Abstract
The paired-end ditagging (PET) technique has been shown to be efficient and accurate for large-scale transcriptome and genome analysis. However, as with other DNA tag-based sequencing strategies, it is constrained by the current efficiency of Sanger technology. A recently developed multiplex sequencing method (454-sequencing™) using picolitre-scale reactions has achieved a remarkable advance in efficiency, but suffers from short-read lengths, and a lack of paired-end information. To further enhance the efficiency of PET analysis and at the same time overcome the drawbacks of the new sequencing method, we coupled multiplex sequencing with paired-end ditagging (MS-PET) using modified PET procedures to simultaneously sequence 200 000 to 300 000 dimerized PET (diPET) templates, with an output of nearly half-a-million PET sequences in a single 4 h machine run. We demonstrate the utility and robustness of MS-PET by analyzing the transcriptome of human breast carcinoma cells, and by mapping p53 binding sites in the genome of human colorectal carcinoma cells. This combined sequencing strategy achieved an approximate 100-fold efficiency increase over the current standard for PET analysis, and furthermore enables the short-read-length multiplex sequencing procedure to acquire paired-end information from large DNA fragments.
INTRODUCTION
A major challenge facing us in this post-genomic era is how to extract maximum information from completed genome sequence assemblies (1), so as to address basic questions in gene annotation, expression profiling, gene regulation and genome variation.
The sequencing approach has clear advantages over microarrays by elucidating the exact nucleotide content of target DNA sequences. However, a major constraint has been its higher cost and lower data-generation speed relative to microarrays. As an improvement on methods involving one template per read, serial analysis of gene expression (SAGE) was developed (2,3). This strategy utilizes short DNA tags representing an entire DNA fragment, and the concatenation of these tags for efficient sequencing enables the characterization of whole transcriptomes and genomes. However, the mapping of short single tags to the genome often results in positional ambiguities. This drawback was partially addressed in recent modifications that specifically extracted 5′ terminal signatures of cDNA (4,5), but it was the simultaneous tagging of both 5′ and 3′ terminal signatures that provided an ideal solution. To achieve this, we initially developed an intermediate approach that separately extracted 5′ and 3′ terminal tags from cDNA fragments for sequencing (6). Subsequently, we developed gene identification signature (GIS) analysis, in which the 5′ and 3′ signatures of each full-length transcript were simultaneously extracted, then covalently-linked into paired-end ditag (PET) structures for concatenated high-throughput sequencing and the accurate demarcation of transcriptional unit boundaries in assembled genome sequences (7). An average capillary sequencing read (700–800 bp) of a single GIS-PET library clone would reveal 10–15 PET U, thus representing a 20- to 30-fold increase in annotation efficiency compared to the bidirectional sequencing analysis of full-length cDNA (flcDNA) clones.
We have also successfully applied this PET-based DNA analysis strategy to characterize genomic DNA fragments enriched for specific target sites by chromatin immunoprecipitation (ChIP), and these chromatin immunoprecipitation-PET (ChIP-PET) analyses have provided a global overview of p53 transcription factor binding sites in the human genome (6), as well as Oct4 and Nanog targets in the mouse genome (8).
The PET concept can conceivably be applied to other DNA sequence analyses that will benefit from paired-end characterization, including the study of epigenetic elements and genome scaffolding. One point to note is that while the number of sequencing reads (∼50 000) required for a comprehensive GIS-PET or ChIP-PET analysis is miniscule for most genome centers with state-of-the-art Sanger capillary sequencers, and within the reach of core facilities in university laboratories, the final cost of each PET experiment can be significant. Hence, we are continually seeking ways to improve the efficiency and cost-effectiveness of PET analysis.
Recently, a novel, highly-parallel multiplex sequencing-by-synthesis method based on pyrosequencing in picolitre-scale reactions (454-sequencing™) was reported, in which ∼300 000 DNA templates were simultaneously sequenced in a single 4 h machine run to a read-length of ∼100 bases, with an accuracy of 99.6% (9). Although this multiplex sequencing approach, as described, potentially yields a remarkable 100-fold increase in throughput compared with current Sanger capillary sequencing technology, its obvious weaknesses are the short-read length that limits wider application to many genome sequencing projects, and its inability to obtain paired-end information.
Another recent advance is the Polony sequencing technology (10) that has as its chief advantages low sequencing cost, and the ability to produce paired-end reads of DNA fragments at a raw data acquisition rate reportedly an order of magnitude faster than conventional Sanger sequencing. In its current manifestation, however, the technology suffers from a lower-than-predicted throughput (∼140 bp/s) and raw base-calling accuracies poorer than in Sanger sequencing. In addition, an unusual sequencing-by-ligation scheme results in short, discontiguous paired-end tags (each of 13 bases interrupted by an indeterminate gap of 4 to 5 bases) that is insufficient for specific mapping in complex genomes, thus precluding the Polony method from applications involving mammalian genome sequencing.
