Abstract
In Drosophila melanogaster, the Dicer-2/R2D2 complex initiates RNA interference (RNAi) by processing long double-stranded RNA (dsRNA) into small interfering RNA (siRNA). Recent biochemical studies suggest that the Dcr-2/R2D2 complex also facilitates incorporation of siRNA into the RNA-induced silencing complex (siRISC). Here we present genetic evidence that R2D2 and Dcr-2 are both required for loading siRNA onto the siRISC complex. Consistent with this, only the Dcr-2/R2D2 complex, but neither Dcr-2 nor R2D2 alone, can efficiently interact with duplex siRNA. Furthermore, both dsRNA-binding domains of R2D2 are critical for binding to siRNA and promoting assembly of the siRISC complexes.
Keywords: RNA interference (RNAi), Dicer-2, R2D2, siRNA, siRISC
INTRODUCTION
RNA interference (RNAi) is an evolutionarily conserved gene-silencing pathway whereby double-stranded RNA (dsRNA) molecules trigger sequence-specific post-transcriptional silencing of cognate mRNA (Hannon 2002; Tomari and Zamore 2005). In general, the RNAi pathway can be initiated by processing long dsRNA or short hairpin RNA into small interfering RNA (siRNA) or microRNA (miRNA), respectively. Both siRNA and miRNA are 21–25 nucleotide (nt) duplexes with 5′ phosphate and 3′ 2 nt overhang, hydroxyl termini (Tomari and Zamore 2005). However, the majority of miRNA, but not siRNA, duplexes contain mismatches, bulges, or G:U wobble base pairs. Nascent siRNA and miRNA are incorporated into the RNA-induced silencing complex termed siRISC and miRISC. The effector RISC complexes contain single-stranded siRNA or miRNA as guide RNA for sequence-specific cleavage and/or translational repression of complementary mRNA.
Both siRNA and miRNA are generated by Dicer, a family of large multidomain RNase III enzymes (Bernstein et al. 2001). In Drosophila, two distinct Dicer (Dcr) complexes, Dcr-1/Loquacious (Loqs)-PB (also known as R3D1-L [long]) and Dcr-2/R2D2, generate miRNA and siRNA, respectively (Liu et al. 2003; Forstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). Despite sharing extensive sequence homology, Dcr-1 and Dcr-2 enzymes display different ATP requirements and substrate specificities (Lee et al. 2004; Jiang et al. 2005). Dcr-1 is ATP-independent and prefers to process the stem–loop precursor of miRNA (pre-miRNA)(Jiang et al. 2005). On the other hand, Dcr-2 favors long dsRNA as its ideal substrate, and requires ATP hydrolysis for efficient siRNA production (Liu et al. 2003; Jiang et al. 2005). Both R2D2 and Loqs belong to a growing family of dsRNA-binding proteins that function in tandem with specific RNase III enzymes. There are two putative dsRNA-binding domains in R2D2 and three in Loqs. The loqs gene encodes at least two protein isoforms, PA (also known as R3D1-S [short]) and PB, by alternative splicing (Forstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). While R2D2 does not regulate the siRNA-generating activity of Dcr-2, Loqs-PB greatly enhances Dcr-1’s miRNA-generating activity possibly by increasing its affinity for pre-miRNA (Liu et al. 2003; Jiang et al. 2005; Saito et al. 2005).
Recent biochemical studies suggest that the Dcr-2/R2D2 complex also binds duplex siRNA, forms the RISC loading complex (RLC), and facilitates the transfer of siRNA onto Ago2, the catalytic component of siRISC (Liu et al. 2003; Pham et al. 2004; Tomari et al. 2004a). We previously employed an in vitro reconstitution assay to demonstrate that recombinant Dcr-2/R2D2 complex enhances incorporation of siRNA into the effector siRISC complex (Liu et al. 2003). Furthermore, photocross-linking experiments suggest that R2D2 preferentially binds the more stable end of siRNA, whereas Dcr-2 is often found at the less stable end (Tomari et al. 2004b). Thus, R2D2 could serve as the sensor for the thermodynamic asymmetry of siRNA, thereby orienting Dcr-2 to the end that is easier to unwind. In the current study, we describe the generation and analysis of an r2d21 null fly. We show that R2D2 and Dcr-2 are both required for loading siRNA onto the siRISC complex. Only the Dcr-2/R2D2 complex, but neither Dcr-2 nor R2D2 alone, can efficiently interact with the siRNA duplex. Finally, both dsRNA-binding domains of R2D2 are critical for siRNA binding and siRISC assembly. Therefore, Dcr-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes.
