Abstract
RNA–protein interactions are important in many biological contexts. Identification of the networks that connect regulatory proteins to one another and to the mRNAs they control is a critical need. Here, we use a yeast three-hybrid screening approach to identify RNAs that bind a known RNA regulatory protein, the Saccharomyces cerevisiae PUF protein, Mpt5p. The assay selects RNAs that bind in vivo using simple phenotypes and reporter genes. It enables rapid analyses of the affinity and specificity of the interaction. We show that the method identifies mRNAs that are genuinely regulated by the protein in vivo, and that it complements biochemical strategies, yielding a set of mRNAs that overlap with, but are distinct from, those obtained by biochemical means. The approach we describe facilitates construction of protein–RNA linkage maps.
Keywords: RNA, gene networks, RNA–protein interactions, translation, mRNA stability
INTRODUCTION
The identification of protein–protein interaction networks has led to important insights in biochemistry, molecular genetics, and cell biology. The classification of DNA elements and chromosomal regions recognized by transcription factors has provided a greater understanding of transcriptional circuitry. Mass spectrometry and molecular genetic assays have been critical in elucidating these networks. Of genetic approaches, the yeast two-hybrid system has been particularly valuable. Analogous methods are essential to dissect networks of RNA regulation.
RNA–protein interactions are essential in many areas of biology, including development, virology, and neurobiology. A single protein can regulate multiple mRNA targets, and likewise, a single mRNA can be controlled by multiple proteins. The yeast three-hybrid system, which assays RNA–protein interactions through simple phenotypes, has enabled screens to identify proteins that bind known RNA sequences (Bernstein et al. 2002), complementing bioinformatic and biochemical approaches, such as RNA affinity chromatography. Here, we describe a yeast three-hybrid method for identifying mRNAs regulated by a specific regulatory protein. A proof-of-principle experiment, in which yeast Snp1 was used to retrieve its known binding site, has been reported (Sengupta et al. 1999). Here, we focus on the yeast protein, Mpt5p.
Mpt5p is a member of the PUF protein family. We chose to focus on a PUF protein because they bind to and regulate multiple mRNAs, and have important biological consequences (Wickens et al. 2002). PUF proteins control proliferation of diverse stem cells (Lin and Spradling 1997; Forbes and Lehmann 1998; Crittenden et al. 2002; Lamont et al. 2004; Salvetti et al. 2005), and are required for pattern formation (Barker et al. 1992; Murata and Wharton 1995) and memory (Dubnau et al. 2003; Ye et al. 2004) in Drosophila. PUF proteins bind to elements in the 3′ untranslated region (UTR) of their target mRNAs, and reduce expression either by translational repression or mRNA destabilization. The yeast PUF protein analyzed here, Mpt5p, regulates HO mRNA (Tadauchi et al. 2001), and is associated with over 200 mRNAs in a yeast lysate (Gerber et al. 2004). Here, we use a molecular genetic approach to identify mRNAs that are regulated by Mpt5p, and discover biologically significant interactions not revealed by other methods.
RESULTS
Inverse three-hybrid strategy
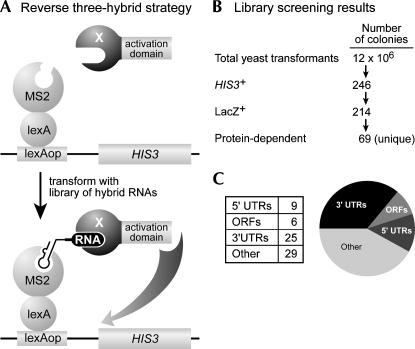
The three-hybrid approach involves expressing a library of naturally occurring genomic sequences as RNA, in a yeast strain that contains the protein of interest, X, linked to a transcription activation domain (AD) (Fig. 1A; Zhang et al. 1997; Bernstein et al. 2002). Each experimental RNA in the library possesses a short stem–loop motif that binds a LexA/MS2 coat protein, and so are tethered upstream of HIS3 and LacZ reporter genes. Interaction between the RNA and protein X activates transcription of the reporter genes, and is detected by simple yeast phenotypes, such as growth on selective media.
FIGURE 1.