It was apparent to us that a melding of technologies would be highly beneficial: the massively-parallel, short-read nature of the new 454-sequencing method lends itself well to enhanced PET analysis: each ∼40 bp PET would compensate for the inherent disadvantages of short-reads by providing paired-end information from long contiguous DNA fragments. Mapping of these PETs to assembled genomes would allow the original sequence to be inferred. Furthermore, by a simple modification of the original GIS-PET procedure, we were able to easily dimerize PETs prior to multiplex sequencing, thereby further increasing data-gathering efficiency. Finally, the very high sequencing throughput of 454-sequencing should allow the global analysis of transcriptomes and genomes at an unprecedented speed.
In this report we describe the utility, efficiency and accuracy of applying this multiplex sequencing of paired-end ditags (MS-PET) analysis to characterize both the human transcriptome and genome.
MATERIALS AND METHODS
Cell lines and drug treatment
MCF7 human breast carcinoma cells were obtained from ATCC and verified to be positive for estrogen receptor expression. They were cultured in DMEM media supplemented with 10% fetal calf serum (FCS), up to passage number 3. The cells were serum-starved for 24 h prior to treatment with 10 nM beta-estradiol for 12 h, then harvested by centrifugation and washed with phosphate-buffered saline (PBS). For the additional sample that was sequenced using the Sanger method, untreated MCF7 cells were harvested as above at log phase growth.
HCT116 human colorectal carcinoma cells (a gift from Dr B. Vogelstein, Johns Hopkins University) were cultured in DMEM with 10% FCS. The cells were treated with 0.1 M 5-Fluorouracil for 10 min to activate p53 and induce target gene expression.
GIS-PET analysis on MCF7 cells
Cells were lysed in Trizol, and the RNA subsequently extracted was subjected to GIS-PET analysis essentially as described previously (7), except for the use of a modified cloning vector pGIS4a (Supplementary Figure 1; vector and sequence available on request). Briefly, flcDNA was prepared by the CapTrapper method (11), and inserted into pGIS4a after the excision of the poly(A) tail with GsuI. This obviates the need for a 3′ adapter-ligation step. Plasmid maxiprep prepared from the flcDNA library was digested with MmeI to excise all of the inserted flcDNA except for the terminal 20 bp signatures. After end-polishing and plasmid recircularization, the recircularized plasmids (each now containing a single-PET of ∼36 bp) were transformed into bacteria to give the GIS single-PET library. A maxiprep of this single-PET library was digested sequentially with BseRI and BamHI to release PETs containing a single BamHI cohesive site. PETs were gel-purified, and ligated to form dimerized PETs (diPETs) of ∼80 bp size. These diPETs were again gel-purified, and subjected to multiplex sequencing (9) using a GS20 sequencer (Roche).
ChIP-PET
ChIP assays with HCT116 cells were carried out as described previously (6). Briefly, cells were treated with 1% formaldehyde for 10 min at room temperature to crosslink proteins and DNA. Formaldehyde was inactivated by addition of 125 mM glycine, and the crosslinked cells were lysed and sonicated. Chromatin extracts containing gDNA fragments of average size ∼500 bp were immuno-precipitated using anti-p53 DO1 monoclonal antibody (Santa Cruz).
ChIP-enriched gDNA fragments were de-linked, size-fractionated on columns to remove excessively small fragments (Invitrogen), blunted using the end-it kit (Epicentre), cloned into pGIS3h (Supplementary Figure 2; vector and sequence available on request) and transformed into TOP10 Escherichia coli (Invitrogen) to give the ChIP DNA library. Similar to the standard GIS procedure, plasmid DNA prepared from the ChIP DNA library was digested with MmeI, end-polished, recircularized and transformed into bacteria to give the GIS single-PET library. Plasmid DNA extracted from the single-PET library was first digested with BseRI, then alkaline phosphatase-treated to inactivate the resulting cohesive site, and finally BamHI-treated to liberate single-PETs. Purified single-PETs were dimerized to form ∼88 bp diPETs and subjected to multiplex sequencing using a GS20 sequencer.
Data collection and analysis-transcriptome analysis of MCF7 cells (library SHC015)
For both the transcriptome analysis of MCF7 cells, and the ChIP-PET analysis of HCT116 cells, PETs were extracted from raw sequences using the PET Tool software designed in-house (K. P. Chiu, C.-H. Wong, Q. Chen, P. Ariyaratne, H. S. Ooi, C.-L. Wei, W.-K. Sung and Y. Ruan, manuscript submitted) and mapped to the human genome assembly (build hg17; http://genome.ucsc.edu). Details of the PET extraction and mapping procedure are found in (7). Details of p53 motif identification are in (6).
Novel gene discovery/gene prediction validation by PCR
Primary PCR was performed using primers that were designed based on PET sequences (sequences available on request). Primer sequences were adjusted to optimize predicted annealing temperatures (based on nearest-neighbor calculations) by incorporating flanking vector sequences as required. Amplification was performed using Qiagen HotStarTaq as per the manufacturer's protocols, for 25 cycles, at an annealing temperature of 55°C. For each reaction, 4 ng of DNA from the flcDNA library was used as template. A total of 5 μl of each 25 μl reaction was visualized on a 1% agarose gel. A total of 5 μl of each primary PCR was then used as template for secondary PCR. For these secondary PCR, primers were designed using Primer3 (12) at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi, based on the genomic DNA sequences spanned by each PET. For convenience, wherever possible secondary PCR products were designed to be ∼400–500 bp in size. The GenScan algorithm (13) at http://genes.mit.edu/GENSCAN.html was used to predict exon locations, to minimize the chances of inadvertently designing primers within introns.