RESULTS
Generation of the r2d21 null fly
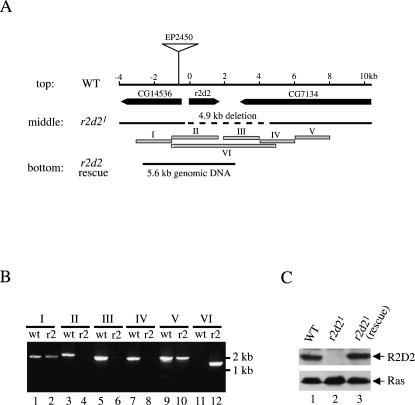
We previously purified the siRNA-generating enzyme from Drosophila S2 cells and discovered that it consisted of DCR-2 and a novel protein that we named R2D2 (Liu et al. 2003). R2D2 contains tandem dsRNA-binding domains (R2) and forms a heterodimeric complex with Dcr-2 (D2). To study the physiological functions of R2D2, we generated r2d2 deletion flies by imprecise excision of a P-element inserted near the r2d2 locus. One such mutant was named r2d21, and was selected for further characterization (Liu et al. 2003). Based on PCR and sequencing, the r2d21 mutant locus contained a ∼4.9-kb deletion, which removed the entire r2d2 open reading frame (ORF) as well as ∼1-kb upstream and ∼3-kb downstream sequences (Fig. 1A,B). Consequently, no R2D2 protein could be detected in homozygous r2d21 fly lysates by Western blot analysis (Fig. 1C). Therefore, we concluded that r2d21 was a null allele. The r2d21 null flies display several developmental abnormalities and female sterility. All of these phenotypes could be completely rescued by a ∼5.6-kb genomic fragment that contained only an intact r2d2 gene (Fig. 1A,C) (S. Kalidas et al., unpubl.). These findings are consistent with the notion that r2d21 null phenotypes are caused by the deletion of the r2d2 gene.
FIGURE 1.
Generation of the r2d21 null mutant. (A, top) A schematic map of the wild-type (WT) r2d2 gene locus. Three open reading frames (ORF) were shown in the 14-kb genomic region. The P-element (EP2450) was inserted ∼0.6 kb upstream of the r2d2 ORF. (middle) A ∼4.9-kb genomic region, represented by the dash line, was deleted in the r2d21 mutant locus. The deletion was mapped by genomic PCR as illustrated below. (bottom) A 5.6-kb genomic DNA fragment that completely rescued all phenotypes of r2d21 mutant flies. (B) PCR mapping the deletion at the r2d21 mutant locus. Six (I–VI) PCR reactions (A) were performed using genomic DNA template isolated from wild-type (wt, odd lanes) or homozygous r2d21 mutant flies (r2, even lanes). The PCR condition used could not allow amplification of the ∼5.9-kb VI PCR product in the wild-type sample. (C) Lysates were prepared from wild-type flies (lane 1) and homozygous r2d21 mutant flies without (lane 2) or with the rescue transgene (lane 3). Western blots were performed with anti-R2D2 and anti-Ras antibodies.
Dcr-2 stabilizes R2D2 in vivo
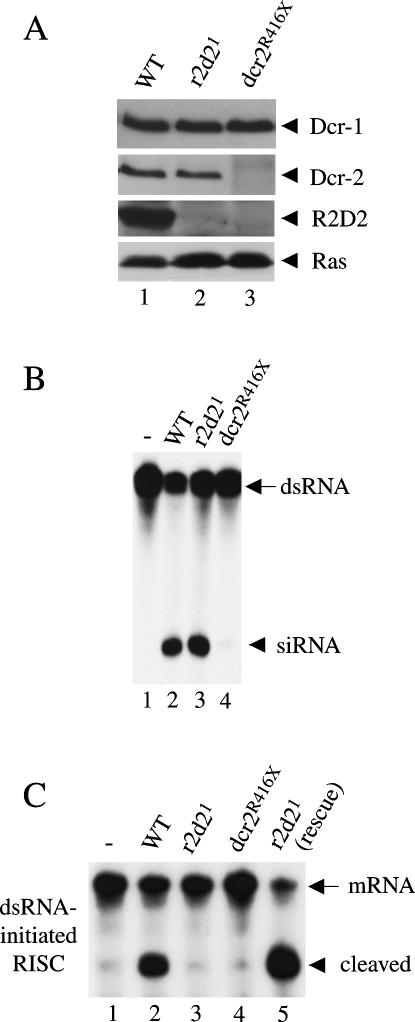
The majority of Dcr-2 and R2D2 proteins form a stable complex in vivo. Both R2D2 and Dcr-2 can be immunodepleted from Drosophila S2 cell extracts using anti-R2D2 antibodies. Moreover, our previous studies show that depletion of Dcr-2 by RNAi also diminishes the level of R2D2 protein, whereas RNAi of R2D2 causes a modest reduction in Dcr-2 protein in S2 cells (Liu et al. 2003). Thus, Dcr-2 and R2D2 may stabilize each other in vivo, and either protein alone is less stable. To confirm this in flies, we measured the levels of Dcr-2 and R2D2 proteins in homozygous r2d21 or dcr-2R416X flies by Western blot analysis. The dcr-2R416X mutant is considered a null allele because it introduces a premature stop codon at the amino (N)-terminus of dcr-2 ORF (Lee et al. 2004). While there was little or no change in the level of Dcr-2 in r2d21 mutant extract, neither Dcr-2 nor R2D2 were detected in dcr-2R416X mutant extract (Fig. 2A). Therefore, Dcr-2 is required for the stability of R2D2, but the stability of Dcr-2 does not depend on R2D2 in the fruitfly.
FIGURE 2.