An inverse three-hybrid assay detects RNAs that bind yeast Mpt5p. (A) Strategy. A yeast strain containing HIS3 and LacZ reporter genes under control of the LexA operator is transformed with RNA binding protein (X) fused to the GAL4 transcription activation domain (AD). Transformation with plasmids encoding a library of hybrid RNAs promotes recruitment of protein X to the reporter genes (e.g., HIS3) and so stimulates their expression. (B) Summary of the screen. Transformants were screened successively for growth under HIS3-selective conditions, activation of the LacZ gene, and dependence of activation on the X/AD plasmid. (C) Distribution of apparent RNA ligands among regions of the yeast genome and transcriptome. “Other” includes both apparent antisense and intergenic transcripts.
The library of RNAs was constructed by cloning short fragments of yeast genomic DNA into a three-hybrid vector (see Materials and Methods). Because the average insert size is ∼100 nt, a large number of yeast transformants are needed to cover all mRNA sequences. In our screen for RNAs that bound Mpt5p, 12 million transformants were analyzed (Fig. 1B). Of these, 246 grew on media selecting for activation of the HIS3 reporter gene. Two hundred fourteen (86%) of those also activated a second reporter gene, LacZ. RNA hybrid plasmids from these HIS3 + LacZ+ transformants were recovered, transformed into Escherichia coli, and sequenced. Plasmids were reintroduced into yeast to verify that they bound Mpt5p specifically (see below). From the 110 recovered plasmids, 69 unique hybrid RNAs were identified that activated transcription in the presence of Mpt5p/AD. Several RNAs were identified in multiple independent isolates, suggesting that the screen approached saturation. In addition, certain RNA plasmids activated reporter transcription on their own. Such RNAs, initially referred to as “RNA activators” (Sengupta et al. 1999; Saha et al. 2003), and later, “riboactivators” (Buskirk et al. 2003), were not analyzed further.
The hybrid RNA library represented all portions of the yeast genome, including sequences not present in mature RNAs: introns, intergenic, and antisense sequences. Plasmids carrying RNAs corresponding to 3′UTRs represented 36% of the total number of positives. This frequency is high, given that yeast 3′UTRs are typically 50–150 nt in length (Graber et al. 2002) and so represent <10% of total genomic sequence. The largest class of positives, comprising 42% (29/69) of the RNAs, apparently lay in antisense or intergenic regions (defined as sequences >500 nt from the nearest open reading frame) (Fig. 1C). A complete list of the unique RNAs recovered is presented in Supplemental Figure 1 (http://www.biochem.wisc.edu/wickens/publications.html). We focused on the 3′UTR RNAs, since, to date, all known actions of PUF proteins require binding to elements in 3′UTRs (Wickens et al. 2002). However, RNAs from other regions may well be genuine targets.
Specificity
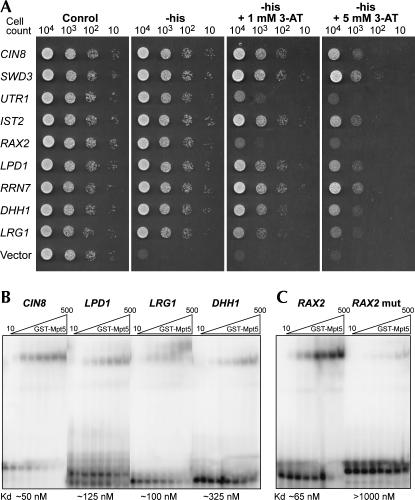
To confirm that the RNAs interacted with Mpt5p, plasmids carrying RNAs were reintroduced into yeast containing either the Mpt5p/AD fusion protein or the AD protein alone; a representative set is shown (Fig. 2A). Of the 69 unique plasmids tested in this fashion, 66 (95.7%) required Mpt5p/AD to activate HIS3, and did not activate in the presence of AD alone. Transformants differed in their resistance to 3-aminotriazole (3-AT), a competitive inhibitor of the His3p enzyme, suggesting differences in affinity for Mpt5p (Hook et al. 2005).
FIGURE 2.
Affinities tested in secondary tests and in vitro. (A) Three-hybrid tests. Serial dilutions of cells containing Mpt5p/AD and the indicated RNA segment were plated under increasing stringency, selecting for varying levels of expression of the reporter gene, HIS3. (Growth at higher concentrations of 3-aminotriazole [3-AT] requires higher levels of His3p.) (B) In vitro binding. Radiolabeled 3′UTR segments were incubated with purified, recombinant Mpt5p and analyzed using an electrophoretic mobility shift assay (EMSA). Dissociation constants (bottom) were determined by Sigma plot analysis of bound and free radiolabeled RNA. (C) RNA binding specificity. Wild-type or mutant forms of the RAX2 3′UTR segment were incubated with purified Mpt5p in vitro, and analyzed by EMSA.
