Abstract
More than 90% of human genes are rich in intronic latent 5′ splice sites whose utilization in pre-mRNA splicing would introduce in-frame stop codons into the resultant mRNAs. We have therefore hypothesized that suppression of splicing (SOS) at latent 5′ splice sites regulates alternative 5′ splice site selection in a way that prevents the production of toxic nonsense mRNAs and verified this idea by showing that the removal of such in-frame stop codons is sufficient to activate latent splicing. Splicing control by SOS requires recognition of the mRNA reading frame, presumably recognizing the start codon sequence. Here we show that AUG sequences are indeed essential for SOS. Although protein translation does not seem to be required for SOS, the first AUG is shown here to be necessary but not sufficient. We further show that latent splicing can be elicited upon treatment with pactamycin—a drug known to block translation by its ability to recognize an RNA fold—but not by treatment with other drugs that inhibit translation through other mechanisms. The effect of pactamycin on SOS is dependent neither on steady-state translation nor on the pioneer round of translation. This effect is found for both transfected and endogenous genes, indicating that SOS is a natural mechanism.
INTRODUCTION
One of the surprising realizations of the human genome project was that a relatively small number of human genes give rise to almost an order of magnitude larger number of proteins (1,2). Bioinformatic as well as biochemical and genetic analyses have revealed that most of this complexity of the proteome can be achieved by alternative pre-mRNA splicing (3,4). One route for generating alternative mRNA isoforms is using alternative 5′ splice sites. Mammalian 5′ splice sites of U2 spliceosomes are characterized by a consensus sequence of 8 nt, AG/GTRAGT, where R denotes purine and ‘/’ denotes the splice junction (5,6). Based on alignment of expressed sequence tag databases, alternative 5′ splice site selection can account for only ∼8% of the alternative splicing events that are conserved between the human and the mouse genomes (7). Evidently, this estimate is by far smaller than an estimate derived from genomic sequence analyses. This discrepancy has been reflected in a survey of a database comprising 446 human multi-exon annotated genes, where we found that the abundance of 5′ splice sites that conform to the consensus but are not involved in splicing (hence termed latent 5′ splice sites) largely exceeds the number of authentic 5′ splice sites that are used for both regular and alternative splicing (8,9). The survey revealed 10 490 intronic consensus 5′ splice site sequences that are located within 1496 constitutive introns, and 551 such sites were found within exons. This means that an average of 4–6 latent splicing events at latent 5′ splice sites can be expected for every splicing event at an authentic site. Nonetheless, splicing events at latent 5′ splice sites have not been reported thus far, although the splice signal motive (10) of the latent splice sites does not differ from that of the latent sites (8). It thus follows that compliance with the consensus is not sufficient to define a given sequence as a functional 5′ splice site.
This survey also showed that almost all latent 5′ splice sites (95.8%) are preceded by a stop codon that is in the reading frame of the upstream exon. We have previously hypothesized that the necessity to avoid the inclusion of premature termination codons within spliced mRNAs may serve as a criterion that differentiates authentic from latent 5′ splice sites. According to this hypothesis, a nuclear scanning mechanism brings about the suppression of splicing (SOS) at latent sites when they are preceded by an intronic in-frame stop codon (11,12), thereby serving as a protection mechanism against aberrant protein synthesis. The SOS hypothesis was evaluated by a statistical analysis showing that the abundance of in-frame stop codons upstream of latent 5′ splice sites is significantly higher than that in introns that do not harbor latent 5′ splice sites and significantly higher than that expected by chance alone (8,9).
An experimental clue for SOS was provided by mutational analyses, whereby it was shown that sense mutations of intronic in-frame stop codons upstream of latent 5′ splice sites were sufficient to render such latent sites as active in splicing (12). Furthermore, eliminating in-frame stop codons by frame-shift mutations elicited latent splicing, and silent mutations (e.g. TGA to TAA or TAG) maintained the suppression of latent splicing. These observations excluded the possibility that interference with splicing control elements [for references see Ref. (13)] may have led to latent splicing in mutant constructs not having in-frame stop codons (12).
We also excluded the possibility that splicing at latent 5′ splice sites might normally occur, but the nonsense mRNAs thus obtained are rapidly and efficiently degraded by the nonsense-mediated mRNA decay (NMD) pathway (14–18). To this end, we conducted a comprehensive study in which we have shown that SOS is a novel mechanism distinct from the known RNA surveillance mechanisms (19). First, SOS is distinct from NMD because it is not abrogated by translation inhibitors and it is not affected by siRNA-mediated downregulation of hUpf1 and hUpf2—two key components of the NMD pathway. Second, SOS is distinct from nonsense-associated alternative splicing (NAS) (17,20–26), because a mutant of hUpf1, which was shown to abrogate NAS (26), did not activate latent splicing.
The apparent linkage between the necessity to maintain an open reading frame (ORF) in mRNAs on one hand, and pre-mRNA splicing on the other hand, is presumably brought about by a general SOS mechanism that suppresses splicing events involving intronic latent 5′ splice sites—if they are preceded by a stop codon in the reading frame of the upstream exon. It therefore follows that for its effective operation the SOS mechanism must recognize a reference point that defines the reading frame. Potential candidates to serve as such reference points are translational start codon sequences. Here we confirm this possibility by examining the effect of mutating ATG sequences on eliciting latent splicing in pre-mRNAs transcribed from constructs encoding the CAD gene. In particular, we show that the translation initiation codon sequence is necessary, but not sufficient, to sustain SOS in a manner that is distinct from its role in protein synthesis.
MATERIALS AND METHODS
Plasmids
CAD1, CAD2 (a variant of CAD1), CAD1-Mut9, CAD2-Mut1 (constructs in which all in-frame stop codons were eliminated) were prepared as described previously (12). CAD constructs with mutated ATGs (Supplementary Table 1S) were prepared from CAD1 by the QuickChange PCR-based mutagenesis method (Stratagene) (see Supplementary Data). Wild-type and mutant IDUA mini gene constructs were described (12). All constructs were confirmed by DNA sequencing.
pcDNA-3HA-4E-BP1, a plasmid carrying the human 4E binding protein1 (27,28), was kindly provided by Dr Nahum Sonenberg (McGill University). pMT2-HA-eIF2α plasmids (29) were kindly provided by Dr Paul Anderson (Brigham and Women's Hospital). A wild-type β-globin construct (pmCMV-Gl-Normal) and a construct expressing a PTC-containing β-globin mRNA (β-globin Ter39) (30) were kindly provided by Dr Lynne E. Maquat, (University of Rochester).