Quantitative PET analysis by real-time RT–PCR
PCR primers (sequences available on request) were designed using Primer3, based on the transcripts corresponding to each of the 12 selected PETs. For real-time PCR analysis, 100 ng of mRNA was reverse-transcribed using SuperScript II (Invitrogen) in a final volume of 20 μl, and 2 μl of this reverse-transcribed mRNA was amplified in a LightCycler (Roche) for 36 cycles using the FastStart DNA Master SYBR Green I kit. Each PCR was performed in triplicate and threshold cycle (Ct) values for each amplicon of interest were normalized against that of Actin as a reference.
Data mining details
PET mapping to genome assembly
More details are found in (7), but briefly, PET sequences were extracted based on the expected characteristic arrangement of bases in each diPET, viz. (34–40 bp PET)-GTCGGATCCGAC-(34–40 bp PET). Extracted PETs were mapped to the human genome assembly using a Compressed Suffix Array-derived algorithm that is much faster than BLAST, while maintaining similar accuracy (H. S. Ooi, Q. Chen, K. P. Chiu, C. K. Wong, T.-W. Lam, C.-L. Wei, Y. Ruan and W.-K. Sung, manuscript submitted). For transcript analysis, we mandated a minimum 16 base contiguous match for the 5′ signature and a minimum 14 base contiguous match for the 3′ signature of each PET when mapping to the genome; for ChIP analysis, we set a minimum 17 base contiguous match for both signatures. Moreover, we required that the 5′ signature not extend beyond position 19, and the 3′ signature not start before position 18. Finally, each 5′ signature was matched with its cognate 3′ signature against existing genome sequence data, the criteria being that both the 5′ and 3′ signatures must be present on the same chromosome, be on the same strand, in the correct orientation (5′ to 3′), and within a predefined genomic distance (1 Mb for transcripts, 4 kb for the ChIP-enriched gDNA fragments). PETs that failed to meet these criteria were considered ‘unmappable’, and archived separately for additional analysis. Clustering of mapped PETs into putative genes is based on both 5′ and 3′ mapping distance: for transcriptome analysis purposes, within a cluster either of the two ends should be within 1 kb of other members in the same cluster. For ChIP-PET analysis, as long as the PETs overlap with any member we include them in the same cluster.
Mapping accuracy analysis
Based on the counts of single-locus PETs mapped to data in any of the RefSeq, KnownGene, GenBank mRNA or MGC sub-bases (representing known genes) within the UCSC database, PETs in the top 20 most abundant clusters were selected and the mapping of the 5′ and 3′ signatures against documented termini was examined. Mapping accuracy was determined by arbitrarily setting a 50 bp cutoff. This procedure was subsequently extended to the larger set of all mapped single-locus PETs that corresponded to known genes in the UCSC database.
Gene categorization
Mapping statistics of each PET was downloaded from the T2G browser (http://t2g.bii.a-star.edu.sg) mentioned previously (7), and sorted into various categories using databasing software based on the following definitions: (i) known gene: PET sequence maps in the vicinity of data in any of the sub-bases RefSeq, KnownGene, MGC or GenBank mRNA; if both signatures map within ±50 bp of both termini, the match is deemed ‘accurate’, otherwise it is considered a splice variant with an alternative TSS and/or PAS; (ii) novel: within the PET span, there is no existing information in any database category; (iii) gene prediction: there is existing information within any of the Ensembl EST, Twinscan, Genscan or SGP Gene databases (overlapping data in any of the other databases will take precedence over gene predictions); (iv) ESTs: there is existing information in only the EST sub-base (overlapping data in any of the known gene sub-bases takes precedence); (v) putative bidirectional promoter regulated: two genes directly adjacent to each other are expressed in opposite directions, originating from an intergenic region not >1 kb in length; (vi) antisense genes: PETs were first sorted into positive strand and negative strand mappings, and clustered by location to indicate putative genes. Overlapping (either complete or partial) of gene clusters from opposite mapped orientations indicates putative antisense genes.
Unmapped PET recovery
We decided to restrict the recovery of initially-unmapped PETs to those that could be subsequently mapped after allowing a single-base insertion (of the same base) or deletion within only homopolymer regions (defined as stretches of two or more identical bases) in the PET sequence. This restriction was imposed to reduce mapping artifacts (noise) caused by allowing insertions/deletions anywhere within the PETs. Within the recovery pipeline, a single-base deletion was permitted within the entire set of unmapped PETs, which were subsequently remapped to the genome; only PETs that now mapped to single loci were retained: these would be considered the result of apparent base over-calling. The remaining PETs were permitted a single-base insertion, remapped, and again only those that could be mapped to single loci were retained: these PETs would be considered the result of base under-calling. All remaining ditags were considered ‘unmappable’. Validation of the approach was performed by determining the fraction of recovered PETs that mapped within 200 bp of known gene annotations in the UCSC database.