To test whether the sequences identified bound directly to Mpt5p, we performed electrophoretic mobility shift assays using purified, recombinant Mpt5p protein with five of the RNA segments identified in the screen (Fig. 2B,C). These RNAs were selected because they cover a range of apparent affinities, as inferred from HIS3 expression levels. The RNAs bound with apparent dissociation constants ranging from 50 to 325 nM. Binding was RNA sequence-specific: a mutation of the UGU sequence, known to be critical for the binding of PUF proteins to other RNAs (Wickens et al. 2002), reduced binding of the RAX2 site to undetectable levels (Fig. 2C).
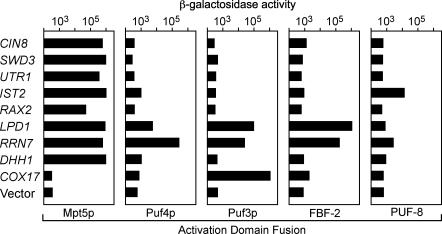
The three-hybrid system enables rapid assessments of sequence specificity. We determined the binding specificity of several 3′UTR RNAs by testing them against multiple PUF proteins: yeast Puf3p and Puf4p, and Caenorhabditis elegans FBF-2 and PUF-8. To do so, we assayed β-galactosidase activity in extracts of yeast that had been expressing various RNA and protein combinations (Fig. 3). β-Galactosidase activity is directly related to Kd in this range (Hook et al. 2005). Most of the RNAs bind tightly only to Mpt5p. Two RNAs, LPD1 and RRN7, are promiscuous, binding several PUF proteins. The COX17 3′UTR, a target of Puf3p (Olivas and Parker 2000), bound only that protein and was not identified in our screen. Interactions of a single RNA segment with multiple PUF proteins is consistent with immunoprecipitation analyses of yeast PUF proteins (Gerber et al. 2004). FBF-2 and PUF-8 bound different RNAs, as expected (Opperman et al. 2005).
FIGURE 3.
Protein specificity of RNA targets Plasmids encoding the RNAs indicated were tested in combination with plasmids encoding AD fusions of several PUF proteins. Mpt5p, Puf5p, and Puf3p are derived from S. cerevisiae, and FBF-2 and PUF-8 from C. elegans. β-Galactosidase activity, which is proportional to Kd from at least 10–100 nM Kd (Hook et al. 2005), is displayed for each combination of RNA and protein. Assays were performed in cell lysates, normalized to cell number (Hook et al. 2005).
Targets in vivo
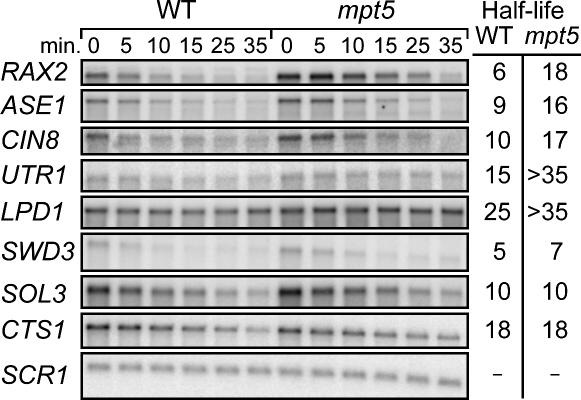
To be considered bona fide targets of Mpt5p, the protein not only must bind the RNA but regulate it in vivo. PUF proteins can control mRNA stability, translation, or localization (Wickens et al. 2002; Gu et al. 2004). We focused on effects on mRNA turnover. To quantify mRNA half-lives, we performed “transcription shut-off” experiments. The drug, thiolutin, was used to inhibit RNA polymerase II rapidly in intact yeast (Jimenez et al. 1973; Tipper 1973; Grigull et al. 2004). It was added to a logarithmically growing culture, and RNAs were isolated at various times and analyzed by Northern blotting. Of the 15 mRNAs tested, seven were stabilized in mpt5 mutants (Fig. 4; not shown). For example, the half-life of RAX2 mRNA increased two- to threefold in mpt5 mutants. CTS1 and SCR1 RNAs, which do not interact with Mpt5p, were unaffected. We conclude that Mpt5p binds to and enhances the degradation of several of the RNAs identified in the screen.
