Abstract
Many important interactions of proteins are facilitated by short, linear motifs (SLiMs) within a protein's primary sequence. Our aim was to establish robust methods for discovering putative functional motifs. The strongest evidence for such motifs is obtained when the same motifs occur in unrelated proteins, evolving by convergence. In practise, searches for such motifs are often swamped by motifs shared in related proteins that are identical by descent. Prediction of motifs among sets of biologically related proteins, including those both with and without detectable similarity, were made using the TEIRESIAS algorithm. The number of motif occurrences arising through common evolutionary descent were normalized based on treatment of BLAST local alignments. Motifs were ranked according to a score derived from the product of the normalized number of occurrences and the information content. The method was shown to significantly outperform methods that do not discount evolutionary relatedness, when applied to known SLiMs from a subset of the eukaryotic linear motif (ELM) database. An implementation of Multiple Spanning Tree weighting outperformed two other weighting schemes, in a variety of settings.
INTRODUCTION
Many protein interactions are facilitated through short, linear motifs (SLiMs). Such motifs have been implicated in many fundamental biological processes, including sub-cellular targeting [e.g. The KDEL Golgi-to-Endoplasmic Reticulum retrieving signal (1)], post-translational modification [e.g. The C- Mannosylation site WxxW (2)] and protein–protein interactions [e.g. The LxCxE ligand motif for the B-domain of the retinoblastoma proteins (3)]. Over a hundred different eukaryotic SLiMs have been identified so far (4) and it has been estimated that hundreds have yet to be discovered (5). When eubacterial, archaebacterial and viral motifs are also considered, the true number of unknown functionally important linear motifs is likely to be huge. Given the fundamental roles these motifs play in the basic functions of proteins and cells, identifying these motifs is of crucial importance for all biological disciplines.
While identifying domains in proteins is relatively straightforward [see (6,7) for reviews] with methods such as PRATT (8), TEIRESIAS (9) and MEME (10) efficiently discovering protein family signatures and other conserved regions, identifying SLiMs presents an inherently greater challenge. Web servers, such as eukaryotic linear motif (ELM) (4) and QuasiMotiFinder (11) employ various methods, such as domain masking and evolutionary filtering respectively, to discover new occurrences of previously known motifs. However, the web-based LMD method (5) became the first method to explicitly attempt novel SLiM discovery. The majority of SLiMs are between 3 and 10 amino acids in length and most have one or more ambiguous (variable) or wildcard (totally variable) residues. These two factors make real SLiMs difficult to distinguish from the background distribution of randomly occurring false positive motifs. Evolutionary conservation in orthologs is frequently used for finding larger domains but is of less utility in SLiM discovery since, due to the degenerate nature of many SLiMs, similar non-functional motifs of the same complexity can show similar levels of conservation in closely related organisms. The short and degenerate nature of SLiMs makes them evolutionarily plastic and particularly amenable to convergent evolution (12). Rather than looking for similarities between evolutionarily related sequences therefore, a potentially powerful way to discover novel SLiMs is to look for motifs that are shared between functionally related proteins that otherwise have little or no sequence similarity.
Here, we present a new motif discovery method, SLiMDisc (Short Linear Motif Discovery), to find shared motifs in proteins with little or no primary sequence similarity from a group of proteins with a common attribute—be it biological function, sub-cellular location or a common interaction partner. The method builds on the basic pattern discovery abilities of simple motif discovery tools, such as the TEIRESIAS (9) algorithm, applying a number of filters to the returned motifs to up-weight those present in apparently unrelated sequences and down-weight those primarily arising due to common evolutionary descent. A key feature of this method is that it requires no pre-filtering of the dataset for evolutionarily conserved sequences and does not suffer from the potential loss of information (and SLiMs) incurred by arbitrarily retaining a single representative of any given group of homologous proteins. Furthermore, a number of filtering options are provided, giving the user a great deal of control over the type of motif returned. We have applied SLiMDisc to a benchmarking dataset from the ELM database (4) and demonstrate that it significantly outperform methods that do not account for evolutionary relationships between the searched proteins.
MATERIALS AND METHODS
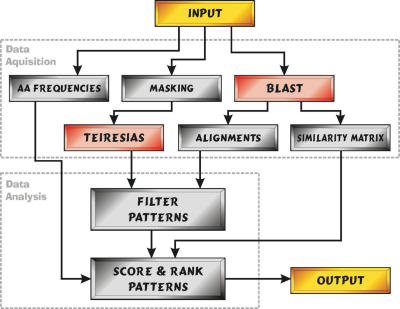
The method was implemented in a Python program, SLiMDisc, which outputs a ranked list of putatively interesting SLiMs from an input dataset of proteins. An overview of the method is given in Figure 1. First, TEIRESIAS is used to identify all motifs with user defined parameters for minimum support (the number of occurrences of a returned motif), motif length and number of non-wildcard positions, and which potential ambiguities are to be allowed. For analyses using ambiguity, the default TEIRESIAS ambiguity codes for amino acids were used (AG, FY, KR, DE, LIVM and QN). Motifs were filtered according to a number of optional criteria, including the evolutionary relatedness of the proteins containing the motif, information content and surface probability. Finally, the motifs are ranked according to information content (13) and normalized support among unrelated sequences, as described below. The SLiMDisc program is available on the SLiMDisc website at http://bioinformatics.ucd.ie/shields/software/slimdisc/.
Figure 1.
Simplified graphical representation of the SLiMDisc method. The steps completed by SLiMDisc are in green, those which occur outside the program are in red. The input dataset is given to the TEIRESIAS algorithm for pattern discovery and the BLAST algorithm to establish the evolutionary relationships of the parent proteins. The returned motifs are then filtered according to a number of user defined criteria. Finally, the motifs are ranked using information content (based on amino acid frequencies) and evolutionary relatedness.