RESULTS
Preparation of diPETs for multiplex sequencing
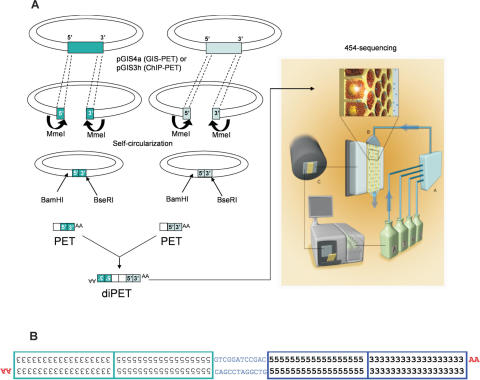
We constructed an flcDNA library from estradiol-treated MCF7 cells, which is believed to be a useful in vitro model for estrogen-responsive breast cancer (14), following a procedure essentially identical to our published GIS analysis protocol (7) with minor modifications to the cloning vector (pGIS4a; Supplementary Figure 1). Subsequently, the plasmid clones of this flcDNA library were subjected to Mme1 digestion, followed by plasmid self-ligation to form a single-PET library, in which each plasmid contained a single-PET flanked by a BamH1 site on its 5′ side and a BseR1 sites on its 3′ side (Figure 1). BseRI digestion would then release the 3′ end of the insert with an AA dinucleotide overhang, and subsequent BamHI digestion would excise a 43 bp asymmetric PET with only one compatible cohesive site (BamHI) at the insert 5′ terminus. Ligation of single-PETs resulted in their dimerization into ∼80 bp diPETs. The collection of diPETs was then subjected to multiplex sequencing analysis (Figure 1) using a GS20 sequencer. From a single GS20 machine run of a full picotiter plate, a total of 462 626 PET units were generated, which were collapsed into 313 983 unique PET sequences (Supplementary Table 1).
Figure 1.
Schematic overview of the MS-PET sequencing strategy. (A) The outline procedure showing the construction of diPETs, which were subjected to multiplex sequencing. (B) Structural details of a diPET. The numbers 5 and 3 represent bases within the 5′ and 3′ signatures, respectively, in each PET component. The orientations of cDNAs are indicated by the ‘AA’ remaining after poly(A) tail removal.
Similarly, we tested the use of the MS-PET sequencing strategy for enhanced ChIP-PET analysis. From the original ChIP DNA used for whole-genome mapping of putative p53 binding sites in HCT116 human colon cancer cells (6), we constructed a p53 ChIP DNA library in an improved vector (pGIS3h; Supplementary Figure 2) and converted this library into ∼88 bp diPET templates for MS-PET analysis. A partial (1/16) picotiter plate run generated 23 283 PET units, which were collapsed into 22 687 unique p53 ChIP-PET sequences for further analysis.
Mapping of GIS-PET sequences to the human genome
High-quality PET sequences derived from the GIS-PET library were mapped to the human genome sequence assembly build hg17 (http://genome.ucsc.edu). Based on the same empirically-determined criteria used in GIS analysis for mapping PETs to genome (7), viz. 5′ and 3′ signatures must contain minimum contiguous 16 and 14 bp matches, respectively, to the genome; they must be in the correct orientation (5′ to 3′); and within an arbitrary genomic span of one million base pair, we found that of the 313 983 unique PET sequences, 157 697 (50.22%) could be mapped on the first-pass to hg17 (Table 1). The 5′ signatures displayed contiguous matches to the genome ranging from 16 to 19 bases, with a modal match of 18 bases, while the 3′ signatures displayed matches from 14 to 21 bases, with a modal match of 16 bases.
Table 1.
Mapping of PETs to the human genome
| PETs | Percentage | |
|---|---|---|
| Total PETs in GIS-PET library analyzed by MS-PET | 313 983 | 100.00% |
| Initial mapped PETs | 157 697 | 50.22% |
| PETs mapped to a single-locus | 136 612 | 86.63% (of mapped PETs) |
| PETs mapped to multiple loci | 21 085 | 13.37% (of mapped PETs) |
| Initial unmapped PETs | 156 286 | 49.78% |
| Single-locus PETs recovered after homopolymer error analysis | 56 194a | 17.89% |
| homopolymer over-call errors (+1 base) | 35 523 | 11.31% |
| homopolymer under-call errors (−1 base) | 27 047 | 8.61% |
| Final mapped PETs | 213 891 | 68.12% |
| PETs mapped to a single-locus | 192 806 | 90.14% (of mapped PETs) |
| PETs mapped to multiple loci | 21 085 | 9.86% (of mapped PETs) |
| Final unmapped PETs | 100 092 | 31.88% |
aBecause the same PET sequence can contain both over-call (+1) and under-call (−1) errors, each category of recovered PETs is not mutually exclusive. Thus, the total PETs recovered is not a simple summation. See Supplementary Data for details on the error-distribution analysis performed.