Transfections and RNA isolation
Human 293T or Syrian hamster 165-28 fibroblast cells were grown to 50% confluency in tissue culture plates, transiently cotransfected with the appropriate CAD (2–10 μg per 5 × 106 cells), 4E-BP1 or eIF2α (5 μg per 1 × 106 cells) DNA constructs and GFP-EA1 DNA (1 μg per 5 × 106 cells), and total RNA was extracted 24 h post-transfection as described previously (12). Treatment with pactamycin (3–10 μg/ml) (kindly provided by Pfizer Inc), CHX (20 μg/ml), hygromycin B (20 μg/ml) or tetracycline as a bacterial antibiotic control (20 μg/ml) was for 2–4 h at 24 h post-transfection, and total RNA was extracted.
Small interfering RNA-mediated downregulation of hUpf1
RNAi of hUpf1 using siRNA (Dharmacon) was performed as described previously (19) according to Mendell et al. (26), using siRNA targeted at firefly luciferase as a control. 293T cells were grown to 30–50% confluency in 6 cm plates in DMEM medium without antibiotics, and siRNA was transfected using oligofectamine (Invitrogen). After 48 h, the medium was replaced by a medium with antibiotics, and the cells were cotransfected with the appropriate CAD or β-globin and GFP constructs for 24 h.
Western blots
Total proteins were analyzed by SDS–PAGE, blotted and probed with antibodies against hUpf1 (kindly provided by Drs Joshua Mendell and Harry Dietz, Johns Hopkins), ADAR1 (kindly provided by Dr Kazuko Nishikura, Wistar Institute), CBP80 (kindly provided by Dr Ian Mattaj, EMBL) and 4E-BP1 (kindly provided by Dr Nahum Sonenberg, McGill University).
RT–PCR
Total RNA was treated with RNase-free DNase I (50 U/ml; Promega) and cDNA was synthesized from 2 μg RNA for CAD and GFP, using dT15 primer and MMLV reverse transcriptase (Gibco BRL). PCR (20 μl) contained cDNA synthesized from 0.2 μg of total CAD RNA, 10 pmol of the CAD or β-globin or β-actin primers, 1 pmol of the GFP primers (for primers list see Supplementary Data), and 1.0 U of Taq DNA polymerase (Promega). Amplification was carried out in a Mastercycler (Eppendorf) for 30–35 cycles, at an annealing temperature of 63°C for CAD primer pair a/b and the β-globin primer pair; 62°C for CAD primer pair c/b; 59°C for CAD primer pair j/k; 58°C for GFP; 60°C for β-actin and 58°C for hUpf1. PCR for hUpf1 were carried out in 2 mM MgCl2.
RT–PCR analysis of the endogenous IDUA gene was carried out with the IDUA primer pair b/c as previously described (12). The amplified DNA was diluted 1:20 and 2 μl were used for a second PCR with primer pair b/c for the authentic mRNA, and primer pair d/c for the latent mRNA. Primer c was radioactively labeled with 32P-phosphate at its 5′end. All the PCR of IDUA were carried out in 1.5 mM MgCl2 and 5% DMSO. The amplified products were analyzed by electrophoresis in 5 or 10% polyacrylamide/7 M urea gels. The assignment of the bands observed was confirmed by sequencing of the DNA extracted from the gel. Results are representative of at least three independent experiments.
Real-time RT–PCR
Three sets of specific primers were designed, enabling the amplification of a single specific product at a time (see Figure 3a). The sense primers were ‘Pre-mRNA’, ‘Authentic mRNA’ and ‘Latent mRNA’ (see Supplementary Material). The antisense primer used in all sets was CAD primer b. Total RNA was prepared as described above, and cDNA was synthesized from 0.2 μg of total RNA. cDNA (5 μl of a 1:20 dilution) was mixed with 10 μl of 2× SYBR MIX (ABgene), the appropriate forward and reverse primers were added to 150 nM in a final volume of 20 μl. Amplification was carried out using an ABI PRISM 7700 sequence detector (Applied Biosystem). The cycling conditions comprised 15 min of polymerase activation at 95°C, 40 cycles at 95°C for 10 s, 65°C (for Authentic and Pre-mRNA), or 68°C (for Latent mRNA) for 20 s, and 74°C for 15 s. The amplification cycles were followed by a melting curve cycle. The final products were subjected to electrophoresis in 10% polyacrylamide/7 M urea gels to validate the production of a single amplicon per reaction. Each assay included (in duplicate) a no-template-control and a mock-transfection RNA. Results from at least three independent experiments were averaged, and the standard errors are indicated.
Data analysis
Data of the amplified splicing products corresponding to pre-mRNA, authentic mRNA, and latent mRNA, were exported as a tab-delimited text file to a Microsoft Excel spread sheet for further analysis. The efficiencies of the reactions were determined by a linear regression method as described in Ref. (31) using the LinReg program. A midpoint threshold was then established, as suggested in Ref. (32), for each of the three amplicons arising from a given cDNA. The expression levels (Q) were then determined according to the equation , where E is the reaction efficiency and Ct is the cycle threshold. To verify that all transfections were done under similar conditions, we analyzed two reference genes (SDHA and β-actin) using the Genorm program (33). The amplification efficiency of the reference genes was determined using the standard curve method.
RESULTS
Mutating all AUG codons upstream of a latent 5′ splice site elicits latent splicing
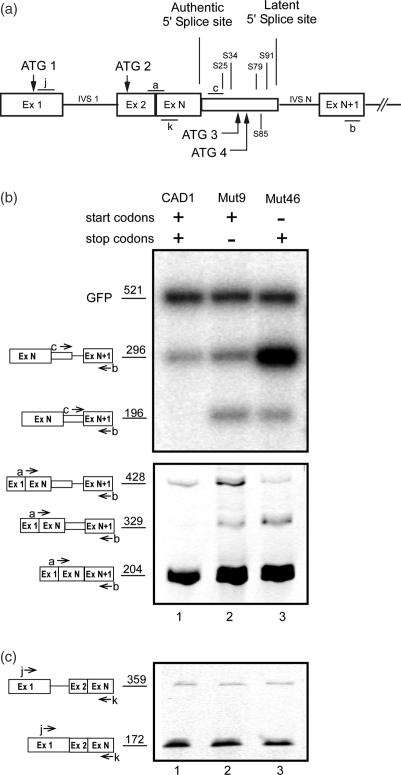
To test for the effect of removal of AUG codons on SOS, we generated mutant minigene constructs derived from the previously described (12) wild-type CAD1 construct (Figure 1a), in which different ATGs were mutated (see Supplementary Table 1S). CAD1 contains a latent 5′ splice site in intron N (IVS N). It is preceded by four stop codons (S25, S34, S79, S91) in reading frame 0 which initiates at ATG1. Reading frame 0 contains an additional ATG sequence (ATG2). Reading frame +1 contains two ATG sequences (ATG3 and ATG4) and a downstream stop codon (S85).
Figure 1.