Dcr-2 stabilizes R2D2 in vivo, and R2D2 is required for RNAi. (A) Lysates were prepared from wild-type (lane 1) and homozygous r2d21 (lane 2) or dcr-2R416X (lane 3) flies. The levels of Dcr-1, Dcr-2, R2D2, and Ras proteins were measured by Western blots with the corresponding antibodies (Dcr-1/Dcr-2 or R2D2/Ras used the same blot). (B) The dsRNA-processing assays were performed in buffer alone (lane 1) or with 8 μg wild-type (lane 2), r2d21 (lane 3), and dcr-2R416X (lane 4) whole fly lysates. (C) The dsRNA-initiated RISC assays were performed in buffer alone (lane 1) or 32-μg ovary lysates prepared from wild-type (lane 2), r2d21 (lane 3), dcr-2R416X (lane 4), and r2d21 mutant flies carrying the rescue transgene (lane 5).
R2D2 is dispensable for siRNA production
To determine if R2D2 is required for siRNA production in vivo, we performed dsRNA-processing assays using whole fly or ovary extracts prepared from wild-type and homozygous r2d21 or dcr-2R416X flies. As shown previously, the majority of siRNA-generating activity was removed in dcr-2R416X extract (Fig. 2B; Liu et al. 2003; Lee et al. 2004; Pham et al. 2004). The remaining activity was likely contributed by Dcr-1 (data not shown). By contrast, r2d21 mutant extract was as efficient in siRNA production as wild-type extract (Fig. 2B and data not shown). Thus, R2D2 is dispensable for siRNA production in vivo. These results are consistent with our previous finding that recombinant R2D2 does not directly regulate the ability of recombinant Dcr-2 to recruit or process dsRNA in vitro.
R2D2 is required for loading siRNA onto siRISC
To determine if R2D2 is essential for RNAi, we performed long dsRNA-initiated RISC (sequence-specific mRNA cleavage) assays using wild-type, r2d21, and dcr-2R416X ovary extracts. As shown in Figure 2C, dcr-2R416X extract was completely defective in this assay. Interestingly, this was also the case for r2d21 extract. However, unlike dcr-2R416X extract, r2d21 extract could efficiently process dsRNA trigger into siRNA. This defect of r2d21 mutant could be fully recovered by the r2d2 rescuing transgene. Thus, R2D2 is a bona fide component of the Drosophila RNAi pathway. Moreover, these data suggest that R2D2 functions downstream of siRNA production, for instance, by facilitating siRNA transfer from Dcr-2 to siRISC.
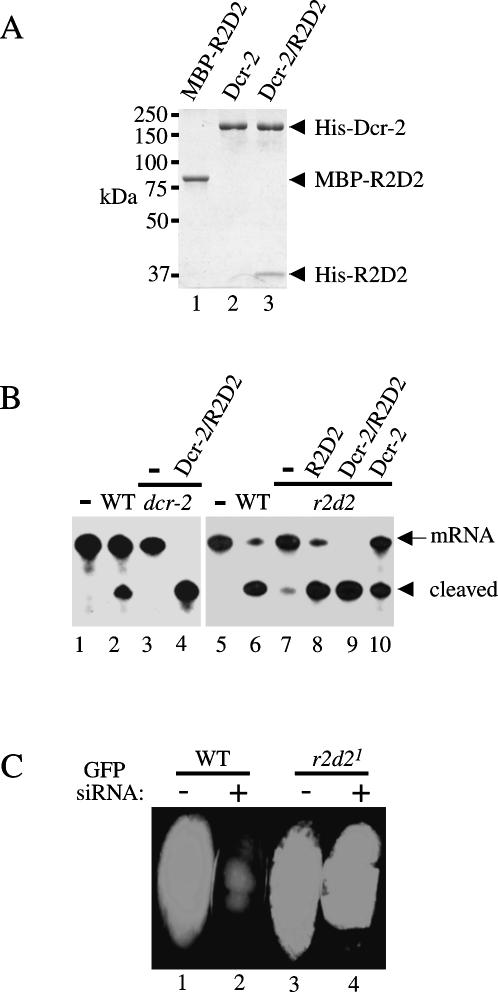
To determine if R2D2 is required for loading siRNA onto the siRISC complex, we performed (duplex) siRNA-initiated RISC assays by using wild-type, r2d21, and dcr-2R416X ovary extracts with or without polyhistidine (His)-tagged recombinant Dcr-2 or the Dcr-2/R2D2 complex produced from insect cells (Liu et al. 2003). We also generate a free and soluble maltose-binding protein (MBP)-R2D2 fusion protein in Escherichia coli BL21. MBP–R2D2 recombinant proteins were purified by an Amylose affinity column (NEB) followed by SP- and Q-Sepharose chromatography (Fig. 3A). As shown previously (Lee et al. 2004; Pham et al. 2004), the siRNA-initiated RISC activity was abolished in dcr-2R416X extract and was efficiently rescued by supplementing the recombinant Dcr-2/R2D2 complex (Fig. 3B). Likewise, we observed little siRNA-initiated RISC activity in r2d21 extract, which could be rescued partially by recombinant Dcr-2, or completely by MBP–R2D2 or the Dcr-2/R2D2 complex. These results indicate that Dcr-2 can promote inefficient assembly of the siRISC complex in the absence of R2D2. However, R2D2 greatly facilitates Dcr-2’s role in loading siRNA onto the siRISC complex.