FIGURE 4.
Candidate target mRNAs are destabilized by MPT5 in vivo Northern blot analysis of putative MPT5 target mRNAs at various times following transcription inhibition in wild-type and mpt5 deletion strains. A representative set of RNAs is shown. Total cellular RNA was analyzed. mRNA half-lives are provided to the right. All experiments were performed in triplicate, including CTS1 and SCR1 RNAs, neither of which binds Mpt5p. These RNAs provide controls for RNA recovery as well as sequence specificity.
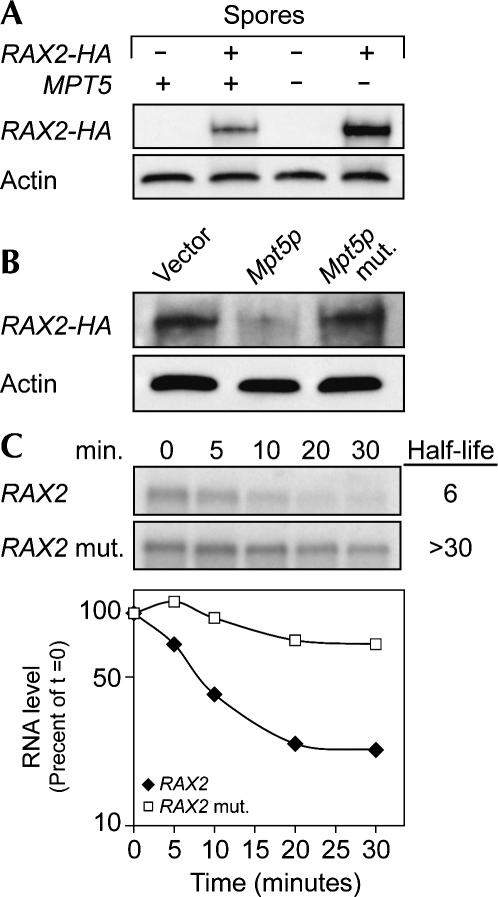
To determine independently whether RNAs identified are indeed regulated by Mpt5p in vivo, we focused on RAX2 mRNA. Three experiments corroborate that RAX2 mRNA is specifically controlled by Mpt5p. First, Rax2p protein levels were elevated in mpt5 mutant strains. Haploid strains were mated to create the heterozygous diploid, MPT5:mpt5 RAX2:RAX2-HA. After sporulation, Rax2p protein levels were assayed in haploid strains derived from the four spores of a single tetrad (Fig. 5A). Rax2-HA protein was two- to threefold more abundant in the mpt5 vs. MPT5 wild-type strain, while actin protein levels were unaffected (Fig. 5A). Second, repression in mpt5 mutants was restored by overexpression of wild-type Mpt5p protein, but not by overexpression of an RNA-binding defective point mutant form of the protein (Fig. 5B; Goldstrohm et al. 2006). The steady-state level of mRNA was comparably affected (not shown). Third, replacement of the RAX2 3′UTR with that of ADH1 in the normal RAX2 chromosomal locus (RAX2 mut) stabilized the mRNA, even though Mpt5p was present (Fig. 5C). Thus, Mpt5p regulation of RAX2 mRNA is contingent upon the protein's RNA binding activity and the 3′UTR of the mRNA.
FIGURE 5.
Sequence specificity of RAX2 control. (A) Mpt5p reduces Rax2p protein abundance. Western blot analysis of Rax2p levels in haploids derived from a sporulated, heterozygous, RAX2:RAX2-HA MPT5:mpt5 diploid. The presence of wild-type MPT5 and the HA-tagged allele of RAX2 were determined by Western blotting, as indicated above the lanes. HA antibody was used to detect the HA-tagged RAX2 allele. Actin protein levels were used as a protein loading control. (B) Expression of wild-type Mpt5p is sufficient to decrease Rax2p abundance. Plasmids containing no insert (“vector”), Mpt5p, and or an RNA-binding defective mutant in Mpt5p (Mpt5p mut) were introduced into an mpt5 deletion strain. HA-Rax2p was detected by Western blotting. (C) RAX2 3′UTR is required for repression. The abundance and decay of RAX2 mRNA bearing its own 3′UTR (“RAX2”) or that of ADH1 (“RAX2 mut”) was analyzed by Northern blotting following thiolutin addition. Below, semi-log plot of transcript levels, normalized to t = 0 for each RNA.