Input
SLiMDisc accepts input in FASTA (14) or UniProt (15) download format. UniProt annotations may be automatically used to restrict analysis to certain domains, or to eliminate certain domains. The user may therefore edit the annotation to customize the protein search space as desired. Areas, such as transmembrane regions, protein domains and inaccessible residues can be masked since, for certain purposes, they are areas which may have a lower likelihood of containing motifs. Alternatively, the user may wish to identify specific regions of the proteins in which to confine the search, e.g. the cytoplasmic regions of a set of proteins, or may have prior knowledge of a region possibly containing a functional motif. This allows easy incorporation of experimentally-derived knowledge without lengthy post-processing of results.
Motif discovery
Putative motifs are returned by the TEIRESIAS algorithm. TEIRESIAS is a flexible algorithm which allows the user to specify several settings controlling the class of motif returned (fixed/ambiguous patterns, minimum number of fixed positions, etc.) and guarantees to return all maximal motifs of that class in the input dataset (9). The number of motifs returned is typically related to the input database size and the relatedness of the proteins in the dataset (the more closely related sequences the dataset contains, the longer the search will take). Motifs which have a complexity below a user defined threshold can also be removed. Multiple hits between regions of low complexity, (such as poly-glutamine repeats) are a common problem in motif discovery searching so this filter may be useful for some datasets. However, it is worth noting that some important biological motifs have low complexity, e.g. the NR box nuclear receptor binding motif LxxLL (16). The Emini method (17) for predicting surface probability has also been implemented, so that motifs where less than a user defined percentage cut-off of residues are calculated to be on the surface of the protein can be optionally removed. However, this method is not as accurate as 3D structure-based methods, such as DSSP (18) and should be used with caution.
Information content
A motif M can be defined as:
M = R1, x(g1), R2, x(g2), R3, x(g3), …, Rn-1, x(gn-1), Rn
where R is a fixed/ambiguous position and x(g) is a gap/wildcard of length g, there are n non-gap positions.
The Information Content is altered to give infrequent amino acids a higher score, their scores are up-weighted by modifying an element of the classical Information Content (13):
The overall altered Information Content IC is calculated using the formula below:
where Ri is the set of amino acids at position i, pRi is the probability of Ri, pa is the probability of a, S is the set of all amino acids, w is a gap weight and gk is the length of the gap at position k.
Normalized number of occurrences of motifs
The number of occurrences of each motif is down-weighted according to the similarity of the sequences containing the motif. Ideally, the normalized support should have the following characteristics:
(i) If n motifs are found in identical proteins, the normalized support is 1; (ii) If n motifs are found within completely dissimilar proteins, the normalized support is n and (iii) the normalized support must increase from 1 towards n as similarity of the parent proteins decreases. We implemented three methods of varying stringency to normalize the score, each of which may be most applicable in different biological contexts.
(1) Minimum spanning tree (MST) normalization
The default method is to use a MST method (19) (Supplementary Figure 1), which groups together closely related proteins down-weighting their overall contribution to the normalized support. The MST is the network of edges connecting all nodes, which has the minimum sum of edge lengths. The MST is calculated using a slightly altered version of Prims algorithm (19) allowing for the weighting of edges in a biologically useful way (20). Here, the MST is calculated based on a distance matrix of the sequence similarity of the proteins containing the motif, normalized to a value between 1 and 0 (where 1 is no similarity). Weighting of the edge lengths, shortening the more closely related branches more harshly than the distantly related branches while still keeping the score between 1 and n (where n is the number of proteins containing the motif) is also possible.
Sequence similarity was calculated using the GABLAM (Global Analysis from BLAST Local AlignMents) method (Supplementary Figure 2), which gives a global percentage similarity for any pairwise comparison of proteins by mapping local BLAST alignments onto the two proteins. Since the GABLAM method calculates a separate % identity for each of two sequences, we took the minimum of the two % identities in each case. As a percentage of the protein, this score is normalized for sequence length (unlike BLAST scores and E-values). Furthermore, multiple hits to the same region (due to multiple domains or low complexity regions, for example) do not artificially inflate scores. GABLAM is not so sensitive to alignment artefacts when comparing unrelated proteins and, unlike pairwise sequence alignment (21), will give them a similarity score of zero. GABLAM will also account for domain rearrangements, which pairwise alignment does not.
(2) Unique homologous segments (UHS) normalization
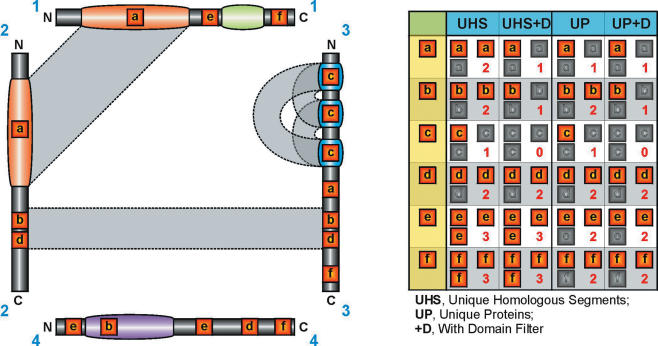
The MST method has the apparent disadvantage that it ignores whether or not a motif lies within the evolutionarily conserved or unconserved region of the protein, as detected by BLAST alignment. For this reason, we considered an alternative weighting scheme. This defined a UHS weighting, defining the support as the number of occurrences within all sets of aligned and unaligned regions (Figure 2). In the case where three occurrences occurred within regions where A aligned with B, and B aligned with C, but A did not align with C, UHS considered this to represent only a single sequence in calculating the support.
Figure 2.