FIGURE 3.
Both R2D2 and Dcr-2 are required for loading siRNA onto siRISC. (A) A coomassie-stained SDS-polyacrylamide gel (PAGE) showing purified recombinant MBP-R2D2 from E. coli (lane 1) and His-tagged Dcr-2 (lane 2) or Dcr-2/R2D2 complex (lane 3) proteins from insect cells. (B) The siRNA-initiated RISC assays were performed with buffer alone (lanes 1 and 5); 32-μg wild-type (lanes 2 and 6) ovary extract; 32-μg dcr-2R416X ovary extract alone (lane 3) or supplemented with 3-pmol recombinant Dcr-2/R2D2 complex (lane 4); 32 μg r2d21 ovary extract alone (lane 7) or in combination with 12.5-pmol recombinant MBP-R2D2 (lane 8), 5-pmol Dcr-2/R2D2 complex (lane 9), and 3-pmol Dcr-2 complex (lane 10). (C) Images include a wild-type or r2d21 mutant green fluorescent embryo injected with buffer control (lanes 1 and 3), or GFP siRNAs (lanes 2 and 4). In total, 32 of 36 wild-type embryos and 0 of 40 r2d21 mutant embryos showed GFP silencing.
To determine if R2D2 is critical for siRISC assembly in vivo, we crossed the r2d21 mutant flies with Ubiquitin (Ub)–GFP flies to derive homozygous r2d21; Ub–GFP mutant flies (Liu et al. 2003). We then collected wild-type or homozygous r2d21 mutant green fluorescent embryos for microinjection experiments as described (Liu et al. 2003). Previously, we injected long dsRNA for GFP into embryos to show that R2D2 is required for RNAi in the fruitfly. Here we used a mixture of GFP siRNAs that were generated from long dsRNA for embryo injection experiments. The GFP siRNAs could efficiently silence expression of green fluorescent proteins in wild-type but not r2d21 mutant embryos (Fig. 3C). These experiments, together with previous studies, convincingly demonstrate that R2D2 and Dcr-2 are both required for loading siRNA onto the siRISC complex (Liu et al. 2003; Lee et al. 2004; Pham et al. 2004; Tomari et al. 2004a).
Dcr-2 and R2D2 coordinately bind the siRNA duplex
Previous biochemical studies have suggested that the Dcr-2/R2D2 complex binds the siRNA duplex, forms the RISC loading complex (RLC), and facilitates incorporation of siRNA into the siRISC complex (Liu et al. 2003; Pham et al. 2004; Tomari et al. 2004b). To determine if either R2D2 or Dcr-2 could bind the siRNA alone, we performed native gel-shift assays by incubating the 5′ radiolabeled synthetic siRNA duplex with recombinant R2D2, Dcr-2, or the Dcr-2/R2D2 complex proteins. As shown in Figure 4, neither Dcr-2 nor R2D2 could efficiently interact with siRNA in this assay. A distinct mobility shift was observed only when Dcr-2 and R2D2 were combined in the reaction. Because of the MBP-tag, the siRNA shift generated by Dcr-2/MBP-R2D2 was slightly higher than that of the Dcr-2/R2D2 complex. These results suggest that Dcr-2 and R2D2 coordinately bind the siRNA duplex to promote assembly of the siRISC complex. Indeed, both Dcr-2 and R2D2 can be efficiently photocross-linked to radiolabeled siRNA by ultraviolet light (Liu et al. 2003; Tomari et al. 2004b).
FIGURE 4.
