–RNA polymerase interactions (Fig. 5E,F).
FIGURE 5.
S1 interacts with transcription complexes. (A) Top: the A280 profile resulting from size-separation of an in vitro transcription reaction mixture on a Superdex 200 HR 10/30 column. The supercoiled pCPGλtr2 DNA template (Reynolds et al. 1992) used in this set of experiments carried the T7A1 promoter and λtr2 terminator. The individual components of the reaction were identified as described in Materials and Methods. Bottom: S1 and RNA polymerase subunits inthe fractions were also identified by Western blot analysis using S1-specific and core RNA polymerase-specific antibodies. (B,C) The in vitro transcription reactions were similar to those described in Materials and Methods, with the exceptions that 0.24× (12 μL) reactions were utilized here, and the reactions contained 0.2 μM purified native S1. The reaction ingredients are indicated at the right. S1 in the Superdex 200 HR 10/30 column fractions is visualized here and below by immunoblotting with S1-specific antibodies. (D) 0.2 μM S1 plus pCPGλtr2 supercoiled DNA (0.089 mg/mL). (E) In vitro transcription reactions similar to those described above were carried out and, after a 30-min incubation at 37°C, 4 U of RNase-free DNase (Promega) was added to the mixtures to digest the DNA template. Following another 30-min incubation at 37°C, the reactions were deproteinized by phenol extraction, and the aqueous phase was precipitated with ethanol. The RNA pellet was redissolved in 12 μL of 1× TB containing 0.2 μM purified native S1, and the binding experiments were performed as described in Materials and Methods. The substitution of DNase with a DNase/RNaseH mixture in the above purification procedure produced similar S1–RNA binding patterns, suggesting that DNA–RNA hybrids are unlikely to play a major role in the interaction of S1 with transcription complexes (data not shown). (F) 0.2 μM S1 plus 1.3 μM RNAP holoenzyme. In a similar binding assay, Δ335–556 S1 showed no detectable complex formation with the polymerase. (G) 0.24 μM S1.
The transcription-stimulatory activity of S1 is more than a mere sequestering of RNA in the in vitro transcription system
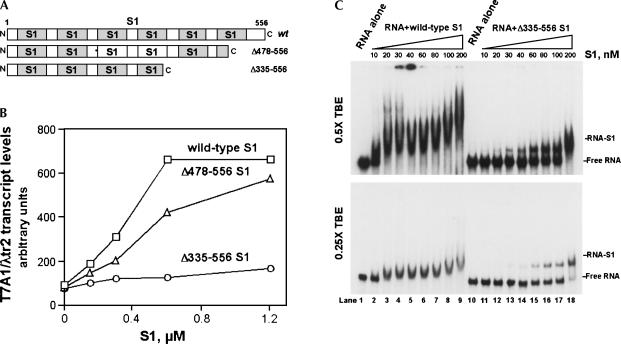
Because it was suggested that the C-terminal RNA-binding modules in S1 participate in mRNA binding (Sillers and Moore 1981) and the N-terminal RNA-binding modules inrRNA binding (Giorginis and Subramanian 1980; Subramanian 1983), we mutagenized the C-terminal part of S1, sequentially deleting the C-terminal 78 and 221 amino acids (Fig. 6A; see Materials and Methods) to test if, and to what extent, alteration of the transcription-stimulatory activity of S1 would correlate with RNA binding. The transcription-stimulatory properties of purified wild-type S1 and the two mutant proteins were tested. The disruption of a single C-terminal RNA-binding module in S1 produced only a marginal effect on its ability to activate transcription in vitro (Fig. 6B). However, the loss of an additional protein segment, encompassing the second C-terminal RNA-binding domain, nearly abolished the transcription-stimulatory activity of S1 (Fig. 6B). Because Δ335–556 S1 showed a near-complete loss of transcription-stimulatory activity even when taken in excess of 1 μM, wetested wild-type S1 and Δ335–556 S1 back-to-back in EMSA assays with purified T7A1/λtr2 RNA (Fig. 6C). The titrations of the purified T7A1/λtr2 RNA with wild-type S1 and Δ335–556 S1 indicated that the 50% saturation of RNA with protein was reached at ∼10–30 nM and 100–200 nM, respectively (depending on the running conditions and thetype of gel apparatus). Furthermore, the Δ335–556 S1mutant demonstrated S1–RNA binding patterns that were distinctly different from those of the wild-type S1 (Fig.6C).
FIGURE 6.