Graphical representation of UHS and UP normalization techniques. Four proteins, labelled 1–4, are shown with annotated domains marked as coloured regions. Regions of homology as detected by BLAST are shown as grey boxes linking the sequences. Sequences 1 and 2 share a large homologous (orange) domain. Sequences 2 and 3 also share a homologous region but this is not annotated as a domain. Three other domains are specific to proteins 1 (green), 3 (blue) and 4 (purple). All motifs a–f have three occurrences in the dataset but have different support (shown in the table on the right) after filtering. a.→Motif a occurs in a shared region between 1 and 2, which is reduced by UHS to a single occurrence. The third occurrence in sequence 3 is not in an homologous region to 1 or 2 and is treated as a separate occurrence by UHS. However, proteins 2 and 3 share a homologous region and so UP will cluster sequences 1, 2 and 3, reducing the number of occurrences to 1. Filtering domains reduces the support to 1 in either case. b.→Motif b occurs in a shared region between 2 and 3, which is reduced by both UHS and UP to a single occurrence. This time, the third occurrence lies in the totally unrelated protein 4 and is counted with either filter. Filtering domains removed the occurrence in 4, reducing the support to 1. c.→Motif c lies purely within a repeated domain in protein 3. This is reduced to a single occurrence by both UHS and UP (the protein is homologous with itself). Although, whole-protein self-hits are ignored by UHS, the additional local BLAST hits between different domains (shown in grey) will still cause motif c to be filtered by UHS. Domain filtering removes it completely. d.→Motif d is the same as motif b, except that none of the occurrences lie in domains and so domain filtering makes no difference. e.→Motif e lies in non-homologous regions of protein 1 and 4. UHS therefore keeps all three occurrences. Whole-protein self-hits are ignored during the UHS filtering, and so both occurrences of motif e in protein 4 are counted. In contrast, UP clusters sequence 4 with itself and reduces the support to 2. No occurrences lie in domains and so domain filtering makes no difference. f.→Motif f is found in proteins 1, 3 and 4. None of these regions are homologous and so UHS gives a support of 3. UP, however, will group proteins 1 and 3; even though they do not directly share homology, they both share homology with common protein 2. UP therefore reduces the support to 2.
Normalizing the support of a motif based on the number of UHS allows the proteins to be treated as modular entities (Figure 2). This method adequately deals with similarities both between proteins and within proteins. Given that many proteins in higher eukaryotes are multi-domain, and many domains exist in multiple copies within a protein, this is an important consideration. The number of UHS containing the motif then provides the normalized support for that motif.
(3) Unrelated proteins (UP) normalization
In certain circumstances, BLAST homology may tend to underestimate the degree of evolutionary relatedness, and may only define certain regions as being homologous, while many more regions, or the entire proteins, may be so. In this circumstance (e.g. in searches restricted to single domain proteins, such as cytokines) it may be appropriate to treat two occurrences in proteins sharing any BLAST similarity whatsoever as being a single occurrence (UP in Figure 2). This is the strictest normalization method available in SLiMDisc. Essentially, given a list of proteins containing a pattern, the technique clusters together proteins which have significant level of homology to each other even if inferred through another protein. For example, if a motif occurs in three sequences, where p1 has significant homology to p2 and p2 has significant homology to p3, but BLAST failed to detect similarity between p1 and p2, the inferred number of normalized occurrences is then estimated as 1 (Figure 2). UP clustering produces one or more groups of proteins with no detectable homology between groups (with the BLAST parameters used). The normalized support is the number of groups. By adjusting the BLAST parameters, this setting can be tailored for very stringent or very relaxed clustering.
Ranking
Having established a large number of motifs, the objective is to rank them according their information content and frequency of occurrence, adjusted for evolutionary relationships. Given IC and one of the above measures of the normalized support N, a motif score is then estimated as:
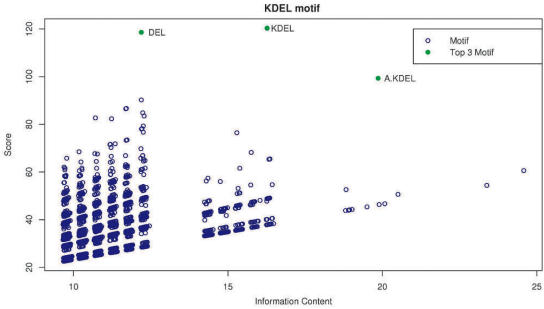
as suggested by Jonassen et al. (20). In practice, this provides a reasonable means of identifying the motifs of most interest. Scattergrams of information content versus the score (Figure 3) provide a visual approach to identify the degree to which the score of the motif of interest stands out from other returned motifs of equivalent IC, which may in part reflect the background distribution of false positives, although care needs to be taken to distinguish false positives from sub-motifs or alternative representations of the most interesting motif.
Figure 3.
Scattergram of the information content versus the score for the KDEL (see Table 2) retrieving motif. Each blue point on the scattergram is a motif which has been considered by SLiMDisc. The points in green are the top three motifs ranked by the method. The actual SLiM for this dataset is the motif described by the regular expression [KRHQSAP][DENQT]EL.