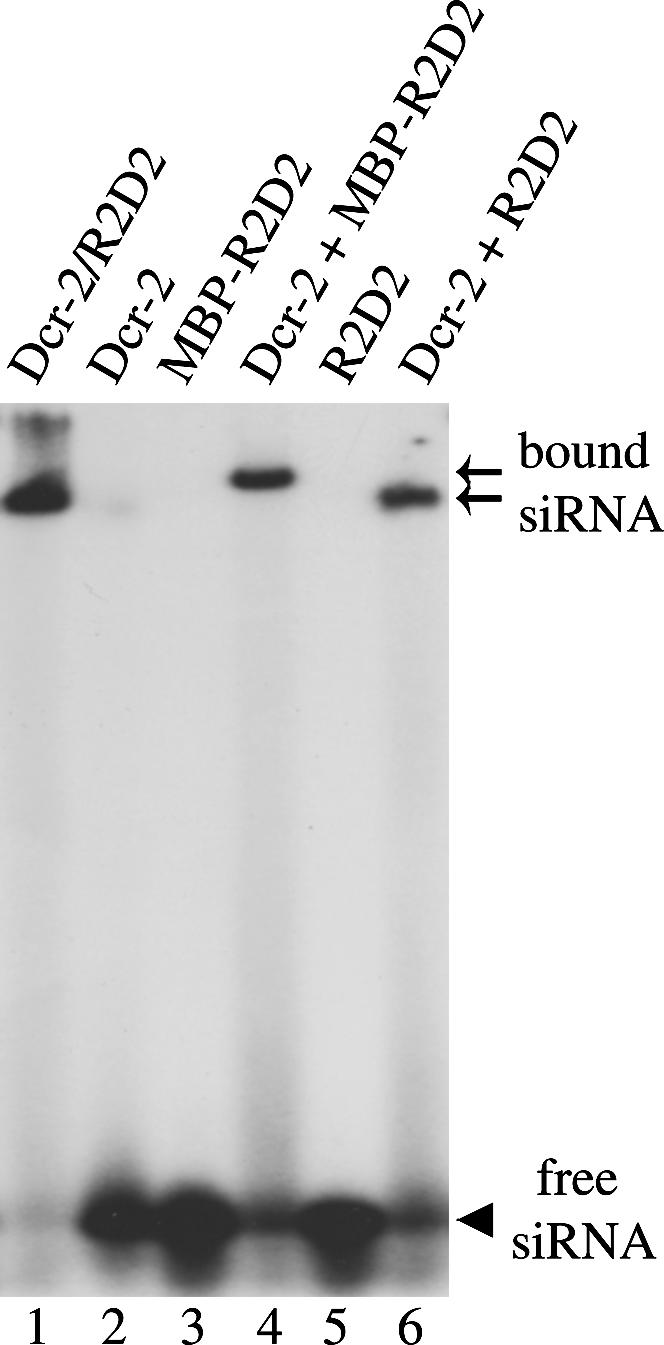
Dcr-2 and R2D2 coordinately bind siRNA duplex. The gel-shift assays were performed by incubating 5′ radiolabeled siRNA duplex and 10-pmol recombinant Dcr-2/R2D2 complex (lane 1), Dcr-2 (lane 2), MBP-R2D2 (lane 3), Dcr-2 + MBP-R2D2 (lane 4), R2D2 (lane 5), and Dcr-2 + R2D2 (lane 6). The arrowhead points to free siRNA. Two arrows refer to the Dcr-2/MBP-R2D2/siRNA and Dcr-2/R2D2/siRNA complexes, respectively.
Both dsRNA-binding domains of R2D2 are critical for siRNA binding
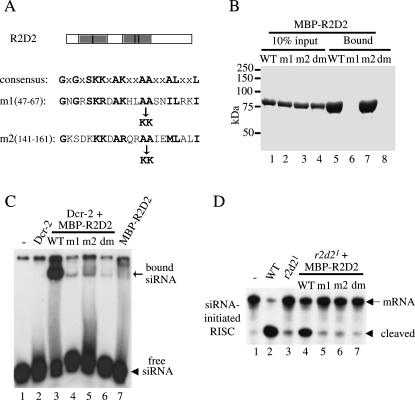
R2D2 contains two putative dsRNA-binding domains (dsRBDs) at the amino (N) terminus. To further analyze these domains, we generated three mutant R2D2 proteins by replacing two highly conserved alanine (A) residues with lysines (K) in the first (m1), second (m2), or both (dm) dsRBDs of R2D2 as previously described (Fig. 5A) (Liu et al. 2003). These mutant forms of R2D2 were also produced and purified as MBP fusion proteins from E. coli. To determine if these R2D2 mutants could bind long dsRNA, we used poly (inosine) (I):poly (cytosine) (C) dsRNA-conjugated agarose beads (Amersham) to pull down purified wild-type or mutant MBP–R2D2 proteins. As shown in Figure 5B, while R2D2m2 was precipitated as efficiently as wild-type R2D2, neither R2D2m1 nor R2D2dm could interact with long dsRNA. These results indicated that the first, but not the second, dsRBD of R2D2 was capable of binding long dsRNA. Alternatively, the second dsRBD of R2D2 might be covered by the C-terminal region of unknown function, thereby rendering it inaccessible to long dsRNA. However, the latter scenario was unlikely because we obtained the same results after removing the C-terminal 107 amino acids from wild-type and mutant MBP–R2D2 (data not shown).
FIGURE 5.
Both dsRNA-binding domains (dsRBDs) of R2D2 are required for binding siRNA and promoting siRISC assembly. (A) A schematic diagram of R2D2 domain structure. The shaded boxes refer to the tandem dsRBDs of R2D2. Part of the consensus dsRBD motif was shown below. Two highly conserved alanine (A) residues were replaced by lysines (K) in the first (m1, A59–60K), second (m2, A153–154K), or both (dm, A59–60K, A153–154K) dsRBD of R2D2. (B) The poly (I): poly (C) long dsRNA-conjugated beads were used to precipitate purified wild-type (WT) and mutant MBP-R2D2 recombinant proteins. 10% input (lanes 1–4) and 100% bound proteins (lanes 5–8) were resolved by SDS-PAGE followed by coomassie staining. (C) The gel-shift assays were performed in buffer alone (lane 1); or with 3.5-pmol recombinant Dcr-2 alone (lane 2) or in combination with 12.5-pmol wild-type or various mutant MBP-R2D2 (lanes 3–6); 12.5-pmol wild-type MBP-R2D2 alone (lane 7). (D) The siRNA-initiated RISC assays were performed with buffer control (lane 1); 32 μg of wild-type ovary extract (lane 2); 32 μg r2d21 mutant extract alone (lane 3), or supplemented with 12.5-pmol wild-type or mutant MBP-R2D2 proteins (lanes 4–7).
