Abstract
We have implemented a genome annotation system for prokaryotes called AGMIAL. Our approach embodies a number of key principles. First, expert manual annotators are seen as a critical component of the overall system; user interfaces were cyclically refined to satisfy their needs. Second, the overall process should be orchestrated in terms of a global annotation strategy; this facilitates coordination between a team of annotators and automatic data analysis. Third, the annotation strategy should allow progressive and incremental annotation from a time when only a few draft contigs are available, to when a final finished assembly is produced. The overall architecture employed is modular and extensible, being based on the W3 standard Web services framework. Specialized modules interact with two independent core modules that are used to annotate, respectively, genomic and protein sequences. AGMIAL is currently being used by several INRA laboratories to analyze genomes of bacteria relevant to the food-processing industry, and is distributed under an open source license.
INTRODUCTION
Around 10 years ago, the first prokaryotic genomes were sequenced and annotated (1). Each project represented a milestone for biology and was often carried out by a consortium of laboratories with substantial resources, including adequate bioinformatics support.
The subsequent decade has seen enormous advances in sequencing technologies, with the result that small teams within individual laboratories are now able to sequence their favorite prokaryotes. Information gleaned from such studies has accelerated the pace of both fundamental and applied biology. However, for many small laboratories, a bottleneck in their progress has been finding bioinformatics expertise to allow them to annotate their genomes of interest. It is now clear that the highest quality annotation arises from manual annotation by experts in the particular organism. So, without bioinformatics support available, one of the key roles of an annotation system is to enable expert biologists to annotate raw genomic data themselves.
Genome annotation is a complex process and involves a number of different dimensions. A substantial aspect is simply coherent data management. Any sequencing project will result in the large numbers of contigs being sequenced and combined, over time, into different assembled versions of the genome. Each assembly requires the application of large numbers of specialist bioinformatics tools, resulting in information about different regions of the genome or proteins expressed therein. The management of such data throughout this complex process is daunting; it is essential that such details are automatically handled by the annotation system.
After basic data management, a second aspect is the overall interpretation of bioinformatics results to form manual annotation. Generally, the expert biologist needs to examine all the automatic analysis and, combined with his/her own knowledge of the organism, form an overall decision on the nature of the different genomic elements, such as genes. This is where bioinformatics expertise is often required, interpreting the results, understanding appropriate thresholds for the different scores, etc. Without bioinformatics support available, it is essential that the analysis system facilitates the annotator to interpret the different analysis results.
Yet another factor to consider is the rapid progress that is also being made in the field of bioinformatics. New tools and databases are constantly under development leading to more accurate predictions. Often the incorporation of complete systems would help annotation, e.g. the inclusion of information about genomic sequences overlaid onto KEGG metabolic pathways (which was incorporated into the current system after initial deployment). Annotation systems which are unable to evolve, possibly due to having a fixed architecture or inflexible annotation strategy, will result in annotation which becomes inferior over time.
Considering all these different aspects, it is not surprising that small sequencing teams, without locally available bioinformatics expertise, are left struggling. It is with these objectives in mind, that the AGMIAL consortium [AGMIAL is a French acronym for Analyse de Génomes Microbiens d'Intérêt Agro-aLimentaire], consisting of a bioinformatics group collaborating with a number of small sequencing teams, developed the AGMIAL system to satisfy these particular needs.
This article is organized into three parts. We first describe a suitable annotation strategy for prokaryotes. An overview of existing annotation platforms is then provided, based on key features of interest. The AGMIAL system is then described as a practical system, focusing on the aspects above and also drawing together the best ideas from these other annotation systems.
ANNOTATION STRATEGY
Annotation, taken in its broadest sense, is the process of extracting biological knowledge from rather cryptic raw data, the nucleotide sequence. Annotation is a complex task that requires the integration of many data sources, such as the results from bioinformatics analysis tools, data extracted from generic and specific databases, biological knowledge accumulated in the literature over the years and results of genome-wide experiments, such as transcriptomics or proteomics experiments.
Two stages can be identified in the annotation of genomic data (2). The first stage corresponds to a static view of the genome where one describes the fundamental objects that it contains: the genes, their associated cis-regulatory regions and the proteins. This stage is necessary but not sufficient to obtain an in depth understanding of the complex relationship between the genome and the biological properties. A more dynamic view must be adopted, i.e. one must consider the multiple ways for genes and proteins to interact so as to create ‘functional modules’ (metabolic paths, signaling cascades, regulatory loops, etc.), which underlie the biological properties. This second stage constitutes a real challenge that the biology community is beginning to address (3). Transcriptomics and proteomics technologies, together with other high-throughput approaches, are playing a vital role in gaining this systems-level understanding of biology.
The first stage, being the foundation of subsequent analyzes, needs to be addressed properly. With the rapid increase of available genomic data, and the development of new bioinformatics methods to analyze these data a great deal of information relevant to the annotation can be extracted automatically. We think that it is important to go beyond the usual variants of the ‘best BLAST hit’ strategy and provide the annotators with the most diverse and comprehensive set of data regarding the genes and proteins to be annotated. In the next section we present the different types of information and the corresponding tools we consider pertinent for the annotation process and we have included in our annotation system.
Identification of genes and other genetic elements
Features of interest in the DNA sequence consist of genes coding for proteins or various types of RNA (tRNA, rRNA, etc.), ribosome binding sites (RBS), terminators, insertion sequences, specific signals (e.g. the CHI site), promoter regions, horizontally transferred regions, repetitions, etc.
A number of bioinformatics tools have been developed to identify these features, in particular much effort has been devoted to gene detection. In prokaryotes, coding sequences represent about 90% of the genome and are not split into exons/introns. These two characteristics allow the development of methods, based for instance on Hidden Markov Models (HMM), that provide very accurate results.
Overall, we think that the detection of genes and other genetic elements for prokaryotes is relatively straightforward, using current tools, with the possible exception of promoters. The subsequent ‘Contig Analysis Manager’ (CAM) section provides a brief description of the software used within Agmial to identify genetic elements.
Protein functional annotation
The notion of function
Compared to actually identifying a gene, assigning a function to its product is a more challenging task. To begin with, the concept of function is hierarchical, it needs to be described at different levels (4):
the molecular function that describes the biochemical role of the protein, whether it is a particular enzyme, transporter, repressor, structural protein, etc.
the cellular function that describes the role of the protein in the cell, e.g. whether it is involved in a particular pathway, a signaling cascade, etc.
the phenotypic function that describes the effect of the protein on general properties of the organism, e.g. if it is involved in the bacterium gliding ability, in the sporulation process, etc.
In addition, a number of proteins have been shown to possess multiple functions within the cell [the so-called moonlighting proteins (5)]. For instance in Escherichia coli a protein which, as a monomer, has a dihydrolipoamide dehydrogenase activity is also found as a subunit of pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase and the glycine cleavage complex (6).