Creating the benchmarking dataset for the method
We adopted the same set of 22 benchmarking ELMs from the ELM database as the LMD method to facilitate comparison with their method. The web-based LMD method (5) is a recently published SLiM discovery method that deals with problems of common evolutionary descent by eliminating all but one representative of a group of homologous segments. This allows LMD to use a probabilistic scoring scheme (based on raw motif occurrences in a random dataset), which is suitable for motif occurrences that can be considered to be independent, when the random dataset approximates well the underlying background motif distribution of the particular group of proteins or protein regions under consideration. For the SLiMDisc method, given the inclusion of non-independent (related) proteins in datasets generating motifs, a simpler scoring scheme based on information content was applied. After testing the validity of their method on the benchmarking test set, the LMD method was applied to interaction datasets, where direct binding assays of synthetic oligopeptides revealed that predicted novel motifs bound the target proteins as expected (5). To compare like with like, we similarly restricted motif discovery to regions outside of documented protein domains (5) but also carried out searches without domain filtering. The benchmark set of 22 validated motifs comprises those considered to have greater than three motifs outside of protein domains, in what appeared to be unrelated proteins (5). Of these, we omitted the Groucho/TLE binding motif (LIG_EH1) (22) (no instances available at www.elm.eu.org), the Mannosylation site motif (MOD_CMANNOS) (2) (occurs in the annotated Thrombospondin type 1 domain), the TRAF2 binding motif (LIG_TRAF2_1) (23) (all instances available on web site are evolutionarily related) and the WRPW motif (LIG_WRPW_1) (24) (all instances available are from the Runt and Hairy protein families and the motif is easily discovered by looking down a multiple alignment of the dataset), leaving 18 motifs for comparison. SLiMDisc was used to discover motifs using several normalization techniques described in the Materials and Methods section, taking as input for each ELM in turn the set of sequences documented to contain the motif. The rank of the highest ranking motif matching the regular expression given for the documented ELM motif was then recorded for each ELM. All datasets used in this analysis are available on the SLiMDisc website at http://bioinformatics.ucd.ie/shields/software/slimdisc/.
RESULTS
ELM dataset
The ELM benchmarking dataset is based on a dataset proposed to validate the performance of the LMD program (5) in which every sequence contains the known motif. It is not clear if taking a set of sequences, all of which contain a known ELM, is a good validation of the process of discovering novel motifs, since most real discovery datasets will contain sequences that do not contain the motif. Nevertheless, it provides one potential comparison and, in the absence of a suitable alternative, represents the best validation resource. We consider the performance of SLiMDisc with other datasets, which do not have 100% support below. The default SLiMDisc method implemented was MST weighting, with two other algorithms, UHS and UP also explored (see Materials and Methods for details).
An overview of the results is presented in Table 1. This demonstrates a clear improvement of SLiMDisc over basic TEIRESIAS, regardless of whether fixed or ambiguous, and regardless of whether ELMs occurring in domains were filtered out. Typically, there was a very substantial improvement in motif rank, and a clear improvement in the number of identified sequences (support). In comparison with MST, UHS performed similarly, but not quite as well. The stricter UP algorithm, which might be expected to eliminate evolutionarily related sequences where the relationship was defined by a region that does not span the motif occurrences, performed more poorly, presumably because it erroneously excluded proteins sharing common domains that were irrelevant to the motif. However, it still clearly outperformed the basic TEIRESIAS method in terms of rank, while support was lowered by the exclusion of a greater number of sequences.
Table 1.
Summary performance of the different normalization techniques on ELM benchmark dataset
| Ambiguity | Domain filter | MST | UHS | UP | TEIRESIAS | ||||
|---|---|---|---|---|---|---|---|---|---|
| Rank | Support % | Rank | Support % | Rank | Support % | Rank | Support % | ||
| No | No | 18.06 | 73.9 | 24.83 | 69.8 | 50.56 | 51.5 | 62.17 | 64.3 |
| No | Yes | 16.61 | 70.5 | 20.83 | 66.4 | 49.83 | 48.1 | 66.61 | 59.6 |
| Yes | No | 41.61 | 70.9 | 56.56 | 69.6 | 69.11 | 57.9 | 84.22 | 52.6 |
| Yes | Yes | 28.39 | 72.5 | 48.61 | 64.0 | 53.56 | 56.6 | 57.33 | 62.6 |
Comparison of the average rank of the motif matching the regular expression given in the ELM database and the average percentage support of the top ranked pattern for the ELM benchmark dataset between the three different normalization techniques and the TEIRESIAS algorithm with and without domain filtering and ambiguity. Rank is calculated using the arbitrary value of 200 when the motif of interest is not found in the top 100 motifs returned. Support for each ELM is the percentage of proteins in the dataset containing the returned ELM.
Looking at the results in more detail for each of the 18 motifs of the ELM dataset, SLiMDisc (MST) returned known ELMs in the top 100 ranked motifs for all motifs with the exception of one. A total of 10 (58%) were the top ranked motif and 15 (84%) were in the top 10 ranks (Table 2), compared to only 8 for TEIRESIAS. These results are similar to the LMD results reported previously (5), with both methods returning in first position the true ELM in 10 of the 18 cases. In our experience, we could not obtain such good performance applying the same datasets to the web version of LMD (5). This may reflect some differences in the test dataset applied here to SLiMDisc, or details of the web server implementation of LMD. The dataset is probably too small to comment in detail on differences between SLiMDisc and LMD. However, SLiMDisc missed only one ELM [the sparse N-glycosylation site (MOD_N-GLC_1) (25)] within the top 100, while the published evaluation of LMD missed three (5). A more detailed comparison would require running both methods on a larger identical test dataset.
Table 2.