It is noteworthy that 156 286 PET sequences (49.78%) remained unmapped to the genome under standard first-pass mapping criteria (Table 1). Although we had expected that a portion of the PETs would remain unmapped for reasons including gaps within the genome assembly, sequence mismatches due to polymorphisms, genome variations in cultured cell lines, and sequencing error, the percentage of unmapped PETs from this library was considerably higher than the range of 20–30% that we consistently find in GIS-PET libraries sequenced by the standard Sanger capillary method [(7) and P. Ng, unpublished data].
To specifically determine what portion of the unmapped PETs in this library was attributable to the unique sequencing characteristics of the multiplex sequencing method, we compared PET sequences generated from the same cell line (MCF7) but using Sanger capillary sequencing (Supplementary Table 2). From 135 757 PET sequences generated by capillary sequencing, 102 660 (75.62%) were mapped to the human reference genome sequence, and the rest (33 097; 24.38%) were unmappable. Therefore, the non-mapping rate of PET sequences by MS-PET analysis was about 25.40% (49.78–24.38%) higher than that of PET sequences generated by the Sanger capillary method, suggesting that about 25% of the 313 983 PETs generated in this MS-PET experiment were probably miscalled by the multiplex sequencing method.
It was previously reported that the GS20 multiplex sequencing is susceptible to base mis-calling (over-call or under-call) within homopolymeric regions (defined as an occurrence within any sequence of two or more identical adjacent bases) (9). We examined the extent to which this phenomenon might affect our analysis. By allowing a sequential one-base removal and addition (but not a substitution) of the same nucleotide base to counteract the possible base over- or under- calling within only homopolymer stretches in the ‘unmapped’ 156 286 PET sequences, we were able to identify apparent 1 base homopolymeric errors (Supplementary Figure 3) and recover 56 194 PETs that could be mapped specifically to a single-locus in the human genome. This resulted in only 100 092 (31.88% of 313 983) PET sequences that remained unmapped, or a total of 213 891 (68.12% of 313 983) mapped PET sequences (Table 1). Because we allowed only single-base errors in this analysis for stringency, the number of recoverable PETs is most likely underestimated. It is apparent therefore that with the correction of such homopolymeric errors, the sequencing accuracy of multiplex sequencing is comparable to the Sanger capillary method, suggesting that homopolymer base-calling errors were indeed a major contributing factor towards the initially higher mapping failure rate of MS-PET sequences, and that mapping algorithms for MS-PET- derived data should take this into consideration. Detailed examination of the error-distribution across different homopolymer lengths (Supplementary Data) showed that these were consistent with earlier published observations that total sequencing errors within homopolymer stretches increased with homopolymer length (Supplementary Figure 4). In addition, the percentage of sequenced bases from long homopolymer stretches within the 56 194 recovered PETs was higher, compared to bases from homopolymer stretches of the same length within the 136 612 single-locus PETs that were mapped on the first-pass (Supplementary Table 3). This is not surprising, given the increased occurrence of sequencing errors with homopolymer length, possibly coupled with decreased PET sequence complexity, both factors contributing to decreased mapping ability.
Mapping of GIS-PET sequences to known human transcription units
All 157 697 PET sequences that mapped to the human genome before homopolymer correction were subsequently clustered into 22 992 groups on the basis of similar (within 1 kb proximity to one another) 5′ and 3′ tag positions. Analysis was performed on data prior to homopolymer correction to increase stringency. These 22 992 clusters may therefore be taken to represent the transcriptome of estradiol-treated MCF7 cells (Supplementary Table 1). A total of 20 864 clusters comprising 136 612 PETs (86.63% of all mapped PETs) were located on unique chromosomal loci, while the remaining 21 085 mapped PETs were found in two or more locations (Table 1), representing potential alignment to expressed pseudogenes, duplicated genes, repetitive regions and possibly non-specific mapping.
To determine if the PET sequences were accurately mapped to known genes annotated in the human genome, we focused first on the top 20 most abundant PET clusters (comprising 10 387 individual PET sequences) that matched to well-characterized known genes. As previously described (7), these top 20 clusters would be expected to represent abundant and well-studied genes (as distinct from an analysis of all clusters in the library, which would be expected to include tentative or less well-studied annotations), and should therefore provide an accurate indication of the library quality in terms of intact transcript sequences, and hence ditag mapping accuracy. The vast majority of PETs in this class (10 083 of 10 387, or 97.07%) were mapped at well-defined 5′ and 3′ boundaries (both termini mapped simultaneously) within an arbitrary ±50 bp of known transcripts (including all documented transcript splice variants) of these genes. This result showed that the library analyzed in this study was of high-quality in terms of full-length transcript content, and also that the MS-PET sequences were of high accuracy.
We then extended this analysis to all 125 986 PETs (within the set of 136 612 single-locus PETs) that matched known transcript information (viz. KnownGene, RefSeq, Human mRNA and MGC in the UCSC genome browser), and found that a total of 93 325 (74.08%) PETs could be mapped within 50 bp of known termini, with both tags matching, respectively the 5′ and 3′ ends of documented termini (Table 2). The remaining PETs therefore potentially represent new transcripts of known genes with alternative 5′ transcription start sites or 3′ polyadenylation sites. This percentage was comparable to the results derived from capillary-sequenced PETs of MCF7 cells (data not shown) and our previous results of other samples (7).