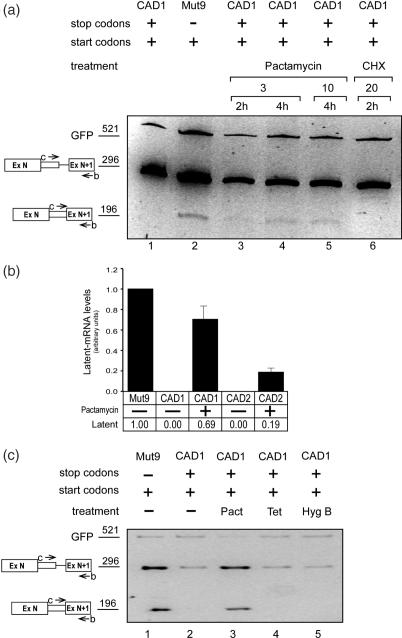
A start codon is required to sustain SOS. (a) A schematic representation of the CAD1 minigene. Start codons (ATG) and stop codons of reading frame 0 and +1 are drawn above and below the scheme, respectively. S designates a stop codon followed by a number that represents its distance in nucleotides from the authentic 5′ splice site. Open boxes, exons; lines, introns; narrow box, latent exon; a, b, c, j and k, PCR primers used to detect the respective splicing products. (b) Eliminating all four AUG codons from a wild-type CAD construct elicits latent splicing. Gel electrophoretic analysis of RT–PCR DNA fragments obtained from CAD mini genes. The presence of wild-type or mutated start and stop codons in each construct is marked by a + or −, respectively (see Supplementary Table 1S). Bands corresponding to precursor and latent CAD fragments amplified with primer pair c/b (upper panel) or corresponding to the precursor, authentic and latent mRNA amplified with primer pair a/b (lower panel) are indicated by schematic drawings on the left. (c) Splicing of IVS1 is not affected upon activation of latent splicing in CAD Mut9 and Mut46. The splicing pattern of IVS1 was analyzed by RT–PCR with primer pair j/k, targeted to sequences flanking IVS1.
The effect of the mutations on latent splicing was analyzed by transient transfections and RT–PCR analyses of the expressed isolated RNAs. PCR analyses of CAD RNAs were performed using an intronic primer, primer c, and primer b, which is complementary to a sequence within the downstream exon N + 1. These primers are expected to amplify the pre-mRNA and latent RNA but not the normally spliced RNA. We also performed RT–PCR analysis using primer pair a/b, where primer a spans the junction between exon 2 and exon N (Figure 1a). These primers amplify the authentic and latent mRNA products, as well as the pre-mRNA. For estimating transfection efficiency, we used GFP DNA, which was cotransfected with the CAD minigene constructs as an internal standard. Transfection with the wild-type CAD1 mini gene construct gave rise to a 296 bp amplicon when amplified with primer pair c/b (Figure 1b, upper panel, lane 1), or a 428 bp amplicon when amplified with primer pair a/b (Figure 1b, lower panel, lane 1), each representing CAD1 pre-mRNA. Notably, no band representing latent RNA (196 bp with primer pair b/c or 329 bp with primer pair a/b) was observed. This result indicates that splicing at the latent 5′ splice site of pre-mRNA transcribed from the wild-type CAD1 construct was suppressed, as expected from the SOS hypothesis for a pre-mRNA transcript having in-frame stop codon(s) upstream of a latent 5′ splice site. RT–PCR analysis of CAD1-Mut9 RNAs, in which all four stop codons had been eliminated by frame shifting (12), gave rise to the amplicons representing latent CAD RNA: 196 bp with primer pair b/c and 329 bp with primer pair a/b (Figure 1b, lane 2, upper and lower panels, respectively).
The involvement of AUGs in the SOS mechanism is shown by eliciting latent splicing in a mutant construct where all four ATGs were mutated, leaving all stop codons unchanged (CAD1-Mut46; see Supplementary Table 1S). Figure 1b, lane 3, shows that the occurrence of latent splicing in this mutant indicates that the stop codons in CAD1-Mut46 are not recognized as such by the SOS mechanism in the absence of a start codon sequence. Notably, the levels of authentic mRNA (the 204 bp amplicon in Figure 1b, lower panel) were not significantly affected by the mutations, indicating that the appearance of latent splicing cannot be attributed to changes in the level of expression of the mutant constructs. Moreover, the splicing pattern of the intron upstream of IVSN is not affected by the mutations (Figure 1c). We can therefore conclude that a start codon sequence may serve as a reference point that defines the reading frame for SOS.
The translation initiation codon sequence is necessary to sustain SOS
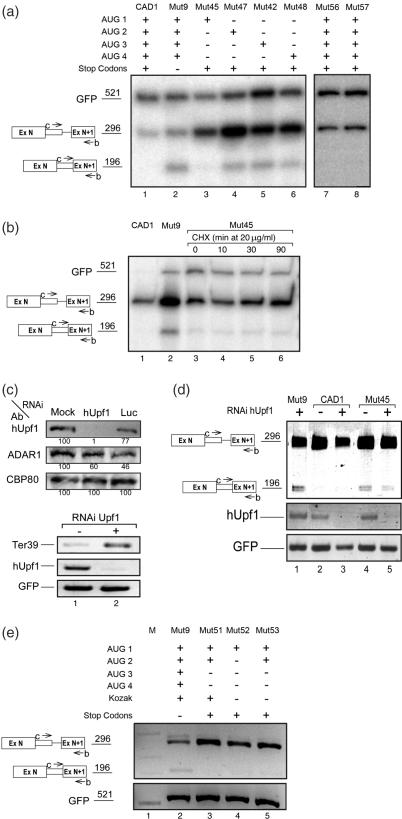
Next we asked which of the four AUG sequences is affecting SOS. To this end we generated four mutant constructs in which all stop codons and only one ATG sequence were retained (Supplementary Table 1S). As can be seen in Figure 2a, latent splicing in the mutant that retained AUG1 (the translation initiation codon sequence) could hardly be detected (CAD1-Mut45; Figure 2a, lane 3; see quantitative PCR below). In contrast, latent splicing was elicited from the remaining mutants, each harboring one of the remaining start codon sequences (AUG 2–4) but lacking AUG1 (CAD1-Muts 47, 42 and 48; Figure 2a, lanes 4–6, respectively; for quantitation see below). To demonstrate that the effect of the mutations on latent splicing was owing to the mutation of the AUG sequence and not the mutation per se, we show that silent point mutations in the neighborhood of AUG1, six codons downstream (Mut56) or six codons upstream (Mut57) of AUG1, did not elicit latent splicing (Muts 56, 57; Figure 2a, lanes 7, 8, respectively, for quantitation see below). Notably, latent splicing was not suppressed in Mut47 although it contains an AUG sequence in frame 0. This observation indicates that an AUG sequence in frame 0 by itself is not sufficient to sustain SOS. Rather, the first AUG sequence, which is the translation initiation codon sequence for the CAD protein, appears to play a predominant role in suppression of CAD latent splicing.
Figure 2.