Comparison of methods based on results from the ELM benchmark dataset
| ELM | TEIRESIAS | SLiMDisc | LMD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial motifs | Rank | Motif | Support | Rank | Motif | Support | Initial motifs | Rank | Motif | Support | ||
| 14-3-3(type 1)-R(SFYW)xSxP | 2232 | 1 | RSxSxP | 4/4 | 1 | RSxSxP | 4/4 | 225 | 1 | RSxSxP | 3/4 | |
| 14-3-3(type 3)-(RHK)(STALV)x(ST)x(PEDSIF) | 11 130 | 6 | SxSxP | 6/6 | 5 | SxSxP | 6/6 | 1656 | 6 | RSxSxE | 7/12 | |
| c-adaptin-(DE)(DES)xFx(DE)(LVIMFD) | 17 339 | 1 | DxFxDFxS | 5/8 | 1 | DxFxDFxS | 5/8 | 392 | 1 | DDxFxxF | 3/4 | |
| Clathrin box-L(ILM)x(ILMF)(DE) | 53 613 | 80 | LxDL | 12/15 | 1 | LxDL | 12/15 | 778 | 1 | LlxLD | 3/5 | |
| CtBP-Px(DEN)L(VAST) | 157 438 | 53 | PxDL | 20/26 | 1 | PxDL | 20/26 | 26 892 | 1 | DxPxDL | 8/25 | |
| Cyclin-(RK)xLx{0–1}(FYLIVMP) | 100 074 | — | — | 0/22 | 7 | KKL | 11/22 | 13 179 | 15 | KRRL | 3/19 | |
| Dynein light chain-(KR)xTQT | 1263 | 1 | SxxKxTQT | 3/4 | 1 | SxxKxTQT | 3/4 | 117 | 1 | KxTQT | 3/4 | |
| HP-1-PxVx(LM) | 10 035 | 2 | PxVxL | 6/6 | 1 | PxVxL | 6/6 | 5287 | 4 | KVPxVxL | 3/7 | |
| NRBOX-LxxLL | 68 944 | 9 | LxxLL | 8/9 | 10 | LxxLL | 8/9 | 27 874 | — | — | 0/18 | |
| PCNA-Qxx(ILM)xx(FHM)(FHM) | 64 978 | — | — | 0/13 | 36 | QxxLxxF | 9/13 | 1505 | 1 | QxxxxxFF | 11/24 | |
| Retinoblastoma-(LI)xCx(DE) | 131 943 | 1 | LxCxE | 18/25 | 1 | LxCxE | 18/25 | 24 581 | 1 | LxCxE | 14/24 | |
| Integrin-RGD | 8351 | 25 | RGD | 9/15 | 1 | RGD | 9/15 | 154 | 2 | R.DV | 3/8 | |
| SH3 (type 2) domain-PxxPx(KR) | 14 380 | 1 | PPxP | 7/9 | 1 | PPxP | 7/9 | 1406 | 1 | PPxxPxR | 4/7 | |
| TRAF6-PxE | 9294 | 18 | QxPxE | 7/8 | 2 | QxPxE | 7/8 | 121 | 1 | PQE | 3/7 | |
| N-glycosylation-NxC | 1982 | — | — | 0/4 | — | — | 0/4 | 53 | — | — | 0/4 | |
| SUMO-1-(VILAFP)Kx(EDNGP) | 132 732 | — | — | 0/29 | 2 | IKxE | 15/29 | 13 722 | — | — | 0/14 | |
| Golgi-to-ER signal-(KRH)(DENQ)EL | 17 236 | 1 | DEL | 11/12 | 1 | KDEL | 8/12 | 8 | 1 | KDEL | 3/5 | |
| Endosome sorting signal-(DER)xxxL(LVI) | 10 201 | — | — | 0/10 | 27 | ExxxLL | 5/10 | 1471 | 22 | ExxxLL | 5/12 | |
Results of the analysis of 18 datasets from the ELM database as proposed by Neduva et al (5). The table compares the first ranked position, support and number of initial motifs between TEIRESIAS (scored using the product of support and information content), SLiMDisc (using default settings) and the LMD method [as described in the LMD paper (5)]. Results in bold are motifs for which the method returns the best rank or equal best rank for that ELM across the three methods.
It is instructive to consider why the alternative weighting schemes do not perform as well as MST (Supplementary Table 1). A particularly striking example is the CtBP motif, which is returned as the top motif by MST, but with a rank of 54 and 50 by UHS and UP. While the overall performance of UP was worse, in one instance it performed better than MST: for the PCNA ligand motif, the true motif was ranked 33 by MST, but third by UP. In this case, the set of proteins may share a degree of weak similarity below the threshold detectable by BLAST which vastly increases the number of background motifs returned by TEIRESIAS. Domain filtering is sometimes recommended for SLiM discovery. With the dataset of 18 ELMs used here, the difference was modest (Supplementary Table 1). However, overall performance was marginally better with domain filtering. The key question is whether this is valid for other motifs outside of those investigated here.
The method can return ambiguous motifs, such as the Golgi-to-ER targeting signal (KRH)(DENQ)EL (1). However our test sets showed that, with this dataset, the ambiguity did not increase the sensitivity of the searches (see Supplementary Table 1). In cases where there was already a highly ranked pattern, ambiguity occasionally returned a more accurate descriptor of the motif [for example the [KR]xTQT motif of the Dynein Light Chain binding motif (23)] and rarely caused the motif to drop down the rankings. On occasions where the motif was poorly ranked without ambiguity, the ambiguity added noise to the dataset causing their rank to worsen. Overall, from our experience with the ELM dataset, application of ambiguity slightly disimproved the performance. TEIRESIAS ambiguity coding is not perfect for amino acid analysis: it does not adequately capture the fact that certain amino acid properties are overlapping, rather than falling into discrete categories. Secondly, for certain motifs, or residues within motifs, the requirements may be very specific for one amino acid, or instead be permissive across the range of ambiguous residues.
Despite the shortcoming of the ELM test dataset that the SLiM was present in all input sequences, in practise there was <100% support when considering the analyses performed without ambiguity: not all occurrences of a given motif had the same residues at the ambiguous positions, effectively generating a number of derived motifs that could be detected, which occur only in a subset of the whole dataset. For example, the SUMO motif recognized for modification by SUMO-1 (26) is described in the ELM database as [VILMAFP]KxE. The returned motif, IKxE, occurs in only 14 of the 28 proteins. This illustrates that it is not necessary for the motif to have very high support to be returned by SLiMDisc. On the other hand, it also highlights the problems of searching for SLiMs with ambiguous positions. Until a good method is developed for introducing ambiguities without generating overwhelming quantities of noise into the results, it may prove most sensible to search without ambiguities and then re-search the dataset for additional motifs that are highly similar to the best SLiMs returned. Only by further experimental validation can the true sequence of a given SLiM be elucidated. The analysis of test datasets with <100% support is considered below.