To determine if either or both dsRBDs of R2D2 was necessary for siRNA binding, we performed siRNA gel-shift assays using recombinant Dcr-2 in combination with wild-type or mutant MBP–R2D2 proteins. As shown in Figure 5C, mutations in either dsRBD abolished the ability of MBP–R2D2 to cooperate with Dcr-2 for siRNA binding. More importantly, none of these mutant MBP–R2D2 proteins could rescue the siRNA-intiated RISC activity in r2d21 null ovary extract (Fig. 5D). It should be noted that Dcr-2 interacted with mutant R2D2 proteins as efficiently as with wild-type control (data not shown; Liu et al. 2003). Together, our results indicate that both dsRBDs of R2D2 are critical for the Dcr-2/R2D2 complex to bind and load siRNA onto the siRISC complex.
DISCUSSION
The current study strengthens the model that R2D2 bridges the initiation step (siRNA production) and the effector step (siRNA loading onto siRISC) of the Drosophila RNAi pathway (Liu et al. 2003). We show that the assembly of the siRISC complex is abolished or diminished when Dcr-2 or R2D2 are genetically removed. The role of the Dcr-2/R2D2 complex to load siRNA onto the siRISC complex is dependent on its ability to bind siRNA. Only the Dcr-2/R2D2 complex, but neither Dcr-2 nor R2D2 alone, could efficiently interact with the siRNA duplex. In addition, both dsRNA-binding domains of R2D2 are critical for binding to siRNA and promoting siRISC assembly. Therefore, Dcr-2 and R2D2 coordinately bind siRNA and constitute the gateway for siRNA loading and RISC activation.
Dcr-2 and R2D2 stabilize each other in vivo
We have previously shown that depletion of Dcr-2 by RNAi also diminished the level of R2D2 protein, whereas RNAi of R2D2 caused a modest reduction in Dcr-2 protein in S2 cells (Liu et al. 2003). Moreover, recombinant Dcr-2 and R2D2 proteins were produced at much higher levels when expressed jointly than individually in insect cells (data not shown). Both results suggest that Dcr-2 and R2D2 proteins stabilize each other in vivo. In the current study, we found that the level of Dcr-2 protein remained the same in r2d21 null flies, whereas both Dcr-2 and R2D2 proteins were missing in dcr-2R416X null flies. One possible explanation for the difference seen in S2 cells and flies is that the level of Dcr-2 protein may be compensated when Drosophila cells permanently lack R2D2. Therefore, we conclude that Dcr-2 stabilizes R2D2, but the stability of Dcr-2 is largely independent of R2D2 in the fruitfly. Consistent with this, His-tagged Dcr-2, but not R2D2, recombinant proteins could be successfully produced in insect cells (data not shown). This is probably because R2D2 becomes unfolded or unstable in the absence of Dcr-2.
How do Dcr-2 and R2D2 bind siRNA?
Our results indicate that Dcr-2 and R2D2 bind siRNA coordinately. Only the Dcr-2/R2D2 complex, but neither Dcr-2 nor R2D2 alone, could efficiently interact with siRNA duplex in the gel-shift assay. R2D2 contains tandem dsRNA-binding domains (dsRBDs). We show that the first, but not the second, dsRBD of R2D2 is capable of binding long dsRNA. However, both dsRBDs of R2D2 are necessary for siRNA binding by the Dcr-2/R2D2 complex. Thus, the second dsRBD of R2D2 is critical for binding to siRNA rather than long dsRNA. It is possible that, without the second dsRBD, the first dsRBD of R2D2 only possesses low affinity for the 21–22-nt siRNA duplex.
We and others have previously shown that R2D2 as well as Dcr-2 could be efficiently cross-linked to radiolabeled siRNA by ultraviolet (UV) light (Liu et al. 2003; Tomari et al. 2004b). Thus, both Dcr-2 and R2D2 are in close contact with siRNA strands. Dcr-2 contains an RNA helicase domain, a DUF283 domain, and a PAZ domain at the N terminus as well as tandem RNase III motifs and a dsRBD motif at the C terminus. It is unclear which of these domains physically contact siRNA. Since neither Dcr-2 nor R2D2 bind siRNA alone, it is possible that siRNA is bound at the interface between Dcr-2 and R2D2. It is also possible that association of Dcr-2 and R2D2 triggers conformational change in either or both proteins, allowing them to bind siRNA cooperatively.
How do Dcr-2 and R2D2 facilitate siRNA loading onto siRISC?