Alteration of the transcription-stimulatory activity of S1 by deletion mutations at the protein's C terminus. (A) Schematic illustrating the modular composition of S1. (B) Effects of C-terminal deletion mutations on the in vitro transcription-stimulatory activity of S1. The transcription-stimulatory activities of the wild-type and mutant S1 proteins are plotted as a function of protein concentration. In vitro transcription reactions were carried out as described in Materials and Methods. (C) The RNA-binding activity of wild-type S1 versus that of Δ335–556 S1. The gels show titrations of purified labeled T7A1/λtr2 RNA with purified recombinant S1 proteins; the S1 concentrations are indicated. EMSA experiments were carried out as described in Materials and Methods.
DISCUSSION
Known properties of the ribosomal protein S1, such as weak, reversible ribosome binding, high-affinity binding at the 5′-termini of mRNAs, its documented role as a component of theQβ RNA bacteriophage replicase, and copurification of stoichiometric amounts and possible interaction of S1 with RNA polymerase, suggest that it functions at the interface of transcription and translation. While previous studies have identified S1 as a translation-related RNA-binding protein, the results presented in this paper point to the possibility that S1 interacts with transcription complexes in a manner that potentially benefits the transcriptional apparatus.
The addition of purified S1 to in vitro transcription reactions greatly increased the yield of RNA transcripts. (1) The sensitivity of the transcription-stimulatory activity of S1 to heparin, (2) its reduction upon increase of the polymerase/DNA template molar ratio, and (3) the lack of transcription-stimulatory effect at early time-points indicate that the transcription-stimulatory effect results primarily from enhanced transcriptional cycling. Our binding studies, meant to determine the specific elements of the transcriptional apparatus targeted by S1, suggested that the transcript and possibly RNA polymerase are targeted, with the relative irreversibility of nonproductive transcription complex formation suggesting a relatively fast engagement by S1 of nascent RNA as it emerges from the RNA channel in RNA polymerase. This engagement could prevent the RNA synthesized de novo from forming RNA–DNA or RNA–RNA polymerase interactions potentially inhibitory to transcription. Purified S1 failed to unwind DNA–RNA duplexes (in the presence or absence of ATP and/or Hfq; the protein also showed a lack of ATPase activity; data not shown), indicating a relatively passive role for S1 in interaction with RNA polymerase–RNA complexes during transcription.
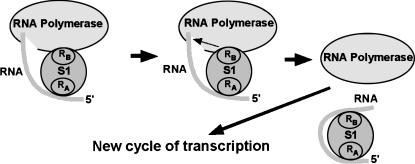
The disruption of a single RNA-binding module at the extreme C terminus of S1 produced only a marginal effect on its transcription-stimulatory activity, whereas the deletion of two adjacent RNA-binding modules virtually abolished it. The solubilities of both S1 mutants were similar to that of the wild-type protein, and the ability of truncated S1 to bind RNA suggested no increased protein misfolding compared with wild-type S1. This near-loss of transcription-stimulatory activity in the Δ335–556 S1 mutant was likely due to altered configuration of the Δ335–556 S1–RNA complex rather than the protein's lack of RNA-binding activity. Although deletion of the two C-terminal RNA binding modules in S1 likely affects the overall configuration of the S1 molecule, the fact that S1 lacking two C-terminal RNA-binding modules is capable binding RNA is important. Other studies also reported RNA binding by truncated forms of S1 (for examples, see McGinnes and Sauer 2004; Okada et al. 2004); taken together, these results suggest relatively autonomous, “domains-on-a-string”-like RNA-binding activity of individual RNA-binding modules in S1. Excess Δ335–556 S1 consistently failed to show the high-magnitude transcription-stimulatory activity of wild-type S1 (typically resulting in >5-fold increase in the yield of promoter-specific transcripts [see Fig. 3B, graph]), suggesting that simple RNA binding does not translate into high-magnitude transcriptional enhancement. The patterns of protein–RNA complexes formed by wild-type S1 in EMSA gels (where theelongated shapes of shifted protein–RNA complexes suggest conformational heterogeneity) are almost certainly due to the engagement of multiple RNA binding modules in protein–RNA interaction. Our results indicate that theregion between amino acids 335 and 478 is critical for manifestation of the transcription-stimulatory activity of S1. We speculate that, in vivo, cooperative interaction of multiple RNA-binding modules in S1 may enhance transcript release from RNA polymerase, alleviating its inhibitory effect on RNA polymerase and possibly the DNA template and enabling the core enzyme for continuous reinitiation of transcription (Fig. 7). The reported complex formation between S1 and the transcription termination factor NusA (Venkatesh and Radding 1993) and the specific binding of S1 to boxA transcriptional antiterminator RNA (Mogridge and Greenblatt 1998) are also consistent with S1 acting at the termination and/or post-termination stage of the transcription cycle.