RECOVERY OF MOTIFS FROM DATASETS WHERE THERE IS <100% SUPPORT
Interaction datasets
A more challenging test for the method would come when noisy biological data are analysed. Large scale interaction databases provide readily available benchmarking datasets, which can be probed to re-discover known SLiMs (5). The datasets provide protein interaction data for a central hub protein known to interact with several spoke proteins. When searching for SLiMs in these datasets, we are looking for SLiMs involved in motif–domain interactions and motif–motif interactions. However, these interactions datasets often contain large numbers of domain–domain interactions, so that the signal of any SLiMs contained in the datasets can be overpowered. Secondly, very large proteins may have multiple domains and motifs involved in interactions, increasing the ‘noise’ around any given motif. Thirdly, high-throughput experimental datasets often contain considerable experimental noise.
To test the performance of SLiMDisc on a dataset with a more realistic level of noise, we took a set of 17 proteins from the literature, which are known to interact with several of their binding partners through ELMs, and also had an entry as a hub protein in the HPRD interaction database (27). We analysed their HPRD-defined binding partners for the presence of interesting motifs using SLiMDisc (default settings). In 7 of these 17 test sets, motifs from known binding sites were re-discovered in the top 10 ranked motifs returned (41% success rate) (Table 3). For these seven returned motifs, there is likely to be considerable noise in the dataset, since between 71 and 96% of the proteins in the datasets do not contain annotated instances of the ELMs (Table 3).When TEIRESIAS motifs ranked based on true support and information content were investigated only 1 of the 17 datasets returned the motifs from known binding sites in the top 100 ranked motifs (LIG_NRBOX motif ranked 96th). To establish how many motifs might be returned at random, we determined the numbers of motifs in the top ten returned for the reversed motif: none of the 17 reversed motifs were returned in the top 100. When restricting datasets to proteins interactions derived from Yeast Two Hybrid experiments (12 datasets), only 2 of these datasets returned the known motif in the top ten ranked motifs (a 17% success rate compared to 41% success rate for all HPRD interactions). This trend is not surprising, given the fairly high level of experimental noise in yeast two hybrid data: however, we would emphasize that the sample sizes are relatively small and the success rates only indicative.
Table 3.
Results from ELM containing HPRD interaction datasets
| Hub protein | HPRD _id | ELM name (annotated motif) | % True annotated motifs | Returned motif (rank) |
|---|---|---|---|---|
| CtBP | 04015 | LIG_CtBP ([PG][LVIPME][DENS]L[VASTRGE]) | 0.29 (9/31) | DLS (6) |
| Clathrin | 00350 | LIG_Clathr_ClatBox_1 (L[IVLMF]x[IVLMF][DE]) | 0.21 (5/24) | LxDL (2) |
| Peroxisome proliferator activated receptor gamma | 03288 | LIG_NRBOX (LxxLL) | 0.14 (3/21) | LxxLL (4) |
| Integrin Alpha 5 | 00627 | LIG_RGD (RGD) | 0.1 (2/21) | RGD (4) |
| Grb2 | 00150 | LIG_SH3_2 (PxxPx[KR]) | 0.04 (6/159) | PxPP (3) |
| 14-3-3- Eta | 00215 | LIG_14-3-3_1 (R[SFYW]xSxP) | 0.06 (2/31) | RSxS (4) |
| Ubiquitin conjugating enzyme E2I | 09045 | MOD_SUMO ([VILMAFP]KxE) | 0.09 (4/43) | IKxE (8) |
Results for the seven datasets which returned true annotated binding motifs in the top 10 ranks for the HPRD interaction datasets. A returned motif is defined as one which is found at the annotated positions of the known instances of the motif (including motifs which account for at least 2 of the residues involved in the ELM interaction). % True annotated motifs is a measure of the extent of anticipated noise in the dataset: datasets with a low % have relatively few of the proteins where the ELM has been annotated.
The introduction of ‘noise’ in the form of false positives (spoke proteins not interacting with the hub through a SLiM) and false negatives (missing proteins) in these datasets clearly degrades performance in comparison to the highly-supported ELM benchmarking datasets. False positives increase the number of stochastically returned motifs, causing the rank of the true motif to decrease. Similarly, missing proteins will reduce the support for a motif, which negatively impacts its score and therefore its rank. Yet the SLiMDisc method appears to more strongly outperform TEIRESIAS (motifs ranked by true support and information content) in noisy biological data compared to the more idealized ELM benchmarking dataset, indicating that correcting for common evolutionary descent is even more critical in typical datasets where motif discovery is preformed.
RGD
The RGD motif (28) interacts directly with integrin extracellular domains and is critical for the cell adhesion of numerous proteins, such as fibrinogen, fibronectin and von Willebrand factor. We searched all 53 human proteins linked to the GO term ‘Integrin binding’, without filtering domains. The RGD motif was returned (MST rank: 4; UHS rank: 64; UP rank: > 100). It has been reported that <15% of SLiMs occur in domains (5). While there can be a substantial increase in sensitivity of a SLiM detection method if domains are removed, this must be used cautiously. Removal of Swiss-Prot annotated domains in this case would lower the pattern support from 23 to 13, significantly lowering the likelihood of RGD being detected.