It is also well known that many proteins consist of several domains that have molecular functions of their own. For instance the genetically mobile SH2 domain binds phosphorylated tyrosines. It is found in proteins that needs to recognize other phosphorylated proteins, for instance within a signaling cascade.
Modular aspect and intrinsic properties of protein sequences
The analysis of a protein sequence should always start with the determination of its underlying substructure, i.e. the identification of the different modules it is made of. This is important for two reasons. The first one is that some regions, such as low complexity regions, can alter subsequent homology searches by causing spurious resemblances between unrelated proteins. The second reason is that ignoring the modular aspect of proteins is the cause of a well known annotation error whereby the function of a protein is transferred to another one that only shares one module, not related to the general function of the protein (7).
One important type of protein module is the globular domain, although a number of other types also exist. These include transmembrane segments and signal peptides, together with regions of structural disorder, low complexity and coiled-coil. A number of databases and tools exist to predict the presence of globular domains, such as genetically mobile domains (8).
Besides modular aspects of proteins, other global properties, such as the molecular mass and isoelectric point are important, particularly in relation to proteomics studies. Protein properties that can be deduced from codon usage are also informative, for instance those concerning protein abundance and whether the gene is in an atypical region of the genome, possibly indicating horizontal transfer.
Homology search
Homology search techniques are the cornerstone of functional annotation. It is well known that methods based on the comparison of a single sequence, such as BLAST (9) or FASTA (10) become inefficient when they reach the ‘twilight zone’, about 25–30% sequence identity. Remote homologue detection can be improved if one uses multiple sequence alignments, either building them on the fly from the query sequence like PSI-BLAST (11) or employing protein family alignments (12) to create a statistical model representative of the family with a HMM.
For remote homologues whose sequences have strongly diverged, and can no longer be detected by sequence comparison techniques, it is possible to search for motifs or functional signatures (13). This is a powerful technique but it requires the motif residues to be more or less contiguous in the sequence. If this is not the case, one can use fold recognition techniques that are based on 3D structure conservation property, to detect remote homologues (14).
Genomic context information
The techniques discussed above consider proteins as isolated entities. With the availability of an increasing number of complete genomes, it now seems appropriate to consider the context of a gene between different genomes to help elucidate its function (15).
Techniques based on the genomic context use the co-localization of genes at various levels of physical proximity. They can be used to obtain information about protein function from chromosomal context but, in addition, they can also provide clues about the functional interactions between proteins thereby providing a first step towards cellular process annotation. Three major types of technique exist: gene fusion, gene neighboring and phylogenetic profiles.
Genomic context techniques provide links, different from the link provided by the homology relationship, between proteins of the genome. When this information is combined with data coming from homology search techniques it permits one to gain insight about protein function. It must be noted that this information is far from being marginal. It has been shown for E.coli K-12 that genomic context techniques allow one to obtain information for a fraction of genes in the genome similar to the fraction for which homology relationship can be found by sequence comparison methods (16).
Subcellular localization
The subcellular localization is an important practical piece of information, in particular in view of subsequent experiments with the organisms. Four localizations can be defined for Gram positive bacteria: cytoplasm, cytoplasmic membrane, cell wall and exterior (for secreted proteins) and five localizations for Gram negative bacteria: cytoplasm, cytoplasmic membrane, periplasm, outer membrane and exterior.
Different techniques are available to predict protein localization. The most straightforward is based on homology. If a protein is homologous to a protein whose localization is known one simply assumes it has the same localization. The second technique involves the identification of the biological mechanism responsible for the addressing of the protein to its localization, for instance signal peptides for secreted proteins, segments characterized by a high content in apolar residues for membrane proteins. The last technique is based on the amino acid composition of protein sequences. This composition shows a slight but detectable bias according to the localization. These techniques can be combined and weighted accordingly to improve the overall localization prediction (17).
Cellular process annotation
Relatively few bioinformatics tools are available to help biologists in this second stage of the annotation. As we mentioned above, genomic context techniques can be considered as a first step towards the study of protein interactions in the cell. High level functional modules must be studied with a number of large scale experiments. It is therefore important to facilitate the integration of annotation data and functional genomics databases.
EXISTING ANNOTATION PLATFORMS
The desirable features of an annotation platform as listed in the introduction lead to the following characteristics for the developed tool:
technical points must not concern human experts, in particular the implementation of the annotation strategy must be fully automated;
human interaction with the results provided by the system must be made as easy as possible.
The latter point can be best implemented through the use of interactive graphic interfaces. Graphic interfaces must allow the visualization of different features at the DNA or protein level, make the bioinformatics analysis results easy to consult. They must provide powerful and flexible ways to manipulate the underlying mechanisms used to query and combine the data, results and annotations, and keep a log of the modifications carried out on the annotations. In addition several annotators must be able to work in parallel on the same data.
We consider that the term ‘annotation’ should be understood in its broadest meaning, as described in the previous section. This requires the implementation and maintenance of a comprehensive annotation strategy, permitting one to extract as much relevant information as possible from the available data. As a consequence, it must be easy to integrate new or improved bioinformatics tools and databases into the system when required, creating a ‘federation’ of tools cooperating together for the purpose of annotating new genomes. The resulting system must be highly modular and robust, based on well tested computer science technologies.
To cope well with most sequencing projects the system must be able to work with draft sequences, in particular it must carry forward, automatically, manual annotations from the previous batch to the new one.
Finally, we firmly believe in the value of open source developments for promoting bioinformatics research in the community. We intend to distribute our system under a GNU Public License and so we only wish to integrate open source, or freely distributed, software into the system.
When we started the AGMIAL project we carried out an analysis of the most salient features of the tools then available. Since then a number of new systems have been developed. Table 1 presents an overview of some of these systems. The list is not meant to be exhaustive, yet, representative of the different features of the tools developed (note that in this table we do not consider systems developed by commercial companies).
Table 1.