FIGURE 7.
Cooperative binding of RNA polymerase-associated transcript RNA by multiple RNA-binding modules in S1 may effectively strip transcript from RNA polymerase, alleviating its inhibitory effect. RA and RB denote independent RNA-binding segments in S1 which may incorporate one or more individual RNA-binding or “S1”-modules (defined as described in Bycroft etal. 1997).
In vivo, a moderate overexpression of wild-type S1 or either of the two deletion mutants from plasmid vectors in the rpsA + background was tolerated by the bacterial cells, although both mutations caused a slight growth defect (data not shown). S1 is an essential protein in E. coli (Kitakawa and Isono 1982; Sorensen et al. 1998; Sukhodolets and Garges 2003), and the problems inherent in the analysis and interpretation of in vivo data meant to distinguish S1 function(s) associated with transcription or translation have been pointed out earlier (Sukhodolets and Garges 2003). Thus, the small differences in growth rates resulting from overproduction of the wild-type and mutant S1 proteins in the rpsA + bacterial strains could very well be the result of altered translation.
In a series of independent experiments we size-fractionated lysates obtained from bacterial cells lysed under mild conditions and monitored the S1 levels in the fractions. These experiments demonstrated that <50% of S1 was associated with ribosomal, high-molecular weight (>240 kDa) fractions (data not shown), suggesting that there is likely tobe a substantial fraction of free S1 that may be availablefor interaction with elements of the transcriptional apparatus.
The previously reported stimulation of transcriptional cycling by the RNA polymerase-associated SWI2/SNF2 homolog RapA (Sukhodolets et al. 2001) mimics the effect of S1, hinting that both S1 and RapA may rely on protein–RNA interaction to enhance transcription. This interpretation does not contradict the previously proposed general model explaining the transcription-stimulatory effect of RapA (Sukhodolets et al. 2001). We speculate that, in vivo, the transcription-stimulatory activities of the two proteins may be manifested under different physiological conditions.
In summary, we propose that free S1 may play a role in minimizing nonproductive mRNA interactions in transcription complexes in vivo. Because S1 is known to bind at the 5′-termini of mRNAs and is critically involved in translation initiation, we also propose that during the cycling process the nascent mRNA–S1 complex recruits S1-free ribosomes (according to the orthodox model, S1 is perceived as an integral part of the ribosome subassembly, rather than an mRNA-associated protein adaptor), while keeping the process coupled to transcription by maintaining its contact with RNA polymerase.
MATERIALS AND METHODS
Proteins
The E. coli RNA polymerase holoenzyme was purified as described (Sukhodolets and Jin 1998) with the exception that the final product was subjected to an additional purification step—gel-filtration on either Superose 6 HR 10/30 or Superdex 200 HR 10/30 columns. Chromatography was performed as previously described (Sukhodolets and Garges 2003). The native and recombinant ribosomal S1 proteins were was also obtained as described (Sukhodolets and Garges 2003).
In vitro transcription
The 16-μL pre-incubation mix in 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 0.5 mg/mL BSA, and 100 mM NaCl (unless indicated otherwise) (transcription buffer, or TB) typically contained 20–30 nM supercoiled or linear DNA template, 120 nM RNA polymerase holoenzyme, and S1 (if present) at the concentrations indicated in the figure legends. Following a 15-min pre-incubation at 37°C, transcription was initiated by the addition of 4 μL of 5× rNTP mix containing 1 mM each of ATP, GTP, CTP, and UTP and 0.02–0.05 μCi of either [α32P] UTP or [α32P] ATP. After incubation at 37°C for 60 min, unless indicated otherwise, 20μL of loading solution (0.25 M EDTA, 50% glycerol, 0.1% Bromphenol Blue) was added to each reaction mixture, followed by the addition of 40 μL of formamide, heating at 90°C for 2 min, and ∼3-min incubation on ice. Aliquots of 6 μL were then analyzed on 8% sequencing gels. The gels were autoradiographed using BioMax MR2 film and scanned on a PhosphorImager (Molecular Dynamics) to quantitate the levels of transcript RNA.