A feature of SLiMDisc is its ability to specify a protein, or even a region within a protein, on which to focus a search (Only SLiMs that occur within this protein/region are then considered). The original discovery of the cell attachment site of fibronectin (29) narrowed it down to a 108 amino acid region. When we searched the ‘Integrin binding’ dataset again, this time requiring the true motif to occur within this 108 amino acid region, RGD was ranked first (MST: 1; UHS: 41; UP: >100; TEIRESIAS >100). Such restricted searches massively cut down search space, reducing both false positives and computational time and are strongly recommended where such information is available.
We then searched all human proteins with the GO cellular component term of ‘Extracellular Matrix’. This dataset consisted of 149 proteins, including the ‘Integrin binding’ dataset as a subset. When searched for interesting motifs (without domain elimination) the known RGD motif was not ranked in the top one hundred motifs. Even when the search was focused on the 108 amino acid region of Fibronectin the motif was only ranked in 32nd position. This illustrates the limits of searches for short motifs on large datasets. When the search was limited to surface accessible regions the ranking was increased to 18th. Protein interaction motifs are often restricted to the surface of proteins. Such motifs will be returned in datasets, and clues as to their nature can be revealed by investigating their positions in the protein, protein structure and by studying multiple alignments.
LPxTG
A dataset of 104 proteins from 37 bacterial species in UniProt (15) containing the keyword ‘Cell Wall’ and term ‘Anchor’ was investigated for its ability to return the known LPxTG motif (30), whose cleavage between Threonine and Glycine permits attachment to the cell wall. SLiMDisc analysis with MST normalization, and filtering out UniProt annotated domains, yielded over a million TEIRESIAS patterns for which 176 889 had an MST-normalized support of 2 or greater. The LPxTG motif occurred in just over half of the proteins present in the dataset, and was returned as the top ranked motif using MST normalization. Ranks of the matching motif using TEIRESIAS, UHS and UP were all >100 when using the default BLAST cut-off of 1e−2. Using a BLAST cut-off of 1e−6, ranks of the motif using TEIRESIAS, UHS and UP were >100, 2 and 61, respectively. This illustrates the sensitivity of the UHS and UP normalization techniques to choice of parameter settings.
Detection of the homologous cytokine receptor WSxWS motif
While the primary objective of this method is focussed on detection of motifs that have arisen by convergent evolution, and distinguishing them from motifs shared by common descent, it is clear that the resolution of the BLAST method will result in the detection of ancient motifs, identical by descent, that have been conserved in otherwise highly diverged proteins. We therefore tested the performance of the method to detect a known motif identical by descent, which defines a distinct signature for a protein family. We searched for motifs among a dataset of 37 human proteins linked to the GO term ‘Hematopoietin/interferon-class cytokine receptor activity’. These are all haematopoietic receptors. The analysis was performed without filtering of domains.
The highest ranked motif to be returned from the dataset was the well known WSxWS motif (31). This motif is not an interaction motif, but a structural motif that is homologous rather than convergent. Running the analysis eliminating UniProt domains would remove this motif, since it lies within a fibronectin domain. The three weighting schemes gave the following ranks for this motif (MST: 1; UHS: >100; UP: >100; TEIRESIAS: >100). In this case UHS and UP ranked the motif poorly as they can detect and down-weight homology via an intermediate protein that MST cannot. Recovering homologous motifs for extremely diverged proteins may give clues regarding homology that may not be detected outside of the motif, or give clues regarding critical functional regions. In this case, the input proteins are too distant to be aligned by Clustalw (32), yet SLiMDisc was able to identify a known functionally important motif.
DISCUSSION
Our implementation of weighting schemes to correct for close relationships shared by larger sequence regions provides a means to routinely discover motifs, without the requirement to discard in advance any sequences from the dataset under investigation. The weighting scheme which performed most strongly in a variety of settings was the MST weighting scheme. It is perhaps a little surprising that this simpler scheme appears the most robust, since it (and UHS) would appear sensitive to the problems of short motif sharing from evolutionary descent in regions that are just outside of a region alignable by BLAST. Perhaps BLAST is typically reasonably efficient at recovering most such short segments as part of a returned alignment. Clearly the datasets tested here are not representative of all possible biological scenarios, and even here we came across one case of a motif (PCNA), which is better recovered by the UP algorithm, which weights against any relationships among parent sequences, regardless of whether they overlap the motif of interest. Thus, these three weighting schemes may represent optimal strategies for particular problems, if enough is known about the domain and homology distributions of the datasets under consideration. The alternative LMD scheme is to simply discard all but one representative homologous segment from a group of related proteins (5); This is not so important for datasets, such as the ELM benchmarking dataset, where one knows that the motif is present in all proteins. In real applications, however, there is a big risk of accidentally discarding a region containing the motif of interest in favour of a related protein region that does not contain the motif. The methods we have presented represent a two-stage process, of motif discovery, followed by score normalization. Ultimately, the development of integrated motif discovery algorithms that incorporate a priori weighting schemes may improve or accelerate motif discovery.
While this method has been implemented here in conjunction with the most rapid motif discovery tool available to us (TEIRESIAS), clearly the same logic may also be applied to other motif discovery tools. Particularly important for certain classes of motifs are tools which permit flexible length gaps. Flexible length gapped motif discovery is of great interest as many biological motifs permit gaps, and the next major advances in SLiM discovery should find efficient approaches to incorporate gapped motif discovery into current techniques. In addition, an improved method for incorporating ambiguity into motifs is clearly desirable for future SLiM discovery tools. While the weighting schemes we propose are useful, the problem of motif discovery remains substantial. Most commonly, a SLiM discovery tool has to find a motif with relatively low support and relatively few fixed positions (low information content). There are problems associated with looking for SLiMs with low support; many motifs which have a biological activity can look less interesting than a pattern which has occurred by chance. Certain motifs which have low support and low information content are almost indistinguishable from random noise in most datasets [e.g. the PCSK cleavage site (33) [KR] R which plays a role in the proteolytic processing of both neuropeptide and peptide hormone precursors or the peroxisomal targeting motif WXXXY/F (34)]. It is vitally important, therefore, to reduce the search space as much as possible, restricting the input to only those proteins (and, where possible, the relevant regions of proteins) that are likely to contain the SLiM of interest. This reduction of search space has the greatest impact on reduction of false positives. The greatest impact in SLiM discovery may come from sensitive handling of biological classifications of proteins that assist in reducing the search space. These can be small scale careful analyses, such as probing a group of proteins which share a function, sub-cellular location or interacting partner; or can be large, genomic scale analyses, such as surveys of entire protein–protein interaction databases (5,27) or GO term analysis (35).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by Science Foundation Ireland. Funding to pay the Open Access publication charges for this article was provided by Science Foundation Ireland.