Table 2.
Mapping of PETs to known gene transcripts
| Top 20 PET clusters | Percentage % | Known single-locus PETs | Percentage % | |
|---|---|---|---|---|
| Total PET sequences | 10 387 | 100.00 | 125 986 | 100.00 |
| Matched to known transcripts | 10 083 | 97.07 | 93 325 | 74.08 |
| Novel extended 5′ termini | 64 | 0.62 | 4742 | 3.76 |
| Novel extended 3′ termini | 24 | 0.23 | 2956 | 2.35 |
| Novel truncated 5′ termini | 36 | 0.35 | 3528 | 2.80 |
| Novel truncated 3′ termini | 169 | 1.63 | 5543 | 4.40 |
| Unclassified | 11 | 0.11 | 15 892 | 12.61 |
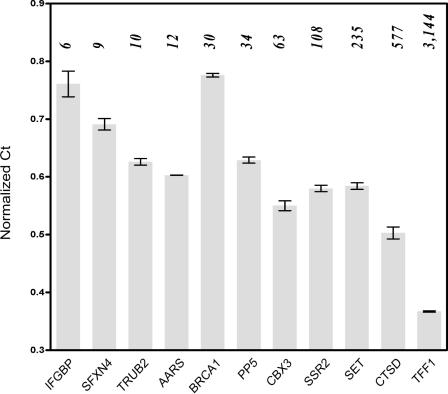
To examine the quantitative nature of MS-PET analysis, an arbitrary selection of 12 PETs with counts ranging from 6 to 3144 was used as the basis for designing PCR primers against each corresponding transcript. Quantitative real-time PCR performed on reverse-transcribed mRNA from the estradiol-treated MCF7 cells indicated that yields of each PCR product (relative to actin as an internal reference) followed a trend that, in general, corresponded to PET counts, viz. PET counts were inversely related to normalized Ct values (Figure 2). This result indicated that the MS-PET method did not excessively distort the relative representation of transcripts within the cells. However, there were obvious outliers (e.g. BRCA1, Figure 2), where poor correlation was observed between the normalized Ct values and PET counts. This is to be expected, as our study focused on PET counts derived only from single-locus-mapped PETs, omitting possible gene duplications (including expressed pseudogenes), which would be expected to contribute to increased PCR yields. Conversely, the presence of internal exon deletions or rearrangements may result in anomalously low PCR yields with certain primer pairs. Finally, there also exists the possibility that some distortion of transcript representation may occur during the various cloning procedures. Nonetheless, the general agreement between PET counts and transcript representation suggests that MS-PET provides a useful semi-quantitative measure of gene expression. The results further indicate that transcripts with PET counts as low as 6/136 612 (44 tags-per-million) can be easily recovered by PCR.
Figure 2.
Validation of MS-PET-identified transcripts by quantitative real-time RT–PCR. Columns represent Mean Ct values (normalized against that of Actin) ±SD of each of 11 amplicons (n = 3). PET counts for each candidate transcript are shown in italics above each column. IFGBP (Interferon-gamma binding protein); SFXN4 (sideroflexin 4); TRUB2 (TruB pseudouridine synthase homolog 2); AARS (alanyl-tRNA synthetase); BRCA1 (breast cancer 1, early onset); PP5 (protein phosphatase 5, catalytic subunit); CBX3 (Chromobox protein homolog 3); SSR2 (signal sequence receptor); SET (SET translocation); CTSD (cathepsin D lysosomal aspartyl peptidase); TFF1 (Trefoil factor 1); Actin (reference), PET counts = 33.
Identification of previously-uncharacterized human transcripts
While the majority (15 163 of 20 860 single-locus gene clusters) of MS-PET-defined transcripts mapped to known genes, a small but significant portion (Table 3) represented full-length transcripts previously defined only by partial EST sequences (3405 PET clusters), de novo predictions (2202 PET clusters), or were mapped to regions devoid of any transcript information, therefore identifying hitherto-unknown genes (1476 PET clusters). Examples are shown in Supplementary Figure 5.
Table 3.
Categorization of transcripts identified by MS-PET sequencing analysis
| PET clusters | PETs | PET counts | |
|---|---|---|---|
| Known genes | 15 163 | 125 986 | 213 026 |
| ESTs | 3405 | 5504 | 7020 |
| Gene prediction | 2202 | 3168 | 4093 |
| Novel genes | 1476 | 1954 | 2240 |
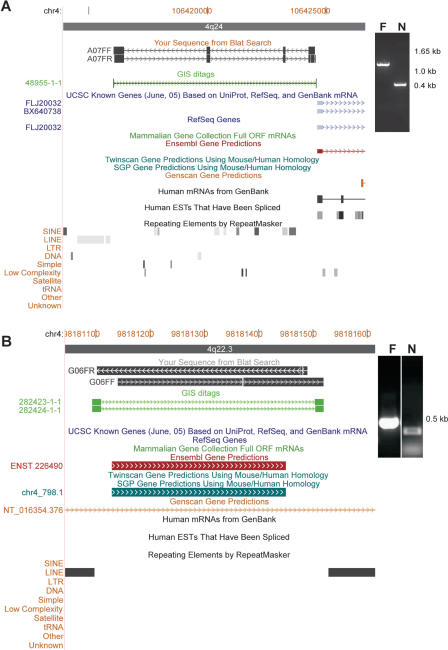
To validate previously-uncharacterized transcripts identified by MS-PET, we performed nested PCR on the original flcDNA library using primers that were designed from the 5′ and 3′ PET signatures and genomic DNA sequence information, and confirmed that 36/48 (75.0%) putative novel genes and 41/48 (85.4%) of the predicted genes were authentic. Sequencing of an arbitrary selection of 25 of these amplicons verified 11/13 (84.62%) of PET-identified novel genes, and 12/12 (100%) of PET-mapped predicted genes. An example from each of these two categories is shown in Figure 3.