SOS depends predominantly on the first AUG. (a) Latent splicing in mutants harboring a single AUG codon. The presence of wild-type or mutated start and stop codons in each construct is marked by + or −, respectively; Mut56 and Mut57 harbor silent mutations upstream and downstream of AUG1, respectively (see Supplementary Table 1S). (b) Cells were transfected with CAD mini gene constructs as indicated. Twenty-four hours post-transfection cells transfected with CAD-Mut45 (lanes 3–6) were treated with CHX (20 μg/ml) for the indicated lengths of time. (c and d) Cells were treated with siRNA against hUpf1 as indicated, or with siRNA against luciferase (Luc) as control. Forty eight hours after RNAi treatment, cells were transfected with β-globin Ter39 (a mutant construct expressing β-globin mRNA having a PTC at position 39) (c, lower panel), or with CAD mini gene constructs as indicated, and the expression of CAD, hUpf1 and GFP RNAs were carried out by RT–PCR analyses (d). Western blot analyses were carried out using antibodies (Ab) against hUpf1, ADAR1 and CBP80, as indicated (c, upper panel). The percentage of hUpf1, normalized to the levels of CBP80 and to the levels of proteins in the mock-transfected lane is indicated below the corresponding bands. (e) Cells were transfected with CAD mini gene constructs as indicated. The presence or absence of start and stop codons is marked by + and −, respectively. Mut52 and Mut53 contain a mutated Kozak sequence (see Supplementary Table 1S). RNA analyses, lettering and symbols are as in Figure 1.
Evidently, however, AUG1 is not sufficient to confer the tight regulation exhibited by SOS in the wild-type CAD construct because latent splicing in Mut45, which contains only AUG1, could be detected, though at a very low level (Figure 2a, lane 3; for quantitation see below). Having four in-frame stop codons and a translation initiation codon sequence, the low level of this RNA could have been attributed to NMD (14). However, treatment with cycloheximide (CHX; an inhibitor of protein synthesis), which is known to abrogate NMD (12,19,34,35), did not increase significantly the level of latent mRNA in Mut45 (Figure 2b). It should be pointed out that NMD is abrogated under these conditions, as evidenced by the upregulation of β-globin Ter39 cotransfected with the wild-type CAD1 construct (12,19) and (Supplementary Figure 1S). We have further validated that the low level of latent splicing in CAD1-Mut45 could not be attributed to NMD by siRNA-directed downregulation of hUpf1. Western blot analyses revealed that RNAi of hUpf1 downregulated its expression to ∼1%, while the expression of CBP80 and ADAR1 (reference proteins in the same molecular-weight range as hUpf1) were not significantly affected (Figure 2c, upper panel). As previously shown (19), downregulation of hUpf1 by RNAi did not elicit latent splicing in CAD1 (Figure 2d, compare lanes 2 and 3). Under these conditions NMD is abrogated, as evidenced by upregulation of β-globin Ter39 (Figure 2c, lower panel). As can be seen in Figure 2d, even when NMD was abrogated by downregulation of hUpf1 by RNAi, the level of latent RNA of Mut45 did not increase (compare lanes 5 with 6). We can therefore conclude that the low level of latent RNA obtained in Mut45 most likely reflects the relative importance of the first AUG sequence in the regulation of SOS. In this context, it should be pointed out that latent splicing in wild-type CAD1 and in CAD mutants having all AUG sequences and variable numbers and locations of intronic in-frame stop codons could not be detected (12). Furthermore, the lack of appearance of latent splicing in such constructs could not be attributed to NMD because treatment with inhibitors of NMD or downregulation by RNAi of the NMD genes hUpf1 and hUpf2 did not elicit latent splicing (12,19) (see also Figure 6a, lane 6, and Supplementary Figure 1S). The reason why a mutant construct having only the first AUG sequence appears to have escaped SOS, though to a low degree, is not yet understood. However, in Mut51, which harbors all four stop codons and in which AUG1 and AUG2 are retained while AUG3 and AUG4 have been removed, the tight regulation of SOS seems to be maintained (Figure 2e, lane 3).
We further asked whether the Kozak consensus sequence (36) may play a role in the definition of the reading frame for the SOS machinery. To this aim we generated two mutant constructs (Mut52 and Mut53) in which the Kozak sequence CCCCATGG flanking ATG1 was mutated to CTCCATGT. Mut52 contains only AUG1 and Mut53 contains both AUG1 and AUG2. As can be seen in Figure 2e, latent splicing in Mut52 and Mut53 could not be detected (lanes 4 and 5, respectively). Taken together, we can conclude that although SOS requires an AUG sequence as a reference starting point, the first AUG sequence is necessary but not sufficient to exert full suppression of latent splicing.
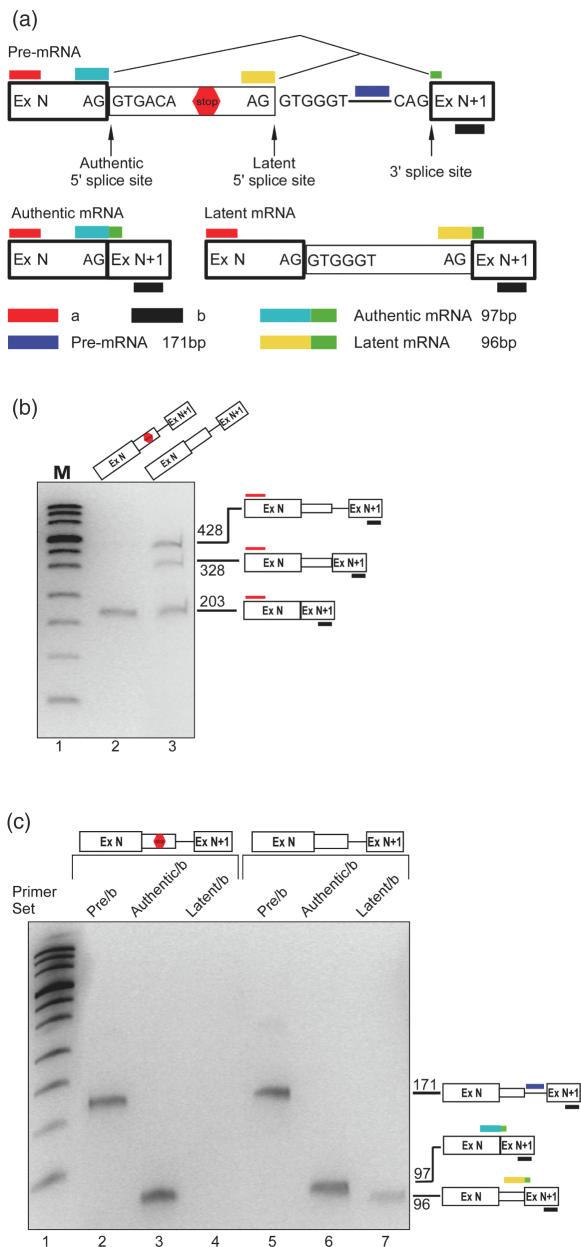
Optimization of product-specific primer pairs for quantitative PCR
For quantitation of latent splicing in the AUG mutant constructs we used real time PCR analysis. For this aim we designed primer pairs such that each should specifically identify only one of the expected splicing products (Figure 3a). The sense primers for the ‘authentic’ and ‘latent’ mRNAs span the respective splice junctions. The sense primer for the ‘pre-mRNA’ is an intronic sequence downstream of the latent 5′ splice site. The antisense primer for all three pairs was primer b in ExN+1. All three species were identified simultaneously by the sense primer a (in ExN) and primer b. To demonstrate the efficacy of this approach we analyzed the splicing patterns of RNAs expressed from the CAD2 and CAD2-Mut1 minigenes. The CAD2 construct has two in-frame stop codons between the normal and the latent 5′ splice site, and CAD2-Mut1 is a construct in which both stop codons have been eliminated (12). As shown in Figure 3b, when RT–PCR analysis was performed using