Characteristics of some annotation platforms
| Method | Reference | Organisms | Graphic interface | Automatic processing of data | Manual annotation | Collaborative annotation | ‘Reasoning’ capabilities | Annotation assessment | Availability |
|---|---|---|---|---|---|---|---|---|---|
| MAGPIE | (40) | Prokaryote | No | Yes | Limited | No | Yes | No | Code available |
| GENOTATOR | (41) | Eukaryote | Yes | Yes | possible | No | No | No | Code available |
| GAIA | (42) | Eukaryote | Yes | Yes | Limited | No | No | No | Web use |
| IMAGENE | (43) | Prokaryote | Yes | Possible | Yes | No | No | No | Availableb |
| GENEQUIZ | (44) | Botha | No | Yes | No | No | Yes | Yes | Web use |
| ARTEMIS | (24) | Both | Yes | No | Yes | No | No | No | GPLc |
| M-AGENTS | (45) | Virus | No | Yes | No | No | No | No | Web use |
| PEDANT | (46) | Both | Yes | Yes | Possible | No | No | No | Web use |
| ENSEMBL | (47) | Both | Nod | Yes | Noe | Yes | No | No | GPL |
| APOLLO | (48) | Both | Yes | No | Limited | Yes | No | No | GPL |
| OTTER | (49) | Both | Yes | No | Yes | Yes | No | No | Code available |
| RICEGAAS | (50) | Eukaryote | Yes | Yes | Limited | No | No | No | Web use |
| GENQUIRE | (51) | Both | Yes | No | Yes | No | No | No | GPL |
| ATUGC | (52) | Prokaryote | No | Yes | No | No | Yes | No | No |
| GENDB | (53) | Prokaryote | Yes | Yes | Yes | No | Yes | No | GPL |
| ASAP | (54) | Both | Yes | No | Yes | Yes | No | No | Web use |
| SABIA | (55) | Prokaryote | No | Yes | No | No | No | No | Code available |
| MANATEE | (see notef) | Both, virus | Yes | Yes | Yes | No | No | No | GPL |
| MAGE | (56) | Prokaryote | Yes | Yes | Yes | Yes | No | No | Web use |
| AGMIAL | Prokaryotes | Yes | Yes | Yes | Yes | No | No | GPL |
See the text for a detailed definition of the column headings.
aGeneQuiz carries out protein sequence analysis only. It has been mostly used to re-annotate prokaryotic genome.
bRequires ILOG licensed libraries.
cGNU public license.
dProvided by APOLLO.
ePerformed with OTTER.
This is not the place to give a detailed analysis of the characteristics of all these systems. In particular, they clearly differ both in the technical and conceptual solutions that have been adopted by the developers. Conceptual solutions concern, for instance, the representation of the biological data and the architecture underlying the system: pipeline, workflow, multi-agent system, various types of interacting ‘layers’, etc. Although conceptual solutions do have an influence on the system capabilities, in the following, we just restrict ourselves to the description of those features that are directly pertinent to the annotators, along the lines described at the beginning of this section.
Features of interest for our purpose are the following. Manual annotation indicates whether the platform is designed to allow human experts to validate the results and provide the final annotation. Automatic processing of data refers to the capability of the platform to carry out bioinformatics analysis without human intervention. Graphics interface indicates whether the interaction between the annotators and the system takes place mostly through the use of a graphical interface, in other words, whether the interface is really central to the process of manual annotation. Thus, the graphical interface must allow the user to inspect the nucleic sequence and associated features, as well as various results coming from bioinformatic tools used during the analysis. This interface must be interactive, allowing the annotator to modify the data presented, to add new information and to provide mechanisms to perform various searches and comparisons. Collaborative annotation refers to the possibility for several teams of annotators to work on the same genome and possibly, when the genome is published, for biologists browsing the data to add new information or propose corrections. Reasoning capabilities refers to the ability of the system to analyze automatically the results produced by the different bioinformatics tools and predict a particular function in consequence. This criterion might appear in contradiction with the emphasis we put on the central role of the human expert during the annotation process. In fact, as we will discuss later, these ‘reasoning’ capabilities are not intended to replace human experts but to assist them in their task. Assessment of annotation describes whether the platform provides some means of quantifying the confidence the annotators have in their annotation. Availability specifies the distribution status of the tool.
Not all the tools described in Table 1 are annotation platforms according to conventional meaning of term. GENQUIRE is an annotation browser/editor, GENEQUIZ is a tool exclusively devoted to protein analysis, APOLLO and OTTER are subsystems of ENSEMBL permitting respectively to browse/visualize DNA features and to manually annotate. The term ‘graphic interface’ covers different realities: from static Web pages (MAGPIE and GENEQUIZ), to interactive applets (GAIA) upto complete, independent, applications (IMAGENE, ARTEMIS, GENQUIRE and APOLLO). These different systems first introduced some of the key features which are described above. MAGPIE was built around two PROLOG daemons allowing some reasoning on the data to be performed. GENEQUIZ generalized this feature with its module GQreason, it was also the first system to provide an assessment of the annotation accuracy. GENOTATOR was the first platform to provide a graphical browser and to stress the importance of visualization. IMAGENE introduced the notion of ‘strategy’, i.e. a series of elementary tasks strung together to accomplish a particular goal, (such as predicting CDS) and one of the strengths of this system was to allow the user to define, build and manipulate these strategies. More recently, genomic institutes have integrated a number of these features into comprehensive annotation platforms, e.g. PEDANT (Institute for Bioinformatics, MIPS), ENSEMBL (Sanger Centre), MANATEE (TIGR) and GENDB (Center for Genome Research).
To summarize Table 1, differences between platforms lie in the way genomic data are processed, the spectrum going from completely automatic systems, such as GENEQUIZ to annotation browser/editors, such as GENQUIRE, the relative weighting between protein-oriented versus DNA-oriented analysis, the annotation strategy implemented through the set of bioinformatic tools used, the emphasis put on the role of human experts and collaborative annotation, and the use or not of some formal representation of biological knowledge (ontology/hierarchical classification of functions).
Several classifications are currently used for (microbial) genome annotation, for instance Riley's functional hierarchy (18), Gene Ontology (19) and MIPS Functional Catalogue (20). It is very important, if one wants to fully benefit from the accumulated data, that a broad agreement emerges from the community regarding the description of biological concepts. From a practical viewpoint, the use of a functional classification enforces the use of a controlled vocabulary that facilitates the comparisons between different annotated genomes.
AGMIAL PLATFORM
Overview
Key to the system are two managers called the protein analysis manager (PAM) and the CAM. The PAM has overall responsibility for managing and analyzing proteins. The CAM is more specialized and is responsible for managing, analyzing and consistently updating batches of assembled contigs within a particular genome sequencing project. Both are independent components, which are complete applications in themselves. They engage in a long term commitment in notifying each other of changes in their respective views of data thus cooperating in the overall task of genome annotation.
Both managers share the same internal architecture. They provide:
a web interface so users can administrate projects and manually annotate;
a suite of bioinformatics methods to analyze the data;
an underlying relational database for storing proteins or contigs respectively, results of analysis tools and annotations;
a mechanism allowing managers to communicate and exchange relevant data when required.
More specific detail of both managers is given in the following sections.
The CAM
Data input and output
Currently the platform is geared toward annotating new genomes and contig sequence data are imported as nucleotide Fasta format files. For re-annotation, where one needs to compare the old annotation with the updated one, we have included a data import mechanism from EMBL/GenBank format files. To publish the data, which requires it to be deposited in public collections, such as GenBank or EMBL, the system outputs in GenBank/EMBL format files, or tabulated flat files used as input by SEQUIN (for submission at genomic centers).
Bioinformatics tools
Bioinformatics tools that have been incorporated in the CAM are listed in Table 2.
Table 2.