Figure 3.
Examples of genes identified by MS-PET analysis. (A) Novel gene discovery. PET_ID# 48 955.1 (green arrowed line) identifies a novel gene transcript on chromosome 4, and is verified by PCR [inset; F, flanking (primary); N, nested (secondary)] and DNA sequencing of the amplicon (black arrowed lines A07FF and A07FR). (B) Validation of a predicted gene. PET_ID# 282 423.1 (green arrowed line) identifies a predicted gene on chromosome 4, and is verified by PCR [inset; F, flanking (primary); N, nested (secondary)] and DNA sequencing of the amplicon (black arrowed blocks G06FF and G06FR).
One putative novel gene transcript was identified by a single copy PET_ID 48 955.1, with a genomic span of 8523 bp on chromosome 4. The clone was subjected to full-length sequencing analysis, whereupon it was revealed that this transcript is 1.2 kb long and contains 4 exons. This transcript is expressed from a putative bidirectional promoter also regulating a known gene transcript (FLJ20032). PET_ID 282 423.1 and PET_ID 282 424.1 identified a single exon gene of 427 bp, which had previously been predicted by at least two independent computational prediction programs. Given its small size, lack of intron/exon organization and location between two LINEs, it may be a non-coding mobile DNA element.
Mapping of ChIP-PET sequences to the human genome
Of the 22 687 unique p53 ChIP-PET sequences obtained, 10 036 (44.24%) were mapped to the human genome sequence using the primary mapping parameters, i.e. without applying homopolymer error correction. Of these, 8896 (88.64%) PETs were mapped to single chromosomal loci. In addition, a total of 1240 PET sequences that were non-identical, but which nonetheless mapped to identical chromosomal locations with only a few (<5) base pair difference were considered to be derived from the same ChIP DNA fragments (details in Supplementary Data and Supplementary Tables 4 and 5). The minor variation was considered a result of artifacts arising from manipulations during molecular cloning and sequencing. Of the 7656 PETs remaining, the majority (7529 of 7656) were mapped as singletons along the genome (i.e. only 1 PET per locus), while 127 PET-defined loci formed 57 discrete clusters, each comprising 2 to 6 PETs. As we had demonstrated in our previous study (6), these PET overlap clusters represented regions enriched by the ChIP procedure. Thus, the genomic loci mapped by the 57 clusters were most likely p53 transcription factor binding sites.
For validation purposes, we compared these 57 PET clusters to the p53 binding sites identified in our previous p53 ChIP-PET analysis, which was capillary-sequenced. We found that 55 of the 57 clusters (96.5%) matched. Furthermore, the majority of these (42 of 55; 76.36%) contained complete p53 consensus binding motifs (Supplementary Table 6), with an additional three regions containing p53 half-sites, when analyzed using optimized weighted matrices p53 motif model, p53PET (6), or by the MatInspector algorithm (15). The high correlation of PET clusters between the two studies suggests that the MS-PET strategy is at least as reliable as standard ChIP-PET experiments for the global identification of transcription factor binding sites.
In this limited MS-PET analysis of ChIP DNA, 127 PETs forming 57 putative p53 binding sites were obtained, against a background ‘noise’ of 7529 singletons. This gave a signal-to-noise ratio of 127/7529 or 1.68%. This is considerably lower than the similar ratio obtained from the larger reference dataset (6), wherein 4302 PETs (in 1766 clusters) were identified against a background of 61 270 singletons, 4302/61 270 = 7.02%. It is likely that the greater sequence coverage of the reference dataset contributes to the higher sensitivity, i.e. the more PET sequences obtained, the larger the number of PET clusters that would be identified.
DISCUSSION
We have established an effective paired-end scheme for the newly-developed short-read multiplex sequencing technology (9), and thereby enabled the characterization of mammalian transcriptomes and genomes using an ultra high-throughput, high information-content, and low-cost sequencing system.
By making possible the extraction of paired-end information from long DNA fragments, we were able to use MS-PET to overcome the short-read obstacle of 454-sequencing in its current format as the GS20 sequencer. We were also able to further increase the analysis efficiency of the existing PET approach (6,7), which is itself already an improvement over conventional sequencing-based genome analysis.