primer pair a/b, CAD2 gave rise to the authentic mRNA and traces of pre-mRNA, whereas CAD2-Mut1 gave rise to both authentic and latent mRNAs, as well as pre-mRNA (lane 3; for a discussion on the expression of CAD2-Mut1 RNA see below). Figure 3c shows the analyses using the specific primer pairs of RNAs expressed from both constructs. In both cases, primer pair ‘pre-mRNA/b’ gave rise to a single PCR product of 171 nt, which corresponds to the pre-mRNA (lanes 2 and 5), and primer pair ‘authentic/b’ gave rise to a single PCR product of 97 nt, which corresponds to the authentic mRNA (lanes 3 and 6). Primer pair ‘latent/b’ revealed a single band of 96 nt, which corresponds to latent mRNA, only in cells transfected with CAD2-Mut1 (lane 7) but none in cells transfected with CAD2 (lane 4). We use hereafter the term ‘fully suppressed’ to indicate that quantitative real time RT–PCR analyses, which could resolve quantities of latent nonsense RNA four to five orders of magnitude lower than that of the latent spliced CAD2 Mut1 mRNA (Supplementary Figure 2S), did not reveal latent splicing in the wild-type constructs. These results validate the specificity of this approach.
Figure 3.
Product-specific real-time PCR. (a) A schematic representation of the underlying design of the quantitation method. Authentic and latent mRNAs are specifically hybridized to PCR primers that flank the junction between a sequence upstream of the respective 5′ splice site and a sequence in the downstream exon. The specificity for the pre-mRNA stems from the location of the sense primer downstream of the latent site. Wide box, exon; line, intron; narrow box, latent exon. (b) Gel electrophoretic analysis of RT–PCR products from transcripts of wild-type CAD2 (lane 2) and the stop-codon-less CAD2-Mut1 (lane 3) constructs. Bands corresponding to precursor (428 bp), authentic (203 bp) and latent (328 bp) CAD fragments amplified with primer pair a/b are indicated by schematic drawings on the right. (c) RNA isolated form cells transfected with CAD2 (lanes 2–4) or with CAD2-Mut1 (lanes 5–7) were analyzed with the specific primer pairs as indicated: The precursor-specific primer pair (lanes 2 and 5); the authentic-specific pair (lanes 3 and 6) and the latent-specific pair (lanes 4 and 7). PCR products were analyzed on a denaturing 7.5% polyacrylamide gel.
Quantitative analyses of the effect of stop codon removal on latent splicing
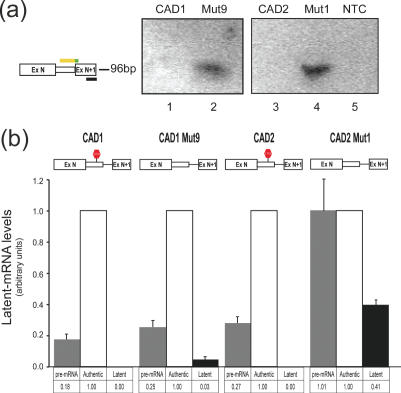
In view of the specificity of the described primer sets, quantitation was performed using real time PCR analysis. We first analyzed the transcripts expressed from CAD1, CAD1-Mut9, CAD2 and CAD2-Mut1. The qualitative analyses of the splicing patterns of these transcripts, using RT–PCR and nuclease S1 mapping analyses, have been described by Li et al. (12). These experiments showed that latent splicing from the constructs harboring intronic in-frame stop codons (CAD1 and CAD2) were fully suppressed. Whereas when the stop codons were eliminated by frame-shifting (CAD1-Mut9) or point mutations (CAD2-Mut1), latent splicing could be readily observed. We have now verified these results using the product-specific PCR. Equal aliquots of cDNA originating from transient transfections of 293T cells with each of the constructs were each challenged with one of the three specific primer pairs described in Figure 3a and subjected to real time PCR analysis. The generation of a single product from each of the primer pairs was verified by performing a melting curve and by gel electrophoretic analysis at the end of each amplification reaction. Using the ‘latent/b’ primer pair, no latent splicing arising from the wild-type CAD1 and CAD2 pre-mRNAs could be detected (Figure 4a, lanes 1 and 3, respectively). However, in CAD1-Mut9 and CAD2-Mut1 where the intronic stop codons had been eliminated, latent splicing was observed (Figure 4a, lanes 2 and 4). The fluorescence readings from the real-time PCR cycles were then used to calculate the relative quantities of the different RNA products (authentic, latent and pre-mRNA). The level of authentic mRNA was defined as 1.00, and used to normalize the fluorescence acquired in the latent and pre-mRNA reactions (Figure 4b). Latent splicing obtained from CAD1-Mut9 was 3% of the authentic splicing event, whereas latent splicing in the wild-type CAD1 pre-mRNA was fully suppressed, in agreement with the qualitative analyses (Figure 1b). No significant change was observed in the levels of the pre-mRNA expressed from CAD1 and CAD1-Mut9 (0.18 and 0.25, respectively). The measurement of latent splicing in the wild-type CAD2 pre-mRNA showed that it was fully suppressed, whereas latent splicing from the stop-codon-less CAD2-Mut1 was 0.41 of the authentic splicing event obtained from that construct. This high level of latent splicing (relative to the 0.03 in CAD1-Mut9) can be attributed to the fact that for generating CAD2-Mut1 the TGA located within the authentic 5′ splice site was mutated to TGG. It should be pointed out, however, that activation of latent splicing in CAD2-Mut1 could not be attributed to this mutation because latent splicing in another construct (CAD2-Mut2), harboring the same mutation but maintaining another in-frame stop codon further downstream, was fully suppressed (12). Also, two mutant constructs both having the first stop codon mutated to TGG and the second stop codon mutated to either TAG or TAA did not elicit latent splicing [see CAD2-Mut10 and CAD2-Mut11 in (12)]. Another consequence of the TGA to TGG mutation within the authentic 5′ splice site is that the level of CAD2-Mut1 pre-mRNA was elevated to 1.01, compared with 0.27 in the wild-type construct CAD2 (Figure 4b; see below a discussion on the level of pre-mRNA in the various mutants). These results reinforce our previous qualitative analyses of latent splicing from these constructs (12), and provide quantitative validation to the tight regulation exerted by the SOS mechanism.