Bioinformatics tools integrated into the CAM
| Method | Description | Reference |
|---|---|---|
| SHOW | Gene detection using a hidden Markov models | http://www-mig.jouy.inra.fr/ssb/SHOW/ |
| tRNAScan | Detection of tRNA sequences | (22) |
| rRNAScan | Detection of rRNA sequences. | |
| PETRIN | Detection of terminator sequences | (23) |
For gene detection and classification we use an in-house system called SHOW (Sequence HOmogeneity Watcher) that is based on HMMs. Gene classification relies on a partitioning of the DNA sequence into regions having a homogeneous composition in words of variable length (21). In the HMM, the detection of genes takes into account: (i) the start and stop codons, (ii) the phased composition of the coding sequence, (iii) the presence of a RBS upstream of a gene and (iv) the possibility for genes to overlap. Parameters of the model are estimated automatically from the DNA sequence to be analyzed, the method does not need special training sets, it adapts itself to the corresponding genomic data. One feature of this approach is that the use of fixed parameters is minimized, such as minimum allowed gene length. The system is thus less biased by arbitrary cut-offs and, for instance, is able to detect very short genes. Some of these putative short genes are interesting and we have on-going collaborations to verify them experimentally. Tests of the method on 839 genes of E.coli whose products are experimentally known (EcoGene dataset) have shown that SHOW is able to correctly identify the precise limits of 93% of these genes.
Other genetic elements which are determined include tRNAs, rRNAs and terminator sequences. Genes coding for tRNAs are predicted with tRNAscan (22). Ribosomal nucleic acid sequences are detected using an in-house system called rRNAscan, which essentially does a BLASTN (9) search against a database containing known rRNA sequences. Finally, terminator sequences are detected using PETRIN (23).
The results of each analysis method are represented in terms of nucleic acid features, similar to GenBank features but with extended qualifier lists to cater for data management and provenance. Annotators are free to define their own features and qualifiers to better describe the characteristics of the nucleic sequence, if they feel the need. However, these user-defined features and qualifiers will not be included in the canonical GenBank/EMBL file produced for genome publication.
The translations for coding regions, or CDS features, are automatically generated using the appropriate genetic code. CAM then sends these translated proteins to PAM, whereupon they are automatically analyzed and an overall functional description is established (see ‘The PAM’ section below).
An important aspect which the CAM needs to handle is when a newly assembled batch of contigs is added to the system. Manually edited annotations from the previous batch are automatically forwarded to the new batch. This allows the user to start a project with an unfinished sequence and to have the different annotations transmitted to new batches.
User interface
Annotators interact with the system exclusively through graphic interfaces. These interfaces provide mechanisms to retrieve and visualize the data stored in the databases, to carry out complex searches on the data and, if required, to edit these data.
Currently CAM has three different graphic interfaces that provide views of genomic data at different scales. The first one is an Artemis client allowing annotators to visualize features on the DNA sequence. Artemis is a fully fledged genome analysis and annotation editor developed at the Sanger Centre (24). We have interfaced it with CAM, permitting the annotator to use its functionalities. For instance it can be employed to browse and to edit parts of contigs within a particular project.
In addition to Artemis, we have recently developed a new graphical interface based on an in-house software package called MuGeN (25) (see Figure 1). Our interest in MuGeN stems from the fact that it is a tool built for navigating through multiple annotated genomes. It thus facilitates cross-genome comparison. In particular for genome re-annotation, it allows the new annotation to be compared side-by-side against the original annotation.
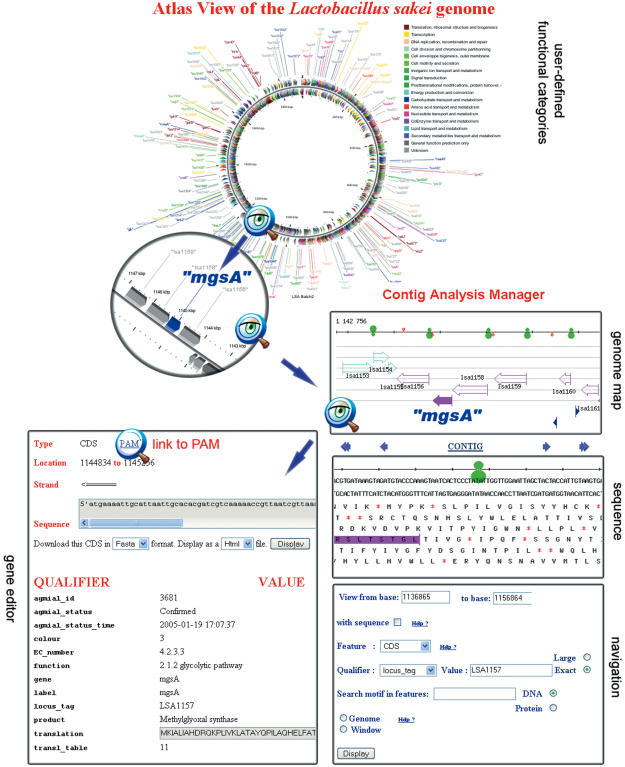
Figure 1.
Views of the DNA sequence at different scales. The upper part of the figure represents an atlas view of the genome obtained with CGView. One can zoom on a particular region of this map, for instance on the area containing the mgsA gene: a methylglyoxal synthase that belongs to the glycolytic pathway. Clicking on this gene will open the MuGeN interface showing its genomic context (genome map frame). It is possible to zoom on this representation to see the DNA sequence and the translation in the six reading frames (sequence frame). The green symbol represents the RBS, the gene sequence is colored as in the previous view. The navigation window allows one to move along the genome, either by entering a range of base numbers, or by looking for a feature with a particular qualifier or by specifying a DNA or protein motif to be searched for in the current window or in the complete genome. The window at the lower left of the figure shows the gene editor. Most fields are automatically filled, in particular the gene annotation qualifiers since, in general, CDS annotation is performed in PAM and then updated in CAM (see Figure 4). Clicking on the link to PAM, indicated by the magnifying glass, will lead the annotator to the PAM interface shown in Figure 2. For clarity the Artemis interface is not shown on the figure.
The last interface consists of the visualization software CGView (26). This software generates high quality, zoom-able maps of circular genomes. It provides very useful views able to summarize various features and properties of the complete genome. These views are particularly suitable as illustrations in publications. We have interfaced CGView with CAM so that it can extract data from the relational database and present it in a circular context (see Figure 1).
The PAM
Data input and output
The PAM can be used, independently of the CAM, to analyze a batch of protein sequences. In this configuration, it imports protein sequences as Fasta format files. As part of the annotation platform, translated protein sequences are directly provided by the CAM using the exchange mechanism between the managers. Annotated proteins can be exported as Uniprot format files.
Bioinformatics tools
We have integrated into PAM a number of tools required to implement the annotation strategy described previously (Table 3).
Table 3.