It has been reported that Dcr-2 is required for siRNA-initiated RISC assembly in vivo (Lee et al. 2004; Pham et al. 2004). Here we present genetic evidence that R2D2 is also required for loading siRNA onto the siRISC complex in vivo. It is possible that the Dcr-2/R2D2 complex helps recruit the siRNA duplex to Ago2 for siRISC assembly (Liu et al. 2003; Tomari et al. 2004b). However, it remains unclear exactly how the Dcr-2/R2D2 complex facilitates incorporation of siRNA into the siRISC complex. While newly synthesized siRNA is double-stranded, siRNA exists as a single-stranded form in an active siRISC complex (Martinez et al. 2002). Thus, the nascent siRNA duplex must be unwound during siRISC assembly. It is reasonable to speculate that the Dcr-2/R2D2 complex facilitates unwinding of the siRNA duplex, thereby promoting incorporation of single-stranded siRNA into the siRISC complex. Dcr-2 is a candidate for the siRNA-unwinding helicase because it carries a putative DExH helicase domain and physically contacts the siRNA end that is easier to unwind (Tomari et al. 2004b). However, two dcr-2 mutations in the helicase domain have been isolated that do not affect the siRNA-initiated RISC activity, suggesting that a functional helicase activity is not required for Dcr-2 to promote siRISC assembly (Lee et al. 2004). In addition, recombinant Dcr-2 or Dcr-2/R2D2 complex cannot unwind the siRNA duplex in vitro (Tomari et al. 2004b). Alternatively, the Dcr-2/R2D2 complex may recruit an unknown helicase to unwind the siRNA duplex. In Caenorhabditis elegans, the DCR-1/RDE-4 (an R2D2 homolog) complex is associated with two highly related RNA helicases, DRH-1 and DRH-2, that are necessary for RNAi (Tabara et al. 2002). Several RNA helicases, such as Armitage and Dmp68, have also been implicated in the Drosophila RNAi pathways (Ishizuka et al. 2002; Tomari et al. 2004a).
Recent studies also suggest an alternative model for separation of siRNA strands and activation of the siRISC complex (Matranga et al. 2005; Rand et al. 2005). After the Dcr-2/R2D2 complex recruits duplex siRNA to Ago2, the PIWI domain of Ago2 cleaves the passenger strand and facilitates the formation of an active siRISC complex containing only the guide strand. It is likely that the orientation of siRNA binding by the Dcr-2/R2D2 complex allows Ago2 to access and cleave only one of the two siRNA strands. Therefore, the Dcr-2/R2D2 complex determines which strand of siRNA duplex becomes the guide strand or passenger strand. The two mechanistic models of siRISC assembly are not mutually exclusive. In either model, the Dcr-2/R2D2 complex plays a critical role in facilitating the strand separation of the duplex siRNA. Since not all Ago proteins possess the slicer activity, there must be more than one mechanism for siRNA loading and RISC activation.
MATERIALS AND METHODS
Generation, analysis, and transgenic rescue of the r2d21 null fly
The r2d21 mutant fly was generated by mobilizing a P-element inserted ∼0.6 kb upstream of the r2d2 ORF in the EP2450 stock (Bloomington stock center). Candidate mutants were selected based on loss of the red-eye marker carried by the P-element, bred to homozygosity, and screened by PCR for lack of the r2d2 ORF. To further map the deletion in the r2d21 mutant, we performed six PCR reactions with the primers listed below using a genomic DNA template isolated from wild-type or homozygous r2d21 flies (Fig. 1). The PCR fragment VI from r2d21 mutant DNA was sequenced to determine the precise boundary of deletion at the r2d21 mutant locus.
P1:5′-GCATCCATCATGCGCTGAAT-3′; P2:5′-GGTTCCCAAAGCTATTTGCG-3′
P3:5′-ACCGACACACCTATGAATCC-3′; P4:5′-ATCAACATGGTGCGAAAATA-3′
P5:5′-CGCACCATGTTGATTTAATGC-3′; P6:5′-AACTGCTTCGCCAATATGCC-3′
P7:5′-AACGGTTGCTCCTGCTGTAC-3′; P8:5′-ACGAGAGAGCGCTCTATCAA-3′
P9:5′-TTGATAGAGCGCTCTCTCGT-3′; P10:5′-CGGATGGATGGAAGTATGTA-3′
(PCR product [primers]: I [P1/P2]; II [P3/P4]; III [P5/P6]; IV [P7/P8]; V [P9/P10]; VI [P3/P8])
To construct the r2d2 rescuing transgene, we cloned into pCasepeR4, a 5.6-kb genomic fragment that only contained an intact r2d2 gene. Because this DNA fragment also carried a large portion of an upstream ORF CG14536, to avoid complication, we introduced a frameshift mutation at the N terminus of ORF CG14536 by filling in a unique SacII site. Transgenes inserted into the third chromosome were crossed into the r2d21 mutant background to perform the rescuing experiments.
Antibodies
The anti-Dcr-2 and anti-R2D2 antibodies were previously described (Liu et al. 2003). The anti-Dcr-1 antibody was a generous gift from Dr. Gregory Hannon, whereas the anti-Ras antibody was purchased from Calbiochem.