Figure 4.
Quantitative analyses of the effect of stop codon removal on latent splicing. RNA isolated from cells transfected with the indicated CAD constructs were analyzed by real-time PCR using the specific primer pairs. (a) Gel electrophoresis of the real time PCR products with the latent-specific primer pair. No band is detected in transfections with CAD1 (lane 1) or CAD2 (lane 3), whereas the mutated constructs lacking the intronic stop codons gave rise to a single band of the expected size (96 bp; lanes 2 and 4, respectively). NTC, no-template-control. (b) Quantitation of the real-time PCR results. Fluorescence reading from the ‘authentic/b’ primer pair was defined as 1.00 and all other readings were scaled accordingly. Note that latent splicing in the wild-type constructs, CAD1 and CAD2, is fully suppressed. Error bars represent SE of three independent experiments.
Quantitative analyses of the effect of AUG removal on latent splicing
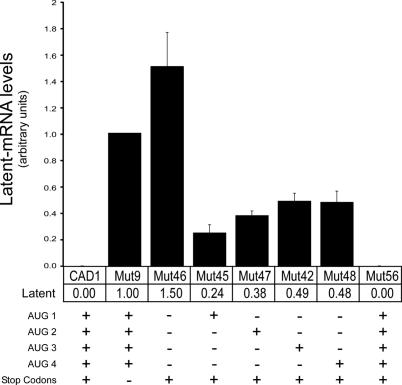
We next used the same real time PCR procedure to quantify the latent splicing events obtained from mutants of CAD1 in which part or all the four ATGs in the construct were mutated, but all stop codons were retained (Figure 5). In order to compare the relative quantities of latent mRNA obtained from each of the mutant constructs, the levels of latent mRNAs were expressed as a percentage of the authentic mRNA obtained from the corresponding construct. These values were then normalized to the level of latent mRNA from CAD1-Mut9. The highest activation of latent splicing was observed in CAD1-Mut46, a construct where all four ATGs were eliminated and all stop codons maintained. This level of latent splicing was 1.5-fold higher than that in CAD1-Mut9, where all ATGs were retained and all stop codons were mutated. Significantly lower levels of latent splicing were observed from constructs in which all but one of the ATGs were mutated. The lowest level (0.24) was in CAD1-Mut45 in which the first ATG was retained. Higher levels (0.38, 0.49, and 0.48) were observed in mutants 47, 42 and 48, where the respective second, third and fourth ATG was retained. To demonstrate that the effect of the mutations on latent splicing was due to the mutation of the ATG sequence and not the mutation per se, we show that when a silent point mutation was made six codons downstream of ATG1 (Mut56), latent splicing was fully suppressed (Figure 5). These results provide a quantitative validation to the qualitative data shown in Figures 1b and 2a. In this context, it should be pointed out that the levels of authentic mRNAs were not dramatically affected in the different ATG mutants (Supplementary Figure 3Sa, b). Furthermore, there appears to be no direct correlation between the occurrence of latent splicing and the level of pre-mRNA (Supplementary Figure 3Sc, d). For example, in comparison to the level of pre-mRNA in wild-type CAD1, which does not show latent splicing, the level of pre-mRNA in CAD1-Mut46 increased by a factor of 1.4 and that in CAD1-Mut48 dropped down to 0.3, while both mutants gave rise to latent splicing. Taken together, our findings demonstrate the crucial role that AUG sequences play in the tight regulation by the SOS machinery. Evidently, however, at least in the case of the CAD gene, each individual AUG sequence on its own was not sufficient to confer tight SOS. On the other hand, the fact that the first AUG sequence was the most effective in suppressing latent splicing, indicates the importance of the first AUG in defining the reading frame used by the SOS mechanism in exerting its activity.
Figure 5.
Quantitative analyses of the effect of start codons removal on latent splicing in the indicated constructs. The presence of wild-type or mutated start and stop codons in each construct is marked by + or −, respectively (see Supplementary Table 1S).. Latent splicing was calculated for each of the indicated constructs as described in Figure 4. Latent splicing from CAD1-Mut9 was defined as 1.00 and all other measurements were scaled accordingly. Error bars represent SE of three independent experiments.
Treatment with pactamycin elicits latent splicing in wild-type CAD1 and CAD2 pre-mRNAs
Pactamycin, a drug known to inhibit translation in bacteria and eukaryotes, has been shown to fold up within the ribosome to mimic a stacked RNA fold (37). It may thus be possible that pactamycin may interfere with putative element(s) involved in the SOS mechanism. In the following experiment, 293T cells were transfected with wild-type CAD1 minigene construct and treated with pactamycin at 24 h post-transfection. Figure 6a shows that increasing amounts or longer treatment with pactamycin resulted in increasing levels of latent splicing in the wild-type construct CAD1 (Figure 6a, lanes 3–5). For a quantitative verification of these results we measured the effect of pactamycin on the splicing patterns of CAD1 and CAD2 pre-mRNAs by the real-time PCR procedure as described above. Cells transfected with each of these wild-type constructs were treated with pactamycin (3 μg/ml for 4 h). This treatment activated latent splicing in both constructs, yet to a different extent (Figure 6b). The measured levels of latent splicing were 0.69 and 0.19, for CAD1 and CAD2, respectively, relative to that obtained for CAD1-Mut9, a mutant lacking all in-frame stop codons, measured in a parallel experiment without pactamycin treatment. Importantly, treatment with CHX, another drug that inhibits translation did not activate latent splicing in wild-type CAD1 (Figure 6a, lane 6). Furthermore, treatment of cells transfected with CAD1 with the translation inhibitor hygromycin B, or with the bacterial translation inhibitor tetracycline as a control, did not elicit latent splicing (Figure 6c, lanes 4 and 5, respectively).
Figure 6.
Pactamycin elicits latent splicing in wild-type CAD RNAs. (a) Cells were transfected with CAD mini gene constructs as indicated. The presence of wild-type or mutated start and stop codons in each construct is marked by + or −, respectively (see Supplementary Table 1S). Twenty-four hours post-transfection, cells transfected with CAD1 (lanes 3–6) were treated with pactamycin (lanes 3–5) or CHX (lane 6), as indicated. RT–PCR products were analyzed on a denaturing 10% polyacrylamide gel. (b) Quantitative analysis of latent splicing was carried out as described in Figure 5 using RNA from cells transfected with wild-type CAD1 and CAD2 and treated with 3 μg/ml pactamycin for 4 h. Error bars represent SE of three independent experiments. (c) Transfection experiments were performed with CAD1 for 24 h. The cells were treated for 2 h with pactamycin (pact; 3 μg/ml; lane 3), tetracycline (tet; 20 μg/ml; lane 4) and hygromycin B (hyg B; 20 μg/ml; lane 5) and RNA was extracted. RT–PCR products were analyzed on a denaturing 5% polyacrylamide gel.