Bioinformatics tools integrated into the PAM
| Method | Description | Reference |
|---|---|---|
| Methods to determine sequence intrinsic properties | ||
| pI | Isoelectric point and molecular mass | |
| SEG | Detection of low-complexity regions | (27) |
| COIL | Detection of coiled-coil structures | (29) |
| SIGSEQ | Detection of signal peptides | (30) |
| MEMSAT | Transmembrane segment prediction | (28) |
| Homology search methods | ||
| RPS-BLAST | Reverse position specific BLAST | |
| PSI-BLAST | Sequenced-based homology search | (11) |
| FROST | Fold recognition method for detecting remote homologues | (14) |
| Miscellaneous methods | ||
| PSORTb | Prediction of subcellular localization | (17) |
| Jalview | Multiple sequence alignment editor | (35) |
| InterProScan | Integrated protein motif detection, including the following software: | (31) |
| Motif and functional signature detection methods | ||
| ProfileScan | Find motifs using profiles | (57) |
| ScanRegExp | Find motifs using regular expressions | (58) |
| FPrintScan | Find multiple motifs | (59) |
| Domain detection using HMM methods | ||
| HMMSmart | Genetically mobile domains from SMART | (8) |
| HMMPfam | Protein family domains from PFAM | (12) |
| HMMTigr | Protein families from TIGR institute | (32) |
| HMMPIR | Protein families from PIR | (60) |
| HMMPanther | Protein families subdivided into functionally related subfamilies | (61) |
| SuperFamily | Proteins of known 3D structure | (62) |
| Gene3D | Protein families in complete genomes | (63) |
| Method to split protein sequences into domains | ||
| BlastProDom | Defines domains in protein sequences | (64) |
| Methods to find sequence intrinsic properties | ||
| SignalPHMM | Prediction of signal peptide | (65) |
| TMHMM | Prediction of transmembrane helices in proteins | (66) |
The first set of tools concerns the partitioning of protein sequences into domains. Non globular domains exhibiting a particular composition in amino acids, such as low complexity region (27), transmembrane segments (28), disordered regions or having some internal organization, such as coiled-coils regions (29), signal peptides (30) are searched for. Globular domains are detected using tools integrated in InterProScan (31).
The second set of tools concerns homology searches. Methods to detect protein family domains in InterProScan, such as PFAM (12) or TIGRFAMS (32), belong to this category. We use PSI-BLAST (11) with a number of databases. Some are generic, such as SWISSPROT, and others are specific, for instance, collections of protein sequences belonging to related organisms chosen by the biologists in charge of the project. Protein sequences of the genome are clustered into paralog families. We integrated the CDD collections (33), that makes use of RPS-BLAST, principally to have access to the cluster of orthologous genes (COG) (34) dataset. Multiple sequence alignments resulting from these analyses can be inspected and edited with Jalview (35) a Java multiple sequence editor. We perform a search for protein family signatures or motifs with PROSITE (13) and PRINTS (36) that are both available in InterProScan. Finally, the fold recognition method FROST (14), can be used to carry out a search for remote homologues.
Protein subcellular localization is predicted using PSORTb (17). PSORTb is based on a multiple classification approach employing different computation techniques to analyze various features: signal peptide, transmembrane helices, homology to proteins of known localization, amino acid composition and motifs.
We are currently in the process of integrating genomic context information. We have carried out a cross comparison of all available microbial genomes (268 at the time of writing) and compiled the required information to analyze gene fusion and gene neighborhood conservation. All the results are stored in a relational database with other relevant information. To allow biologists to fully, and easily, exploit these data we are presently designing an efficient user interface.
User interface
Results provided by the above tools are displayed as web pages that annotators can consult with their favorite web browser (see Figure 2). Results are organized in sections: general properties, homology results, feature results, paralog results, etc. The ordering and appearance of these sections can be parameterized by the user. Multiple cross references to the databases used by the different tools (see Table 4) and to the tools themselves exist allowing annotators to browse relevant information.
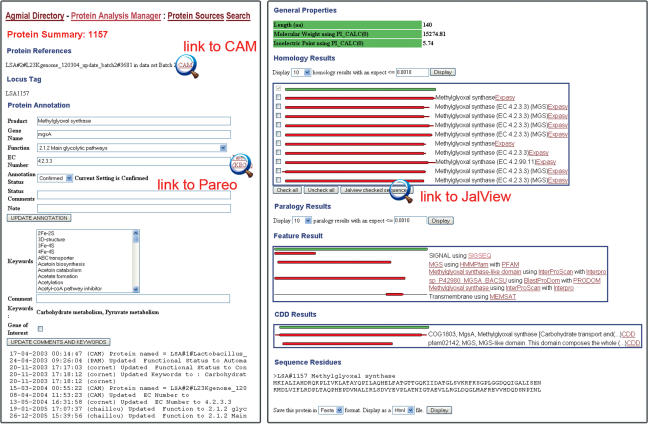
Figure 2.
The right part of the figure shows the results of the different bioinformatic methods applied to the sequence of MgsA. Not all result sections are shown here. In the ‘homology’ section, checking the boxes on the left of the homologous sequences and then clicking on the link to Jalview, below, will show the multiple alignment of the selected sequences and the corresponding phylogenetic tree. The left part of the figure shows the annotation window where the protein annotation is performed. Information entered in this section is forwarded to CAM, the system always makes sure that both managers are synchronized. The bottom of this window shows the annotation history. The link to CAM at the top will lead the annotator back to the CAM interface (MuGeN interface, see Figure 1). The link to PAREO (our relational version of the KEGG database) near the ‘EC number’ box will lead the user to the KEGG interface shown in Figure 3.
Table 4.
Databases used by the PAM tools
| Name | Description | Reference |
|---|---|---|
| UNIPROT | Protein sequences and functions | (67) |
| KEGG | Molecular interaction networks | (37) |
| CDD | Conserved domain database | (33) |
| PDB | 3D structures database | (68) |
| SCOP | Protein 3D domain database | (69) |
| PROSITE | Functional motifs and profiles derived from SWISSPROT | (70) |
| PRINTS | Manually derived functional motifs | (36) |
| SMART | Motifs of genetically mobile domains | (8) |
| PFAM | Protein family domains | (71) |
| TIGRFAMs | Similar to PFAM | (72) |
| SUPERFAMILY | Families of proteins of known 3D structures | (62) |
| GENE3D | Protein families and domain architectures in complete genomes | (63) |
| PANTHER | Protein families subdivided into functionally related subfamilies | (61) |
| PRODOM | Automatically generated protein domain families | (64) |
Homology search methods provide often a large number of homologous proteins. To unravel the complex evolutionary relationship between these proteins and the query protein it is useful to inspect the multiple sequence alignment and to consider the resulting phylogenetic tree. To carry out this task we have interfaced the Jalview multiple sequence editor that allows annotators to browse, and manipulate in a number of ways, the multiple alignment. In addition Jalview can draw the corresponding phylogenetic tree and also cluster the sequences using a principal component analysis technique. This provides a very effective tool for the annotators in their task of assigning a function to the query protein.