Regulation during the cell cycle
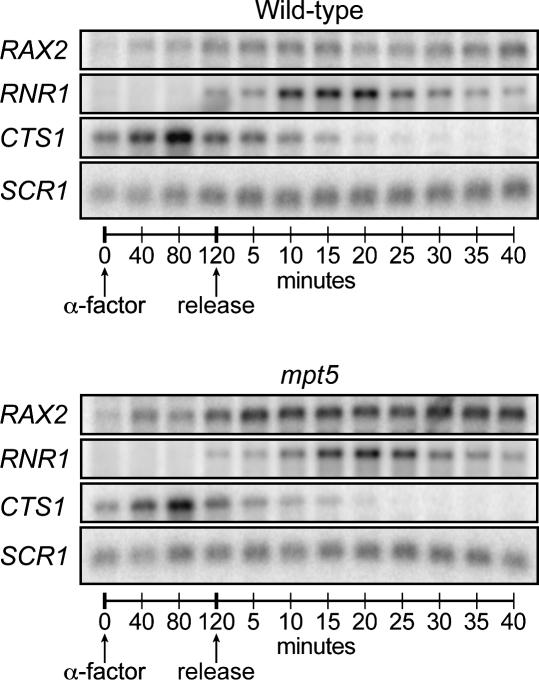
To examine the physiological role of Mpt5p regulation, we first analyzed RAX2 mRNA during the cell cycle. Cultures were arrested with α factor mating pheromone for two hours, the approximate time required for two cell divisions, thus synchronizing cultures in early G1. Upon release from α factor arrest, endogenous RAX2 transcription was activated, leading to a peak of RAX2 mRNA in late G1 of wild-type cells (Fig. 6). In contrast, in mpt5 mutants, RAX2 message abundance was elevated fivefold shortly after release from arrest, and failed to return to low, wild-type levels through S and G2 phases (Fig. 6), as well as M and the following G1 (data not shown). Control transcripts, CTS1 and RNR1, which do not bind Mpt5p, were unaffected by deletion of MPT5 (Fig. 6).
FIGURE 6.
Cell cycle consequences α factor mating pheromone was added to wild-type or mpt5 mutants. After 120 min, cells were released from α factor arrest, and RNAs prepared at 5 min intervals thereafter. RAX2 mRNA levels were determined by Northern blotting, using RNR1, CTS1, and SCR1 as controls. The controls show that Mpt5p affects neither RNR1 (whose abundance peaks in late G1 and early S phases) (Elledge and Davis 1990) nor CTS1 RNA (which accumulates during the α factor block) (Kovacech et al. 1996).
These findings suggested that Mpt5p might regulate other mRNAs that oscillate during the cell cycle. Indeed, Mpt5p regulates HO mRNA, which increases greatly in abundance in mpt5 mutants during G1, after release from α factor arrest (data not shown).
DISCUSSION
Our results demonstrate that the inverse three-hybrid approach yields multiple mRNAs that are regulated by a single protein: in this case, Mpt5p. Most of the RNAs identified in our screen likely bind Mpt5p in vitro (e.g., Fig. 2). Roughly half of the 3′UTRs tested showed Mpt5p-dependent changes in half-life (Fig. 4). This is a minimum estimate of the fraction of mRNAs that are genuine targets, for two reasons. First, in some instances, multiple PUF proteins bind the same mRNA (Gerber et al. 2004; Fig. 3; not shown); in these cases, multiple PUF genes must be deleted to reveal defects in control of that mRNA. Second, RNAs whose stability was unaffected in mpt5 mutants nonetheless may be regulated under specific conditions, or at the levels of translation or localization (Wickens et al. 2002; Gu et al. 2004; Goldstrohm et al. 2006).