Endogenous genes are subject to SOS
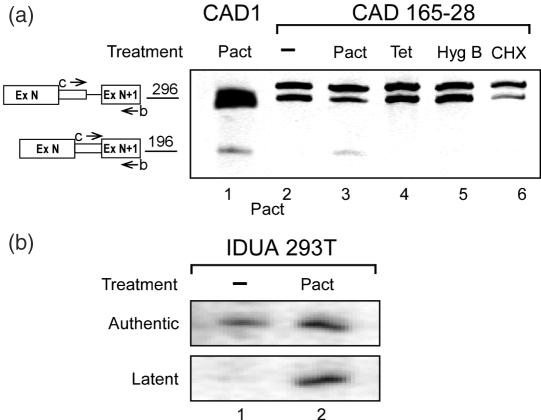
The finding that pactamycin abrogates SOS in transfection experiments, provides us with an opportunity to study whether endogenous, unmutated, genes are subject to SOS. This possibility has already been indicated by Miriami et al. (11), where latent splicing in the endogenous CAD gene product was elicited by heat treatment. Here, we first tested the effect of pactamycin on the expression of the endogenous CAD gene in normally grown Syrian hamster cells. In parallel, we tested the effect of CHX, hygromycin-B, and the bacterial translation inhibitor tetracycline as control. Figure 7a shows analyses by RT–PCR of CAD RNA expressed from the drug-treated cells. Whereas no latent splicing was expressed from the untreated cells, or from cells treated with either CHX, hygromycin-B or tetracycline, treatment with pactamycin elicited latent splicing. These results are similar to those obtained with transfected minigenes. Next, we tested the effect of pactamycin on the expression of the human endogenous α-l-iduronidase (IDUA) gene, minigene constructs of which were previously reported to be under the control of SOS (12,19). Figure 7b shows that, in human 293T cells grown under normal conditions, latent splicing of the endogenous IDUA pre-mRNA was fully suppressed. However, when the cells were treated with pactamycin (3 μg/ml for 4 h), latent splicing was elicited as detected by RT–PCR analysis. These findings indicate that SOS is likely to be a mechanism that operates naturally on endogenous genes.
Figure 7.
Pre-mRNAs expressed from endogenous genes are subject to SOS. (a) Cultured Syrian hamster 165-28 cells were treated with antibiotics as in Figure 6, and RT–PCR analysis of endogenous CAD RNAs was performed using the b/c primer pair. Latent splicing was elicited only by treatment with pactamycin. For comparison we show the expression of latent CAD mRNA in 293T cells transfected with the wild-type CAD1 construct and treated with pactamycin (lane 1). Cloning and sequencing of the PCR products of primer pair b/c revealed that the lower band in the 296 bp region represents the CAD pre-mRNA, while the upper band represents a non-relevant endogenous gene. (b) Human 293T cells were treated with pactamycin and radioactive RT–PCR analysis of the endogenous IDUA RNAs was carried out using IDUA-specific primer pairs (see Primers in Supplementary material) using the 5′ 32P-lebeled antisense primer. The IDUA PCR products were revealed by electrophoresis in a denaturing gel and autoradiography.
SOS is not dependent on translation initiation
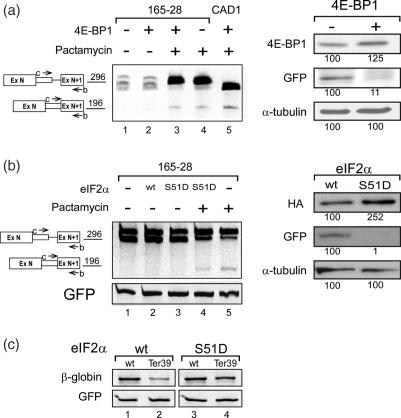
Given that the removal of the AUG sequences and treatment with pactamycin elicited latent splicing, raised the possibility that translation initiation is involved in SOS. To address this possibility we tested the effect of 4E binding protein (4E-BP1) on latent splicing. This protein disrupts an important interaction within the translation initiation complex between the cap-bound eIF4E and the ribosomal-subunit-associated eIF4G, which is required for cap-dependent translation in eukaryotes (38). Transfection of Syrian hamster cells with the 4E-BP1 construct (27,28) together with a GFP construct did not elicit latent splicing in the endogenously expressed CAD pre-mRNA (Figure 8a, lane 2), whereas treatment with pactamycin did elicit latent splicing (Figure 8a, lane 4). Notably, the effect of pactamycin on SOS was independent on translation initiation because the drug elicited latent splicing when translation initiation was inhibited (Figure 8a lane 3). Inhibition of translation under these conditions was exhibited by a 10-fold downregulation of the GFP protein expressed from the cotransfected GFP construct (Figure 8a, right panel).
Figure 8.
SOS is not dependent on translation initiation. (a) SOS is not dependent on steady-state translation. Left panel: RT–PCR analyses of endogenous CAD RNA expressed in cultured Syrian hamster 165-28 cells transfected with 4E-BP1 and GFP constructs, with or without pactamycin treatment as indicated. For comparison, we show the expression of latent RNA from 293T cells transfected with the wild-type CAD1 construct treated with pactamycin (lane 5). Right panel: western blot analysis of total proteins, prepared from cells transfected as indicated, were performed using antibodies against 4E-BP1, GFP, and α-tubulin. The percentage of GFP, normalized to the levels of α-tubulin and to the levels of proteins in the mock-transfection lane, is indicated below the corresponding bands. (b) SOS is not dependent on the pioneer round of translation. Left panel: RT–PCR analyses of endogenous CAD RNA expressed in cultured Syrian hamster 165-28 cells transfected with eIF2α (wt) or with a dominant negative mutant of eIF2α (S51D), and GFP constructs, with or without pactamycin treatment as indicated. Right panel: western blot analyses of total proteins, prepared from cells transfected as indicated, were performed using antibodies against HA, GFP and α-tubulin. The percentage of GFP, normalized to the levels of α-tubulin and to the levels of proteins in the wild-type-transfection lane is indicated below the corresponding bands. (c) NMD is abrogated by eIF2α S51D. Cultured Syrian hamster 165-28 cells were cotransfected with β-globin (wild-type or Ter 39) constructs, GFP construct and with eIF2α (wild-type or S51D) as indicated. RT–PCR analysis of β-Globin mRNA was performed as described in Figure 2c. Abrogation of NMD by eIF2α–S51D is evidenced by the upregulation of β-globin Ter39 mRNA.