At the cellular level it is important to describe pathways responsible for cellular processes. To help annotators studying these pathways and the proteins involved therein, we have developed a graphical tool built on a relational model of the KEGG database (37). It allows annotators to automatically visualize genome proteins involved in specific metabolic pathways. When a related genome is available, this tool also permits an easy comparison of the proteins involved in the same pathways (see Figure 3). Using this tool it is straightforward, for a specific pathway, to identify proteins that are only present in one of the two genomes and proteins that are common to both. This feature allows annotators to quickly pinpoint, in the studied pathway, the major differences and similarities between the two organisms.
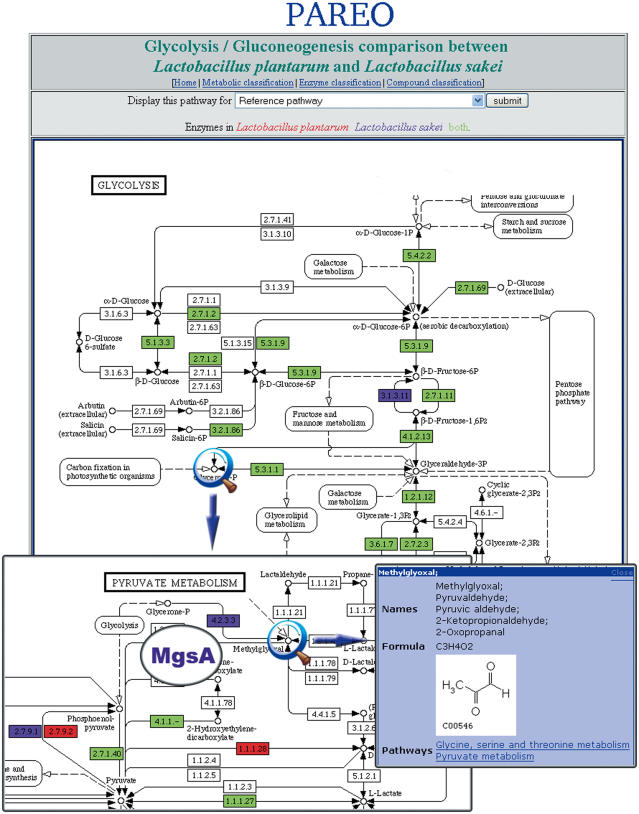
Figure 3.
This figure shows both glycolysis and pyruvate metabolism pathways for Lactobacillus plantarum and L.sakei. As indicated in the legend at the top of the figure, enzymes that are only found in L.plantarum and L.sakei are colored respectively in red and purple. Enzymes found in both organisms are colored in green. The magnifying glasses are used to indicate the role of MgsA in these pathways. This enzyme appears to be involved in a methylglyoxal bypass (reversible reaction) of glycolysis in L.sakei. The figure illustrates well the major difference in glycolysis in L.plantarum and L.sakei. The bottom right box shows the product of the reaction catalyzed by MgsA. A detailed account of L.sakei energy production pathways contributing to meat adaptation can be found in ref. (38).
System usage
The initial stage of the annotation process is entirely automatic. The annotation procedure starts when an authorized user loads in CAM the first batch of assembled contigs for the sequencing project, possibly consisting of hundreds of short contigs. As the genome sequencing and assembly continues, new batches of fewer, longer, contigs will be added to the project.
The contigs are automatically analyzed by the tools in CAM and the results, together with the initial data, are stored in the CAM relational database. CDS features are then translated using the appropriate genetic code and the corresponding protein sequences are sent to PAM whereupon they are automatically processed by the various tools and an overall function description is proposed when possible (for the time being, based on a very simple, automatic, analysis of the homologue list). The corresponding results and data are stored in the PAM relational database.
Human experts take part in the annotation process after the completion of the first stage. Using the interfaces described above they are able to consult the results, visualize the features on the DNA sequence, carry out various types of searches and edit the data stored in the database. In AGMIAL both the contig and protein views are integrated. It is thus possible, while examining genes within a particular DNA region, to switch to the PAM interface to consult the available information for the corresponding proteins. Naturally, the converse is also true, the annotator that examines results for a given protein can visualize the DNA region around the corresponding gene with a simple click of the mouse. In a similar fashion, the annotator, while browsing the general view of the genome provided by CGView, can also click on a particular gene to move to a detailed view of the DNA region around it, in MuGeN.
Genome proteins undergo a change of status during the annotation process. When CAM first transfer a protein sequence to PAM the status is set to ‘original’. Following the automatic analysis of the sequence, PAM may suggest a function for the protein whose status becomes ‘automatic’. After pondering the different results and data available, annotators may confirm the assigned function by changing the status of the prediction from ‘automatic’ to ‘confirmed’. Alternatively, they may decide to modify and update the functional description before changing it to ‘confirmed’.
It is important to note that both managers store every description and status change a protein goes through. So the detailed history of a protein's annotation is kept and displayed by the web interface. In this way PAM provides secure, persistent storage of collections of protein sequences within projects.
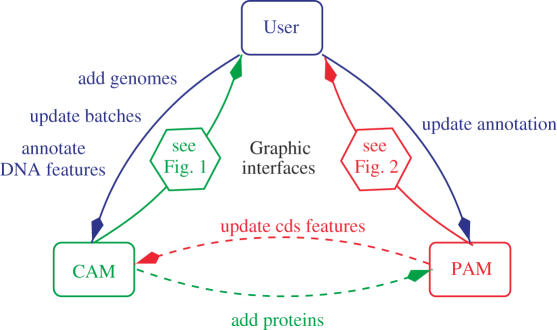
For annotators, it is critical that protein annotations in PAM and the corresponding CDSs in CAM are synchronized. In order to achieve this, both managers establish a dialog. Actions of annotators on the data on any manager automatically result in an exchange of information between this manager and its counterpart so the annotation is kept consistent and up-to-date on both sides. For instance, each time an annotator edits protein features using the PAM web interface, PAM sends the modifications to CAM which updates the corresponding CDS in its database. Similarly, whenever a user edits a CDS feature with CAM, e.g. modifying the start of a gene, CAM sends a message to PAM which updates its database accordingly for the corresponding protein. When the modification results in a new protein the whole battery of PAM tools is automatically applied to the new sequence. The old gene and protein are kept in the databases but marked as disabled. The interaction between the two managers and the user is shown in Figure 4.
Figure 4.
Dashed arrows represent automatic processes between managers, solid arrows represent human interaction with the managers. Graphic interfaces are described in Figures 1 and 2.