Purification of recombinant Dcr-2 and R2D2 proteins
The His-tagged recombinant Dcr-2 and Dcr-2/R2D2 complexes were produced and purified from insect cells as previously described (Liu et al. 2003). The R2D2 ORF was cloned as an NcoI–NotI fragment into pMBP-parallel. Different R2D2 mutants were generated by Quickchange (Stratagene). Both wild-type and mutant MBP–R2D2 proteins were produced and purified from BL21 bacteria. In brief, after the bacteria culture reached OD600 of ∼0.8 at 37°C, IPTG was added to a 1-mM concentration to induce expression of MBP–R2D2 proteins for 5–6 h at 25°C. Cells were harvested and lysed by sonication in Buffer T (20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM β-mercaptoethanol) freshly supplemented with complete protease inhibitor cocktail (Roche). Recombinant MBP–R2D2 proteins were purified using an Amylose–Agarose column (NEB) followed by SP- and Q-Sepharose chromatography (Amersham).
The dsRNA- and pre-miRNA-processing assays
The dsRNA- and pre-miRNA-processing assays were performed essentially as previously described (Liu et al. 2003; Jiang et al. 2005). The uniformly radiolabeled 250-bp dsRNA was made by in vitro transcription using the Riboprobe T7 Kit (Promega). A 61-nt pre-let7 RNA was 5′-end labeled by γ-32P by T4 polynucleotide kinase (NEB) followed by PAGE purification. For dsRNA-processing assay, 105 cpm dsRNA was incubated with fly extacts or recombinant proteins in 1× buffer 12 (100 mM KOAc, 10 mM HEPES (pH 7.4), 2 mM Mg(OAc)2, and 5 mM DTT). The pre-miRNA-processing assay used 4 × 104 cpm pre-miRNA in 1× buffer 16: 100 mM KOAc, 10 mM HEPES (pH 7.4), 10 mM Mg(OAc)2, and 5 mM DTT. Typically, 10-μL reactions were performed at 30°C for 30 min in the presence of an ATP-regenerating system: 1 mM ATP, 30 mM creatine phosphate (Fluka), and 30 U creatine phosphokinase (Sigma). The reaction was stopped by addition of 200 μL 0.3 M NaOAc, phenol/chloroform extracted and ethanol precipitated, resolved on a 16% denaturing (7 M Urea) polyacrylamide gel.
The siRNA gel-shift assay
Both strands of siRNA were radiolabeled at the 5′ end with [γ-32P] ATP by T4 polynucleotide kinase, annealed in 1× annealing buffer (100 mM KOAc, 30 mM HEPES [pH 7.4], 2 mM Mg(OAc)2), and passed through a G25 column (Amersham). In a 10-μL reaction, 5 × 104 cpm siRNA was incubated with fly ovary extracts or recombinant proteins at 30°C for 30 min in 1× buffer 12. After addition of 1.2 μL 50% glycerol, the reaction was resolved on a 5% native PAGE, and exposed to X-ray film.
The dsRNA-, pre-miRNA, and siRNA-initiated RISC assays
All RISC (sequence-specific mRNA cleavage) assays were performed using an mRNA substrate containing a perfect sense or antisense let7 target site as previously described (Liu et al. 2003; Jiang et al. 2005). The mRNA was radiolabeled at the 5′ G cap by gaunylyl transferase (Ambion) followed by PAGE purification. The 130-bp dsRNA trigger contained essentially four repeats of let7 sequence (Liu et al. 2003; Jiang et al. 2005). Typically, 2.5 × 104 cpm mRNA was incubated with fly ovary extracts for 1 h at 30°C in 1× buffer 12 with the presence of ATP-regenerating system. The dsRNA, pre-let7, or let7 siRNA triggers were respectively used at 250 nM, 250 nM, and 25 nM final concentrations.
Embryo injections
A 500-bp GFP dsRNA was transcribed by Megascript (Ambion) and processed by a recombinant Dcr-2/R2D2 complex to generate GFP siRNAs followed by PAGE purification. Wild-type or r2d21 mutant green fluorescent embryos were injected with GFP siRNAs at a concentration of 0.24 μg/μL (Liu et al. 2003). Embryos were viewed and photographed 48 h after injection.
ACKNOWLEDGMENTS
We thank Dr. Gregory Hannon for reagents, Zhengzheng Li, Courtney Karner, and Tim Rand for technical assistance, and Drs. Zain Paroo and Gaya Amarasinghe for discussion and critical reading of the manuscript. Q.L. is a W.A. “Tex” Moncrief Jr. Scholar in Medical Research and a Damon Runyon Scholar, supported by the Damon Runyon Cancer Research Foundation (DRS-43). This work is also supported by a Welch grant (I-1608) to Q.L. and an NIH grant (GM070648) to D.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.101606.
REFERENCES
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi M.C., Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Ye X., Liu X., Fincher L., McKearin D., Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila . Genes & Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Pham J.W., Pellino J.L., Lee Y.S., Carthew R.W., Sontheimer E.J. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila . Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Rand T.A., Petersen S., Du F., Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizuka A., Siomi H., Siomi M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H., Yigit E., Siomi H., Mello C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans . Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Zamore P.D. Perspective: Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du T., Haley B., Schwarz D.S., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004a;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Matranga C., Haley B., Martinez N., Zamore P.D. A protein sensor for siRNA asymmetry. Science. 2004b;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]