S1 mutagenesis
Truncated fragments of the rpsA gene were amplified from pQE31S1 using the primers MS73 (Sukhodolets and Garges 2003) / MS134 (5′-CGTCGATGCATGCGGATTATTACAGGTAACCTTCAACGCCGTCAGCCAGT) (S1Δ478–556) or MS73/MS135 (5′-CGTCGATGCATGCGGATTATTAGATATCCAGAACCATAACTTCCACTACA) (S1Δ335–556). The amplified DNA fragments (digested with BamHI and SphI) were then cloned into the BamHI and SphI restriction sites of the expression vector pQE31 (Qiagen). The protein products expressed from both plasmids (pQE31S1Δ478–556 and pQE31S1Δ335–556) were verified by immunoblotting using S1-specific antibodies, and the mutant proteins were isolated as previously described for recombinant wild-type His-tagged S1 (Sukhodolets and Garges 2003).
Electrophoretic mobility shift assay (EMSA)
S1—at the concentrations specified in the Figure Legends—and the 32P-labeled purified RNA probe (typically 500–1000 cpm/20-μL binding reaction) were mixed in 1× TB. The binding reactions were incubated for 15 min at room temperature, and 5 μL of loading buffer (50% glycerol supplemented with 0.05% Bromphenol Blue) was added to each binding reaction. Aliquots of 2–8μL were then analyzed on 8% polyacrylamide gels in either 0.5× or 0.25× TBE buffer. The gels were dried and exposed to BioMax ML film (typically for 6–18 h at –70°C) using BioMax MS screens.
Purification and analysis of nonproductive transcription complexes
Five microliters of 500 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 1 M NaCl were mixed with 25 μL of purified water, 5 μL RNA polymerase holoenzyme (1.2 mg/mL), 5 μL pCPGλtr2 supercoiled DNA (0.89 mg/mL), 10 μL of 5× rNTP mix containing 1 mM each of ATP, GTP, CTP, and UTP, and 0.01 μCi of [α32P] UTP. Following a 30-min incubation at 37°C, the entire 50-μL reaction mixture was passed through a Superdex 200 HR 10/30 column pre-equilibrated with TGED buffer containing 100 mM NaCl; 0.5-mL fractions were collected. The fractions were concentrated and assayed as described in the legend to Figure 4. To visualize nucleic acids and proteins in the purified transcription complexes, aliquots of ∼125 μL from each fraction were also concentrated to∼10 μL using Microcon-10 concentrators (Amicon); 10 μL of 2× Laemmli sample buffer was then added directly into each concentrator, and the recovered 20-μL aliquots from the indicated fractions were analyzed on silver-stained 10% SDS-PAGEs. Autoradiographs indicating the position of [α32P] UTP-labeled transcript RNA were obtained after exposing the silver-stained protein gels to BioMax ML X-ray film. The ∼277-nt T7A1/λtr2 transcript was visualized by both silver staining and 32P-labeling. The RNA polymerase–DNA complexes thus isolated typically retained 20%–30% of σ70, the presence of which is critical for promoter-specific transcription. The amounts of σ70 retained by the DNA-bound polymerase were estimated from comparison of the quantitated σ70/α ratios in the DNA-bound polymerase with those in the input enzyme. The presence of DNA in the fractions was apparent due to their T7A1 promoter-specific activity in the absence of an added DNA template, as well as other criteria. Three independent, parallel experiments were performed. Experiments performed with or without BSA in the transcription buffer produced similar results.
Gel filtration-based study of the interaction of S1 with RNA or RNA polymerase–RNA complexes
In vitro transcription reactions or mixtures of purified enzymes (or enzymes and nucleic acids) were prepared in a total volume of 12 μL in 1× TB (the final enzyme concentrations are specified in the legend to Fig. 5). Following pre-incubation for 30 min at 37°C, the entire reaction was injected onto a Superdex 200 HR 10/30 column (Amersham Pharmacia) pre-equilibrated with TGED buffer (10 mM Tris-HCl, pH 8.0, 5% glycerol, 0.1 mM EDTA, 0.015 mg/mL dithiothreitol) containing 0.1 M NaCl. The column was run at a flow rate of 0.5 mL/min; 0.5-mL fractions werecollected. After completion of each individual run, the volumes ofthe fractions were reduced to ∼10 μL using Microcon 30microconcentrators (Amicon), each concentrated fraction was mixed with an equal volume of 2× Laemmli sample buffer, and approximately half of the content of each fraction was analyzed bySDS-PAGE, followed by immunoblotting of the fractions with S1-specific or core RNA polymerase-specific polyclonal antibodies, as seen in Figure 5. Each binding experiment was repeated at least twice.