APPLICATIONS
Annotation tool
The AGMIAL system is currently being employed by several laboratories at INRA to annotate different genomes of interest, for instance, Lactobacillus sakei (38), Lactobacillus bulgaricus (39), Flavobacterium psychrophilum (submitted), Staphylococcus xylosus, Propionibacterium freudenreichii, Arthrobacter arilaitensis and the strains JIM8777 and JIM8780 of Streptococcus salivarius.
Re-annotation tool
Besides using the AGMIAL platform to annotate newly sequenced genomes, a number of INRA groups were interested in re-annotating already published genomes, in particular when different strains of some organism of interest, or closely related organisms, were known (e.g. Enterococcus faecalis, Enterococcus faecium, Lactococcus lactis, Bacteroides thetaiotaomicron, Streptococcus thermophilus). The fact that the genomes are processed by the same set of tools and stored under the same relational schema facilitates considerably the comparison of the different strains, or related organisms. It also provides a good framework for data mining techniques or other bioinformatics methods.
Genome database
Once the genomes are annotated or re-annotated the platform can be used as a model organism database. We identify two potential groups of users:
Biologists who will be able to visualize genome features, browse and retrieve corresponding annotations using the graphical interfaces provided.
Bioinformatics groups that can take advantage of the availability of the source code, the use of computer science open standards and of the modular architecture of the platform, to develop new plug in modules to analyze the data.
CONCLUSION
We have implemented a genome annotation system consisting of two distributed and independent components which cooperate, one managing protein sequences and the other managing contig sequences. This tool is currently deployed in a number of laboratories throughout INRA where it is used to annotate microbial genomes.
The general philosophy of the AGMIAL platform is that human experts are central to the process of annotation, the role of computers is to assist them in this complex task. Hence, the two aspects of the platform are, on one hand the automation of the maximum number of tasks that do not require human expertise, and on the other hand the strong emphasis put on the man-machine interface that is intended to help annotators to interact efficiently with the system.
From a computer science viewpoint, the system is built from an open community of distributed and independent components, which can cooperate together. It is thus highly modular, facilitating the integration of new tools. The components are written in Java. The system is based on well established standard computer science technologies (Web services, relational database management systems, Java, etc.) and only integrates open source software allowing us to distribute the platform freely under a GNU public license.
From a user's viewpoint, besides the characteristics mentioned above, the system has, we believe, several interesting features. It is able to handle draft sequences at various level of completion. It permits a collaborative annotation from members of a laboratory by letting them organize the annotation process as they wish and provides a history mechanisms. The latter permits the tracking of all changes occurring at the gene or protein level during the annotation process. The system enforces the use of a functional classification (that can be chosen by the team of biologists at the beginning of the project).
SYSTEM REQUIREMENTS
Being written in Java, the platform can be installed on machines running under different operating systems. So far, we have only tested the platform deployment on Linux machines or a cluster of Linux machines. On the other hand, annotators do not need to be concerned about the type of machine on which the platform is running since they only interact with the system through Web interfaces and Java applets. Applets are programs designed to be executed from within a Web browser that permit the design of dynamic Web pages, enhancing the interaction of the user with the system. Annotators can thus remain in their favorite environment (Windows, Mac OS and Linux) but still interact easily and transparently with the platform running on a remote server.
AVAILABILITY
A public version of the annotated genome of L.sakei can be browsed at the following URL: http://genome.jouy.inra.fr/sakei-agmial. A demo version (‘sandbox’) of the AGMIAL platform is available at the URL: http://genome.jouy.inra.fr/demo-agmial. The complete system, (i.e. the framework) can be downloaded at the URL: http://genome.jouy.inra.fr/agmial and installed locally under the GNU Public License.
Groups interested in annotating newly sequenced genomes with the AGMIAL platform but lacking the manpower to install it locally can contact J.-F. Gibrat (gibrat@jouy.inra.fr) to ask for their data to be analyzed and managed on our machines.
Acknowledgments
The authors are grateful to INRA for funding this work by an award of a Postdoctoral Fellowship to K.B. and R.B., and also to the European Union for awarding an Individual Marie-Curie Fellowship to K.B. The authors thank M. Zagorec for critical reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by INRA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kunst F., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., Bertero M.G., Bessieres P., et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 2.Stein L. Genome annotation: from sequence to biology. Nature Rev. Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- 3.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 4.Bork P., Dandekar T., Diaz-Lazcoz Y., Eisenhaber F., Huynen M., Yuan Y. Predicting function: from genes to genomes and back. J. Mol. Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- 5.Jefery C.J. Moonlighting proteins. Trends Biochem. Sci. 2000;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 6.Riley M. Genes and proteins in Escherichia coli k-12 (GenProtEC) Nucleic Acids Res. 1997;25:51–52. doi: 10.1093/nar/25.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galperin M.Y., Koonin E.V. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. [PubMed] [Google Scholar]
- 8.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic. Acids. Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Pearson W.R. Flexible sequence similarity searching with the FASTA3 program package. Meth. Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C.J., Hofmann K., Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marin A., Pothier J., Zimmermann K., Gibrat J.-F. FROST: a filterbased fold recognition method. Proteins. 2002;49:493–509. doi: 10.1002/prot.10231. [DOI] [PubMed] [Google Scholar]
- 15.Notebaart R.A., Huynen M.A., Teusink B., Siezen R.J., Snel B. Correlation between sequence conservation and the genomic context after gene duplication. Nucleic Acids Res. 2005;33:6164–6171. doi: 10.1093/nar/gki913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynen M.A., Snel B., Mering C., Bork P. Function prediction and protein networks. Curr. Opin. Cell. Biol. 2003;15:191–198. doi: 10.1016/s0955-0674(03)00009-7. [DOI] [PubMed] [Google Scholar]
- 17.Gardy J.L., Laird M.R., Chen F., Rey S., Walsh C.J., Ester M., Brinkman F.S. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 18.Riley M. Systems for categorizing functions of gene products. Curr. Opin. Struct. Biol. 1998;8:388–392. doi: 10.1016/s0959-440x(98)80074-2. [DOI] [PubMed] [Google Scholar]