The three-hybrid system complements biochemical and bioinformatic strategies. In one biochemical approach, a specific protein is immunoprecipitated from a cellular extract, and the RNAs to which it is bound were identified using a DNA microarray. This protocol can successfully identify RNAs that interact with an RNA-binding protein (Tenenbaum et al. 2000; Gerber et al. 2004). However, not all RNAs that are associated with the protein in the extract are associated in vivo: extraneous binding can occur after the cells are lysed (Mili and Steitz 2004). UV crosslinking in vivo can minimize this complication (Ule et al. 2003). Conversely, RNAs that are actually bound in vivo can be missed because the RNA is rare, or because only a small fraction of the target mRNA molecules are bound in vivo (Wreden et al. 1997), or because the mRNA–protein interaction only occurs under specific circumstances (Tenenbaum et al. 2000).
Our results with Mpt5p can be directly compared to those obtained using the same protein in an immunoprecipitation/microarray protocol (Gerber et al. 2004). In that work, 224 RNAs were identified that were bound to Mpt5p in a yeast extract. Ten of these RNAs were identified in our screen; 59 others were identified only in the three-hybrid approach. Of the 25 3′UTRs identified in our screen, seven were detected by immunoprecipitation. Our half-life measurements demonstrate that mRNAs detected only in our screen nonetheless are genuine targets of MPT5 (Fig. 4), as are RNAs identified by both methods. Thus, the sets of targets identified in the two approaches overlap, but are distinct.
Immunoprecipitation-based approaches can detect only those RNAs that are expressed and bound to the protein under the experimental conditions used. In yeast, mRNA abundances are regulated dramatically with carbon source, diauxic shift (DeRisi et al. 1997), α factor concentration (Roberts et al. 2000), and many other conditions (Gasch et al. 2000, 2001; Gasch and Werner-Washburne 2002). Several mRNAs we identified are transcribed only under very narrow circumstances. For example, SPO12 is only expressed during meiosis (Chu et al. 1998). Similarly, physical interactions may occur only under specific conditions, as observed with the dynamic HuB mRNP in mammalian cells (Tenenbaum et al. 2000). The inverse three-hybrid approach largely circumvents these problems. On the other hand, interactions that require multiple components can be detected by immunoprecipitation, but will be missed in the three-hybrid assay.
Ninety-seven percent of the RNA segments we identified possess one or more UGUR sequence (Supplemental Fig. 1, http://www.biochem.wisc.edu/wickens/publications.html). This tetranucleotide is required for all PUF–RNA interactions identified to date (Wickens et al. 2002). Mutations that disrupt the UGUR in several of the RNAs we identified abrogate binding (Fig. 2C; not shown). The consensus derived from the three-hybrid RNAs is UGUAHYHNWN. In immunoprepicipitation/microarray studies, 32% of the targets possess a subset of this consensus, namely UGUAAYAWUA (Gerber et al. 2004). In that work, the PUF protein may have been bound indirectly to the remaining 68% of the RNAs, or recognize a different sequence.
Inverse three-hybrid screens can be used to identify targets of RNA-binding proteins, regardless of their function or origin. In the human genome, ∼10,000 mRNAs appear to contain conserved sites for regulatory proteins in their 3′UTRs (Wasserman et al. 2000; Xie et al. 2005). The identification of the cognate proteins, and of the RNAs that are coregulated by a specific protein, is a critical task. Application of the inverse three-hybrid assay to organisms with large genomes, such as humans, will entail the construction of suitable libraries with reduced complexity, perhaps derived from exclusively expressed RNAs, or alternative RNA expression strategies. The two-hybrid system has complemented mass spectrometry to create maps of protein–protein interactions. Similarly, the three-hybrid system may complement cDNA array-based and bioinformatic approaches to help construct large-scale RNA–protein linkage maps.
MATERIALS AND METHODS
Screen
The RNA library was created by ligation of genomic DNA fragments into the pIII/MS2–2 plasmid (Sengupta et al. 1999). Chromosomal DNA was isolated from yeast strain S288C, digested with restriction enzymes MseI, Tsp50091, AluI, and RsaI, and 50–150-bp fragments inserted into the SmaI site of pIII/MS2–2. The RNA expression library was transformed into strain YBZ-1 (Hook et al. 2005) containing pMPT5-AD by standard yeast transformation methods. Transformants were spread on plates containing selective media (SD–uracil–leucine–histidine + 4.0 mM 3-aminotriazole). Following 5-d growth at 30°C, colonies were picked and patched onto fresh plates.