A single 4 h run of one GS20 sequencer was previously shown to produce more than 25 million bases of high-quality sequence, with an average read-length of 110 bases (9). To fully exploit this ∼100 base read-length, we modified our existing PET procedures by dimerizing asymmetric PETs into diPET templates, such that each MS-PET read would reveal two PET sequences. This immediately doubled the output of MS-PET analysis. As demonstrated in this study, we were able to obtain a total of 462 626 PETs from a single 4 h MS-PET run. With further streamlining, we can now generate over 600 000 PETs from each MS-PET run (C.-L. Wei, unpublished data).
For comparison purposes, in the same 4 h period, a state-of-the-art Sanger 96-well capillary sequencer (ABI 3730 × l) on a rapid-run protocol can produce a total of about 320 000 bases (Phred score >20) from 640 templates (data obtained from http://www.appliedbiosystems.com/products/abi3730xlspecs.cfm), or about 6400 PETs (assuming a best-case average of 10 PETs per clone, 500 bases/read in the rapid-run protocol). MS-PET therefore represents a further 100-fold improvement over PET analysis using Sanger capillary sequencers. It is expected that the prototype MS-PET sequencing methodology can be further improved for increased efficiency and accuracy.
It is known that 454-sequencing is prone to base mis-calling in homopolymer regions (9). While this poses a potential issue for de novo genome sequencing, homopolymer errors can be at least partly resolved in MS-PET analysis because PET sequences obtained from MS-PET experiments are mapped to reference genome sequences. As described in this study, a prototype scheme for correcting over- and under-call homopolymer errors in PET sequences by allowing single-base deletions and insertions, respectively, was validated. The resulting mapping rate is comparable to that of PETs generated by Sanger capillary sequencing.
The current reagent cost is US$5000 for generating half-a-million PETs in a 4 h machine run of GS20 sequencing. The ability to generate one million PET sequences in a single working day for under US$10 000 using MS-PET contrasts with Sanger capillary sequencing, where it would cost at least US$100 000 (estimated US$1 per sequence read of 500 bases per template, rapid-run, revealing 10 PETs; 100 000 reads for one million PETs) and take months to produce the same amount of information. This represents a substantial advance in both cost-reduction and speed-improvement for PET analysis.
The data presented in this work validates the feasibility of using MS-PET for very high-efficiency and comprehensive transcriptome analysis and whole-genome interrogation. Using MS-PET to perform a comprehensive transcriptome analysis, or whole-genome mapping of transcription factor binding sites after ChIP enrichment, would require a single 4 h run at a current cost of ∼US$5000 (9), resulting in the collection of half-a-million PET sequences. These represent sufficient transcripts for a useful profile of the human transcriptome, or of the global localization of transcription factor binding sites in the human genome. The completeness of the profile would depend on the complexity of the system being analyzed.
The PET concept, as exemplified by GIS-PET for transcriptome analysis (7) and ChIP-PET for whole-genome transcription factor binding site mapping (6), can conceivably be applied to other DNA fragment analyses where paired-end sequence information would be useful. These may include the identification of epigenetic elements, mapping chromosomal variations in cancer genomes, and assisting in genome scaffolding. The recent rapid development of new sequencing technologies that are ever faster and cheaper promises to revolutionize the field of genome characterization (16,17). The MS-PET procedure presented here, as well as optimizations in future versions that continue to exploit expected improvements in sequencing read-lengths or throughput, should prove to be of considerable utility in this arena.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Atif Shahad and How Choong Yong for their assistance in optimizing the T2G genome browser, Pramila Nuwantha Ariyaratne for help with data analysis, and Kong Say Li and Dawn Choi for useful discussions on estrogen-induced gene expression and capillary sequencing characteristics, respectively. This work was supported in part by an NIH ENCODE grant #1R01HG003521-01, and by the Agency for Science, Technology and Research (A*STAR), Singapore. Funding to pay the Open Access publication charges for this article was provided by Agency for Science, Technology and Research (A*STAR), Singapore.
Conflict of interest statement. Clotilde Perbost and Lei Du are full-time employees of 454 Life Sciences, the company that owns 454 Sequencing™ Technology. They are encouraged by the company to conduct rigorous research and participate in peer-reviewed publications. The other authors have declared no conflicts of interest.
REFERENCES
- 1.The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 3.Saha S., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E. Using the transcriptome to annotate the genome. Nat. Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- 4.Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto S., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K. 5′ End SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- 6.Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 7.Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A., Wong C.H., et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nature Meth. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- 8.Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 9.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shendure J., Porreca G.J., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 11.Carninci P., Hayashizaki Y. High-efficiency full-length cDNA cloning. Meth. Enzymol. 1999;303:19–44. doi: 10.1016/s0076-6879(99)03004-9. [DOI] [PubMed] [Google Scholar]
- 12.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 13.Burge C.B., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 14.Westley B., Rochefort H. Estradiol induced proteins in the MCF7 human breast cancer cell line. Biochem. Biophys. Res. Commun. 1979;90:410–416. doi: 10.1016/0006-291x(79)91250-6. [DOI] [PubMed] [Google Scholar]
- 15.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector—New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Service R.F. Gene sequencing. The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 17.Chan E.Y. Advances in sequencing technology. Mutat. Res. 2005;573:13–40. doi: 10.1016/j.mrfmmm.2005.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.