- 19.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Guldener U., Mannhaupt G., Munsterkotter M., Mewes H.W. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas P., Bize L., Muri F., Hoebeke M., Rodolphe F., Ehrlich S.D., Prum B., Bessires P. Mining Bacillus subtilis chromosome heterogeneities using Hidden Markov Models. Nucleic Acids Res. 2002;30:1418–1426. doi: 10.1093/nar/30.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.d'Aubenton C.Y., Brody E., Thermes C. Prediction of rhoindependent Escherichia coli transcription terminators. a statistical analysis of their RNA stem–loop structures. J. Mol. Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 24.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 25.Hoebeke M., Nicolas P., Bessieres P. MuGeN: simultaneous exploration of multiple genomes and computer analysis results. Bioinformatics. 2003;19:859–864. doi: 10.1093/bioinformatics/btg101. [DOI] [PubMed] [Google Scholar]
- 26.Stothard P., Wishart D.S. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 27.Wootton J.C. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 28.Jones D.T., Taylor W.R., Thornton J.M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 29.Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 30.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zdobnov E.M., Apweiler R. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 1986;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 32.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer-Marchler A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 36.Attwood T.K., Bradley P., Flower D.R., Gaulton A., Maudling N., Mitchell A.L., Moulton G., Nordle A., Paine K., Taylor P. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaillou S., Champomier-Vergs M.-C., Cornet M., Crutz-Le Coq A.-M., Dudez A.-M., Martin V., Beaufils S., Darbon-Rongre E., Bossy R., Loux V. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23k. Nat. Biotechnol. 2005;23:1527–1533. doi: 10.1038/nbt1160. [DOI] [PubMed] [Google Scholar]
- 39.van de Guchte M., Penaud S., Grimaldi C., Barbe V., Bryson K., Nicolas P., Robert C., Oztas S., Mangenot S., Couloux A., et al. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl Acad. Sci., USA. 2006;103:9274–9279. doi: 10.1073/pnas.0603024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaasterland T., Sensen C.W. Fully automated genome analysis that reflects user needs and preferences. a detailed introduction to the MAGPIE system architecture. Biochimie. 1996;78:302–310. doi: 10.1016/0300-9084(96)84761-4. [DOI] [PubMed] [Google Scholar]
- 41.Harris N.L. Genotator: a workbench for sequence annotation. Genome Res. 1997;7:754–762. doi: 10.1101/gr.7.7.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey L.C., Fischer S., Schug J., Crabtree J., Gibson M., Overton G.C. GAIA: framework annotation of genomic sequence. Genome Res. 1998;8:234–250. doi: 10.1101/gr.8.3.234. [DOI] [PubMed] [Google Scholar]
- 43.Medigue C., Rechenmann F., Danchin A., Viari A. Imagene: an integrated computer environment for sequence annotation and analysis. Bioinformatics. 1999;15:2–15. doi: 10.1093/bioinformatics/15.1.2. [DOI] [PubMed] [Google Scholar]
- 44.Andrade M.A., Brown N.P., Leroy C., Hoersch S., de Daruvar A., Reich C., Franchini A., Tamames J., Valencia A., Ouzounis C. Automated genome sequence analysis and annotation. Bioinformatics. 1999;15:391–412. doi: 10.1093/bioinformatics/15.5.391. [DOI] [PubMed] [Google Scholar]
- 45.Decker K., Zheng X., Schmidt C. A multi-agent system for automated genomic annotation. Proceedings of the fifth international conference on Autonomous agents; ACM Press; 2001. pp. 433–440. [Google Scholar]
- 46.Frishman D., Albermann K., Hani J., Heumann K., Metanomski A., Zollner A., Mewes H.W. Functional and structural genomics using PEDANT. Bioinformatics. 2001;17:44–57. doi: 10.1093/bioinformatics/17.1.44. [DOI] [PubMed] [Google Scholar]
- 47.Hubbard T., Barker D., Birney E., Cameron G., Chen L., Clark Y., Cox T., Cu J., Curwen V., Down T. The ENSEMBL genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis S.E., Searle S.M., Harris N., Gibson M., Lyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A. Apollo: a sequence annotation editor. Genome Biol. 2002;3:1–14. doi: 10.1186/gb-2002-3-12-research0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Searle S.M., Gilbert J., Iyer V., Clamp M. The OTTER annotation system. Genome Res. 2004;14:963–970. doi: 10.1101/gr.1864804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakata K., Nagamura Y., Numa H., Antonio B.A., Nagasaki H., Idonuma A., Watanabe W., Shimizu Y., Horiuchi I., Matsumoto T., Sasaki T., Higo K. RiceGAAS: an automated annotation system and database for rice genome sequence. Nucleic Acids Res. 2002;30:98–102. doi: 10.1093/nar/30.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson M.D., Block D., Crosby W.L. Genquire: genome annotation browser/editor. Bioinformatics. 2002;18:1398–1399. doi: 10.1093/bioinformatics/18.10.1398. [DOI] [PubMed] [Google Scholar]
- 52.Bazzan A.L.C., Duarte R., Pitinga A.N., Schroeder L.F., Souto F.D.A., Ceroni da Silva S. ATUGC—an agent-based environment for automatic annotation of genomes. International Journal of Cooperative Information Systems. 2003;12:241–273. [Google Scholar]
- 53.Meyer F., Goesmann A., McHardy A.C., Bartels D., Bekel T., Clausen J., Kalinowski J., Linke B., Rupp O., Giegerich R. GenDB-an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glasner J.D., Liss P., Plunkett G., Darling A., Prasad T., Rusch M., Byrnes A., Gilson M., Biehl B., Blattner F.R. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 2002;31:147–151. doi: 10.1093/nar/gkg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Almeida L.G.P., Paixao R., Souza R.C., da Costa G.C., Barrientos F.J., dos Santos M.T., de Almeida D.F., Vasconcelos A.T.R. A system for automated bacterial (genome) integrated annotation-SABIA. Bioinformatics. 2004;20:2832–2833. doi: 10.1093/bioinformatics/bth273. [DOI] [PubMed] [Google Scholar]
- 56.Vallenet D., Labarre L., Rouy Z., Barbe V., Bocs S., Cruveiller S., Lajus A., Pascal G., Scarpelli C., Medigue C. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gattiker A., Gasteiger E., Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- 58.Sigrist C.J.A., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M., Bairoch A., Bucher P. PROSITE: a documented data base using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 59.Scordis P., Flower D.R., Attwood T.K. FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics. 1999;15:799–806. doi: 10.1093/bioinformatics/15.10.799. [DOI] [PubMed] [Google Scholar]
- 60.Wu C.H., Nikolskaya A., Huang H., Yeh L.S., Natale D.A., Vinayaka C.R., Hu Z.Z., Mazumder R., Kumar S., Kourtesis P. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergri J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of Hidden Markov Models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 63.Lee D., Grant A., Marsden R.L., Orengo C. Identification and distribution of protein families in 120 completed genomes using Gene3D. Proteins. 2005;59:603–615. doi: 10.1002/prot.20409. [DOI] [PubMed] [Google Scholar]
- 64.Bru C., Courcelle E., Carrere S., Beausse Y., Dalmar S., Kahn D. The ProDom database of protein domain families: more emphasis on 3d. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dyrlov Bendtsen J., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 66.Moller S., Croning M.D.R., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 67.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deshpande N., Addess K.J., Bluhm W.F., Merino-Ott J.C., Townsend-Merino W., Zhang Q., Knezevich C., Xie L., Chen L., Feng L. The RCSB protein data bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. SCOP database in 2004: re.nements integrate structure and sequence family data. Nucleic Acids. Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hulo N., Sigrist C.J., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths J., Khanna A., Marshall M., Moxon S., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–337. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]