ACKNOWLEDGMENTS
We thank Karen Sukhodolets for critical discussion of the experimental results. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, anda Welch Foundation grant (V-0004).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2321606.
REFERENCES
- Arndt K.M., Chamberlin M.J. Transcription termination in Escherichia coli. Measurement of the rate of enzyme release from Rho-independent terminators. J. Mol. Biol. 1988;202:271–285. doi: 10.1016/0022-2836(88)90457-3. [DOI] [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Release of transcript and template during transcription termination at the trp operon attenuator. J.Biol. Chem. 1983;258:1714–1719. [PubMed] [Google Scholar]
- Boni I.V., Isaeva D.M., Musychenko M.L., Tzareva N.V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni I.V., Artamonova V.S., Tzareva N.V., Dreyfus M. Non-canonical mechanism for translational control in bacteria: Synthesis of ribosomal protein S1. EMBO J. 2001;20:4222–4232. doi: 10.1093/emboj/20.15.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft M., Hubbard T.J.P., Proctor M., Freund S.M.V., Murzin A.G. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- Daube S., Hart C.R., von Hippel P. Coupling of RNA displacement and intrinsic termination in transcription from synthetic RNA–DNA bubble duplex constructs. Biochemistry. 1994;91:9539–9543. doi: 10.1073/pnas.91.20.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton C.J., Prosen D.E., Parker K.L., Cech C.L. Kinetic measurements of Escherichia coli RNA polymerase association withbacteriophage T7 early promoters. J. Biol. Chem. 1984;259:1616–1621. [PubMed] [Google Scholar]
- Drolet M., Broccoli S., Rallu F., Hraiky C., Fortin C., Masse E., Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front. Biosci. 2003;8:210–221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- Giorginis S., Subramanian A.R. The major ribosome binding site of Escherichia coli ribosomal protein S1 is located in its N-terminal segment. J. Mol. Biol. 1980;141:393–408. doi: 10.1016/0022-2836(80)90253-3. [DOI] [PubMed] [Google Scholar]
- Gusarov I., Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Inouye H., Pollack Y., Petre J. Physical and functional homology between ribosomal protein S1 and interference factor i. Eur. J. Biochem. 1974;45:109–117. doi: 10.1111/j.1432-1033.1974.tb03535.x. [DOI] [PubMed] [Google Scholar]
- Ivanov S., Alekseev Y., Bertrand J., Malvy C., Gottikh M.B. Formation of stable triplexes between purine RNA and pyrimidine oligodeoxyxylonucleotides. Nucleic Acids Res. 2003;31:4256–4263. doi: 10.1093/nar/gkg443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesch T.R., Williams R.C., Chamberlin M.J. Electron microscopic studies of the binding of Escherichia coli RNA polymerase to DNA. J. Mol. Biol. 1980;136:79–93. doi: 10.1016/0022-2836(80)90367-8. [DOI] [PubMed] [Google Scholar]
- Kitakawa M., Isono K. An amber mutation in the gene rpsA for ribosomal protein S1 in Escherichia coli . Mol. Gen. Genet. 1982;185:445–447. doi: 10.1007/BF00334137. [DOI] [PubMed] [Google Scholar]
- Komarova A.V., Tchufistova L.S., Supina E.V., Boni I.V. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–1147. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N., Kashlev M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. 1998;95:14699–14704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzheva N., Mustaev A., Kozlov M., Malhotra A., Nikiforov V., Goldfarb A., Darst S.A. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- McGinnes K.E., Sauer R.T. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc. Natl. Acad. Sci. 2004;101:13454–13459. doi: 10.1073/pnas.0405521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogridge J., Greenblatt J. Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J. Bacteriol. 1998;180:2248–2252. doi: 10.1128/jb.180.8.2248-2252.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R.A., Artsimovitch I., Landick R. Information processing by RNA polymerase: Recognition of regulatory signals during RNA chain elongation. J. Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan F., Imbach J.L., Rayner B. Comparative stability of eight different triple helices formed by differently modified DNA or RNA pyrimidine strands and a DNA hairpin. Antisense Nucleic Acid Drug Dev. 1997;7:327–334. doi: 10.1089/oli.1.1997.7.327. [DOI] [PubMed] [Google Scholar]
- Okada T., Wower I.K., Wower J., Zwieb C.W., Kimura M. Contribution of the second OB fold of ribosomal protein S1 from Escherichia coli to the recognition of TmRNA. Biosci. Biotechnol. Biochem. 2004;68:2119–2125. doi: 10.1271/bbb.68.2319. [DOI] [PubMed] [Google Scholar]