Conflict of interest statement. None declared.
REFERENCES
- 1.Munro S., Pelham H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 2.Furmanek A., Hofsteenge J. Protein C-mannosylation: facts and questions. Acta. Biochim. Pol. 2000;47:781–789. [PubMed] [Google Scholar]
- 3.Dahiya A., Gavin M.R., Luo R.X., Dean D.C. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 2000;20:6799–6805. doi: 10.1128/mcb.20.18.6799-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puntervoll P., Linding R., Gemund C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D.M., Ausiello G., Brannetti B., Costantini A., et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neduva V., Linding R., Su-Angrand I., Stark A., Masi F.D., Gibson T.J., Lewis J., Serrano L., Russell R.B. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS. Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazma A., Jonassen I., Eidhammer I., Gilbert D. Approaches to the automatic discovery of patterns in biosequences. J. Comput. Biol. 1998;5:279–305. doi: 10.1089/cmb.1998.5.279. [DOI] [PubMed] [Google Scholar]
- 7.Rigoutsos I., Floratos A., Parida L., Gao Y., Platt D. The emergence of pattern discovery techniques in computational biology. Metab. Eng. 2000;2:159–177. doi: 10.1006/mben.2000.0151. [DOI] [PubMed] [Google Scholar]
- 8.Jonassen I., Collins J.F., Higgins D.G. Finding flexible patterns in unaligned protein sequences. Protein. Sci. 1995;4:1587–1595. doi: 10.1002/pro.5560040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigoutsos I., Floratos A. Combinatorial pattern discovery in biological sequences: the TEIRESIAS algorithm. Bioinformatics. 1998;14:55–67. doi: 10.1093/bioinformatics/14.1.55. [DOI] [PubMed] [Google Scholar]
- 10.Bailey T.L., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 11.Gutman R., Berezin C., Wollman R., Rosenberg Y., Ben-Tal N. QuasiMotiFinder: protein annotation by searching for evolutionarily conserved motif-like patterns. Nucleic Acids Res. 2005;33:W255–W261. doi: 10.1093/nar/gki496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neduva V., Russell R.B. Linear motifs: evolutionary interaction switches. FEBS. Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Shannon C.E. The mathematical theory of communication. 1963. MD. Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 14.Pearson W.R. Rapid and sensitive sequence comparison with FASTP and FASTA. Meth. Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 15.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heery D.M., Kalkhoven E., Hoare S., Parker M.G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Emini E.A., Hughes J.V., Perlow D.S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 19.Prim R.C. Shortest connection networks and some generalizations. Bell Syst. Tech. J. 1957;36:1389–1401. [Google Scholar]
- 20.Jonassen I., Helgesen C., Higgins D.G. Scoring function for pattern discovery programs taking into account sequence diversity. Reports in Informatics. 1996 no. 116. [Google Scholar]
- 21.Rosenberg M.S. Evolutionary distance estimation and fidelity of pair wise sequence alignment. BMC Bioinformatics. 2005;6:102. doi: 10.1186/1471-2105-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez G., Verrijzer C.P., Ish-Horowicz D. A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol. Cell. Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye H., Park Y.C., Kreishman M., Kieff E., Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- 24.Fisher A.L., Ohsako S., Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein–protein interaction domain. Mol. Cell. Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vance B.A., Wu W., Ribaudo R.K., Segal D.M., Kearse K.P. Multiple dimeric forms of human CD69 result from differential addition of N-glycans to typical (Asn-X-Ser/Thr) and atypical (Asn-X-cys) glycosylation motifs. J. Biol. Chem. 1997;272:23117–23122. doi: 10.1074/jbc.272.37.23117. [DOI] [PubMed] [Google Scholar]
- 26.Melchior F. SUMO—nonclassical ubiquitin. Annu. Rev. Cell. Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 27.Mishra G.R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T.M., et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Main A.L., Harvey T.S., Baron M., Boyd J., Campbell I.D. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- 29.Pierschbacher M.D., Hayman E.G., Ruoslahti E. Location of the cell-attachment site in fibronectin with monoclonal antibodies and proteolytic fragments of the molecule. Cell. 1981;26:259–267. doi: 10.1016/0092-8674(81)90308-1. [DOI] [PubMed] [Google Scholar]
- 30.Schneewind O., Mihaylova-Petkov D., Model P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO. J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumgartner J.W., Wells C.A., Chen C.M., Waters M.J. The role of the WSXWS equivalent motif in growth hormone receptor function. J. Biol. Chem. 1994;269:29094–29101. [PubMed] [Google Scholar]
- 32.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuller R.S., Brake A., Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl Acad. Sci. USA. 1989;86:1434–1438. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jardim A., Liu W., Zheleznova E., Ullman B. Peroxisomal targeting signal-1 receptor protein PEX5 from Leishmania donovani. Molecular biochemical and. immunocytochemical. characterization. J. Biol. Chem. 2000;275:13637–13644. doi: 10.1074/jbc.275.18.13637. [DOI] [PubMed] [Google Scholar]
- 35.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.