Transcription shut-off
RNA polymerase inhibition was performed as in Duttagupta et al. (2003). Cultures were grown in 50 mL YPAD to an OD600 ∼0.8. Cells were collected by centrifugation and resuspended in 10 mL YPAD. Cultures were grown at 30°C with agitation for 20 min before addition of bathocuproinedisulfonic acid (150 μM final concentration). Cultures were incubated for 2 min before addition cupric sulphate (150 picoM final concentration) and thiolutin (20 μg/mL final concentration) (a gift from Pfizer). Aliquots are taken as specific time points following RNA polymerase arrest, centrifuged, decanted, and frozen in liquid nitrogen for later RNA extraction.
α Factor arrest
Cells were synchronized and described in Price et al. (1991). Cultures were grown in YPAD to OD600 ∼0.8, washed, and resuspended in YPAD containing 10 μg/mL yeast mating pheromone α (Sigma). Cultures were grown for 2 h resulting in >90% cells arrested in G1 as determined under the microscope. Cells were separated by filtration and resuspended in YPAD media. Aliquots were taken at specified points following release from α factor arrest.
RNA extraction and Northern blotting
Cell pellets were resuspended in 500 μL 1:1 acid phenol/chloroform and 500 μL TENS (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.3 M NaCl, 0.2% SDS), heated at 70°C for 5 min, then vortexed for 2 min. The aqueous phase was removed following centrifugation and added to 500 μL 1:1 acid phenol/chloroform and votexed briefly. The aqueous phase was removed again, and added to 500 μL chloroform to remove phenol. Following brief vortexing and centrifugation, the nucleic acids in the aqueous phase were precipitated with 150 mM NaOAc and 2.5 volumes ethanol. Total RNA was loaded (2 μg/sample) into 1.0% agarose, 3.7% formaldehyde, 1× MOPS gel, and run for ∼1.5 h at 80 V. Nucleic acid was transferred by capillary action using 10× SSC for ∼4 h onto a charged nylon membrane (Ambion). Hybridization and subsequent washes were performed as directed (Ambion; Ultra-Hyb). A radioactive signal was detected using Typhoon PhosphorImager (Amersham), and analyzed using ImageQuant (Molecular Dynamics).
Western blotting
Cultures were grown in rich or selective media to a density of OD600 ∼0.8. Aliquots were centrifuged and pellets frozen in liquid nitrogen. Pellets were resuspended in 100 μL ice-cold 1× Complete Protease Inhibitors (Roche). Equal volumes of 2× SDS protein loading buffer (final concentration 62.5 mM Tris HCl (pH 6.8), 2% SDS, 10% glycerol, 0.01% bromophenol blue, 2.5% BME) and glass beads were added. Samples were lysed mechanically by vortexing at 4°C for 5 min. Samples were separated on gradient SDS polyacrylamide gels (Cambrex) and transferred to a PVDF membrane. Primary and secondary antibodies were hybridized using standard laboratory techniques and a signal was visualized using the ECL reagent (Amersham).
Protein purification
MPT5 (amino acids 126–626) was cloned into pGex6P1 (Amersham). Protein expression, purification, and purify were performed as described in Bernstein et al. (2005).
Electrophoretic mobility shift assays
One hundred femtomoles 32P-end-labeled RNA oligonucleotides (IDT) were combined with proteins over a range of protein concentrations. Protein and RNA were incubated at room temperature for 30 min in 10 mM HEPES (pH 7.4), 1 mM EDTA, 50 mM KCl, 2 mM DTT, and 0.02% Tween-20. Two microliters of loading dye (10% [v/v] Ficoll 400,000, 0.05% [w/v] xylene cyanol) were added before loading on a prerun native polyacrylamide gel (6% [w/v] 29:1 acrylamide/bis-acrylamide, 0.5× TBE). Gels were resolved at 200 V for 2 h at 4°C. The fraction of retarded RNA, relative to the total in the incubation, was determined using ImageQuant (Amersham).
β-Galactosidase activity measurements
Quantitation of LacZ reporter expression was performed as described by Hook et al. (2005).