- Potapov A.P., Subramanian A.R. Effect of E. coli ribosomal protein S1 on the fidelity of the translational elongation step: Reading and misreading of poly(U) and poly(dT) Biochem. Int. 1992;27:745–753. [PubMed] [Google Scholar]
- Reynolds R., Bermudez-Cruz R.M., Chamberlin M.J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J.Mol. Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- Richardson J.P. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- Richardson J.P. Loading Rho to terminate transcription. Cell. 2003;114:157–159. doi: 10.1016/s0092-8674(03)00554-3. [DOI] [PubMed] [Google Scholar]
- Richardson J.P., Conaway R. Ribonucleic acid release activity of transcription termination protein rho is dependent on the hydrolysis of nucleoside triphosphates. Biochemistry. 1980;19:4293–4299. doi: 10.1021/bi00559a022. [DOI] [PubMed] [Google Scholar]
- Roberts R.W., Crothers D.M. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- Ross W., Thompson J.F., Newlands J.T., Gourse R.L. E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlax P.J., Capp M.W., Record M.T., Jr. Inhibition of transcription initiation by lac repressor. J. Mol. Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- Selby C.P., Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Selby C.P., Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol. Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J., Agrawal R.K., Frank J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigesada K., Wu C.W. Studies of RNA release reaction catalyzed by E. coli transcription termination factor rho using isolated ternary transcription complexes. Nucleic Acids Res. 1980;8:3355–3369. doi: 10.1093/nar/8.15.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenkov I., Komissarova N., Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- Sillers I.Y., Moore P.B. Position of protein S1 in the 30 S ribosomal subunit of Escherichia coli . J. Mol. Biol. 1981;153:761–780. doi: 10.1016/0022-2836(81)90417-4. [DOI] [PubMed] [Google Scholar]
- Sorensen M.A., Fricke J., Pedersen S. Ribosomal protein s1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- Subramanian A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- Subramanian A.R., van Duin J. Exchange of individual ribosomal proteins between ribosomes as studied by heavy isotope-transfer experiments. Mol. Gen. Genet. 1977;258:1–9. doi: 10.1007/BF00455113. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Jin D.J. RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J. Biol. Chem. 1998;273:7018–7023. doi: 10.1074/jbc.273.12.7018. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Garges S., Jin D.J. Purification and activity assays of RapA, the RNA polymerase-associated homolog of the SWI/SNF protein superfamily. Methods Enzymol. 2003;370:283–290. doi: 10.1016/S0076-6879(03)70025-1. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Cabrera J.E., Zhi H., Jin D.J. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes & Dev. 2001;15:3300–3341. doi: 10.1101/gad.936701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa M., Sen R., Shimamoto N. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 2002;277:15407–15412. doi: 10.1074/jbc.M112481200. [DOI] [PubMed] [Google Scholar]
- Tzareva N.V., Makhno V.I., Boni I.V. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- Venkatesh V.T., Radding C.M. Ribosomal protein S1 and NusA protein complexed to recombination protein β of bacteriophage λ. J. Bacteriol. 1993;175:1844–1846. doi: 10.1128/jb.175.6.1844-1846.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba A.J., Miller M.J., Niveleau A., Landers T.A., Carmichael G.G., Weber K., Hawley D.A., Slobin L.I. Subunit I of G beta replicase and 30 S ribosomal protein S1 of Escherichia coli. Evidence for the identity of the two proteins. J. Biol. Chem. 1974;249:3314–3316. [PubMed] [Google Scholar]
- Walter G., Zillig W., Palm P., Fuchs E. Initiation of DNA-dependent RNA synthesis and the effect of heparin on RNA polymerase. Eur. J. Biochem. 1967;3:194–201. doi: 10.1111/j.1432-1033.1967.tb19515.x. [DOI] [PubMed] [Google Scholar]
- Wilson K.S., von Hippel P.H. Transcription termination atintrinsic terminators: The role of the RNA hairpin. Proc. Natl. Acad. Sci. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell W.S., Roberts J.W. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- Zaychikov E., Denissova L., Heumann H. Translocation of the Escherichia coli transcription complex observed in the registers 11 to 20: “Jumping” of RNA polymerase and asymmetric expansion and contraction of the “transcription bubble”. Proc. Natl. Acad. Sci. 1995;28:1739–1743. doi: 10.1073/pnas.92.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]