ACKNOWLEDGMENTS
We thank Dr. Jeffery Coller (Case Western Reserve) and Craig Stumpf for help with the screen and Dr. Hay-Oak Park (The Ohio State University) for providing the HA-tagged RAX2 strain. We appreciate MPT5 constructs from Dr. Aaron Goldstrohm, and helpful discussions of the work and the manuscript with members of the Wickens laboratory. We thank the Biochemistry Medialab and Laura vander Ploeg for assistance with figures. This work was supported by an NIH research grant to M.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.145306.
REFERENCES
- Barker D.D., Wang C., Moore J., Dickinson L.K., Lehmann R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes & Dev. 1992;6:2312–2326. doi: 10.1101/gad.6.12a.2312. [DOI] [PubMed] [Google Scholar]
- Bernstein D.S., Buter N., Stumpf C., Wickens M. Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods. 2002;26:123–141. doi: 10.1016/S1046-2023(02)00015-4. [DOI] [PubMed] [Google Scholar]
- Bernstein D., Hook B., Hajarnavis A., Opperman L., Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk A.R., Kehayova P.D., Landrigan A., Liu D.R. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Crittenden S.L., Bernstein D.S., Bachorik J.L., Thompson B.E., Gallegos M., Petcherski A.G., Moulder G., Barstead R., Wickens M., Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans . Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Dubnau J., Chiang A.S., Grady L., Barditch J., Gossweiler S., McNeil J., Smith P., Buldoc F., Scott R., Certa U., et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Duttagupta R., Vasudevan S., Wilusz C.J., Peltz S.W. A yeast homologue of Hsp70, Ssa1p, regulates turnover of the MFA2 transcript through its AU-rich 3′ untranslated region. Mol. Cell. Biol. 2003;23:2623–2632. doi: 10.1128/MCB.23.8.2623-2632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S.J., Davis R.W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes & Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- Forbes A., Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Huang M., Metzner S., Botstein D., Elledge S.J., Brown P.O. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate mRNAs. Nat. Struct. Mol. Biol. 2006 doi: 10.1038/nsmb1100. doi:10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Graber J.H., McAllister G.D., Smith T.F. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh S., Pootoolal J., Robinson M.D., Hughes T.R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Deng Y., Zenklusen D., Singer R.H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes & Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook B., Bernstein D., Zhang B., Wickens M. RNA–protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA. 2005;11:227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A., Tipper D.J., Davies J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae . Antimicrob. Agents Chemother. 1973;3:729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacech B., Nasmyth K., Schuster T. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol. Cell. Biol. 1996;16:3264–3274. doi: 10.1128/mcb.16.7.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont L.B., Crittenden S.L., Bernstein D., Wickens M., Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin H., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Mili S., Steitz J.A. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Wharton R.P. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Olivas W., Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman L., Hook B., DeFino M., Bernstein D.S., Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Price C., Nasmyth K., Schuster T. A general approach to the isolation of cell cycle-regulated genes in the budding yeast. Saccharomyces cerevisiae. J. Mol. Biol. 1991;218:543–556. doi: 10.1016/0022-2836(91)90700-g. [DOI] [PubMed] [Google Scholar]
- Roberts C.J., Nelson B., Marton M.J., Stoughton R., Meyer M.R., Bennett H.A., He Y.D., Dai H., Walker W.L., Hughes T.R., et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Saha S., Ansari A.Z., Jarrell K.A., Ptashne M. RNA sequences that work as transcriptional activating regions. Nucleic Acids Res. 2003;31:1565–1570. doi: 10.1093/nar/gkg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A., Rossi L., Lena A., Batistoni R., Deri P., Rainaldi G., Locci M.T., Evangelista M., Gremigni V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- Sengupta D.J., Wickens M., Fields S. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA. 1999;5:596–601. doi: 10.1017/s1355838299002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadauchi T., Matsumoto K., Herskowitz I., Irie K. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 2001;20:552–561. doi: 10.1093/emboj/20.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum S.A., Carson C.C., Lager P.J., Keene J.D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipper D.J. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J. Bacteriol. 1973;116:245–256. doi: 10.1128/jb.116.1.245-256.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Wickens M., Bernstein D.S., Kimble J., Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wreden C., Verrotti A.C., Schisa J.A., Lieberfarb M.E., Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B., Petritsch C., Clark I.E., Gavis E.R., Jan L.Y., Jan Y.N. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M.P. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]