The above results show that steady-state translation initiation is not involved in SOS. We have further tested whether SOS could be affected by the pioneer round of translation, because it has been previously shown that this reaction is not inhibited by 4E-BP1. On the other hand, the pioneer round of translation is dependent on eIF2α, which is an integral part of the pioneer round of translation and steady-state translation (39). To this end we monitored the effect of a dominant negative phosphomimetic mutant of the α subunit of eIF2 (eIF2α S51D), which is constitutively inactive in translation initiation (29). Transfection of Syrian hamster cells with either the eIF2α S51D or the wild-type constructs did not elicit latent splicing in the endogenously expressed CAD pre-mRNA (Figure 8b, lanes 2 and 3), indicating that SOS is independent on the pioneer round of translation in addition to being independent on steady-state translation. Notably, the effect of pactamycin on SOS was independent on the pioneer round of translation initiation, as latent splicing occurred even under conditions inhibiting this translation (Figure 8b, lane 4). Inhibition of translation under these conditions was exhibited by the ∼100-fold downregulation of the GFP protein expressed from the cotransfected GFP construct (Figure 8b, right upper panel). As expected, NMD was abrogated under these conditions, as evident from the upregulation of β-globin Ter39 in cells treated with eIF2α S51D (Figure 8c). We can therefore conclude that translation initiation is not likely to be involved in SOS.
DISCUSSION
Most human genes harbor a substantial number of intronic latent 5′ splice sites (8). Suppression of splicing (SOS) at such sites was shown to be sustained by upstream intronic stop codons in the reading frame of the upstream exon (12). The occurrence of SOS can therefore be rationalized by the necessity to maintain the translatability of mRNAs, thereby eliminating the production of toxic nonsense mRNAs. The recognition of an ORF at the level of pre-mRNA raises several mechanistic questions which are yet unanswered. One crucial question is the requirement for a starting point that defines the reading frame, as without a register to establish a reading frame it would be difficult to understand how SOS could recognize in-frame stop codons. Here we show that AUG sequences are essential for the tight regulation by SOS. Specifically, we show that mutating all AUG sequences upstream of a latent 5′ splice site elicited latent splicing in a CAD construct in which all upstream stop codons were retained. We further show that the first AUG sequence plays a predominant role in sustaining SOS, yet it is necessary but not sufficient to impose the tight regulation by SOS, which is observed in the wild-type constructs. We are aware that this mechanism is difficult to envision because it implies that the reading frame of the mRNA is recognized prior to splicing. Nevertheless, the present study reinforces the concept of SOS and provides an initial mechanistic insight into this mechanism.
The effect of pactamycin on SOS
Our results have shown that SOS and the effect of pactamycin on SOS are not dependent on translation initiation. Therefore, the effect of pactamycin on latent splicing is not linked to its role in protein translation, but rather to a direct effect on SOS. A possible clue for this effect may be inferred from the fact that hygromycin B did not elicit latent splicing in the transfected as well as the endogenous CAD genes (Figures 6c and 7a, lane 5, respectively), and from the difference in the mode of interaction of pactamycin and hygromycin B with the ribosome. While both drugs occupy close locations on the bacterial 30S ribosomal subunit, they differ in their binding characteristics to the 16S RNA (37). In particular, upon binding to the ribosome, pactamycin folds up to mimic a stacked RNA dinucleotide that lies in a position originally occupied by the last two bases of the E site codon in the native structure (37). It is therefore possible that by virtue of the capacity of pactamycin to recognize an RNA-fold, rather than specific bases or sequences, it binds to the pre-mRNA. By doing so, probably by employing molecular mimicry, it may interfere with binding of an element(s) that define the starting point for a putative surveillance machine that underlies the SOS mechanism. This view is strengthened by our observation that the effect of pactamycin on SOS is independent of translation initiation—both the pioneer round of translation and the steady-state translation (Figure 8)—indicating that its effect on SOS is not likely to be related to its effect on protein synthesis.
The relationship between SOS and other RNA surveillance mechanisms
In addition to SOS, two cellular pathways have been described as capable of eliminating the inclusion of premature translation termination codons (PTCs) in mature mRNAs—NMD and NAS. Unlike SOS, which responses to intronic in-frame stop (nonsense) codons and appears to function in splicing of many normal genes, NMD and NAS are responses to somatic or intentional mutations that introduce nonsense codons into bona fide exons. SOS is distinct from NMD in additional important parameters. First, while NMD is dependent on protein translation (20), or at least on a pioneer round of translation (40), SOS is not dependent on protein synthesis. To this end, we have shown that SOS is not affected by the translation inhibitor CHX [Figure 6a, lane 6; Supplementary Figure 1S; see also (12)]. Furthermore, SOS is not affected by reagents that allow read-through of in-frame stop codons, such as G-418 and suppressor tRNAs (12,19). In contrast, all the above reagents were shown to abrogate NMD thereby increasing the levels of PTC-harboring mRNAs (Supplementary Figure 1S and Refs (12,34,35,41,42)). Second, SOS is distinct from NMD in its response to a dominant negative mutant of NMD-associated hUpf1 gene, because expression of this mutant, which was shown to abrogate NMD (43–45), did not elicit latent splicing in wild-type CAD1 pre-mRNA (19). Furthermore, in contrast to NMD, SOS is not affected by RNAi treatment of the NMD genes hUpf1 (Figure 2d) and hUpf2 (19). Although SOS and NMD appear to be two distinct surveillance mechanisms, it is probable that if nonsense latent mRNAs are formed by somehow escaping the SOS mechanism, they might be subjected to the NMD pathway.
Are SOS and NAS functionally related? While NAS occurs infrequently and is observed only with specific exons of particular mRNAs, SOS can be viewed as a mechanism that ensures that only functional mRNAs are produced and it appears to do so in most genes (8). Yet, both mechanisms imply that mRNA reading frames can be recognized in the nucleus prior to splicing. How this task is achieved is yet an unresolved question. Nuclear translation (46) was invoked for occurrences of NAS that were shown to be dependent on protein translation (25), though an alternative mechanism, which relies on cytoplasmic translation in conjunction with shuttling splicing factors, was proposed (47). The dependence of SOS on a translation initiation sequence might have implicated protein translation in this splicing control process, but our data exclude this possibility. Namely, we have shown in this article and in our previous studies (12,19) that SOS is a nuclear mechanism that affects splicing and is not affected by translation inhibitors. Furthermore, we show here that inhibition of translation initiation by the eIF4E-binding protein (4E-BP1), which inhibits steady-state translation (Figure 8a, lane 2), or by eIF2α S51D, which inhibits both steady-state translation and the pioneer round of translation (Figure 8b lane 3), did not elicit latent splicing. In this sense SOS may be related to the NAS in the Igκ gene, which was shown to be independent on protein synthesis (21). It thus remains to be explained how the mRNA reading frame can be recognized in the nucleus prior to splicing, regardless of the unresolved issue of nuclear translation (47). This recognition may not necessitate ribosomes as defined for protein synthesis, but may be brought about by one or more ribosomal components whose nuclear localization is not questionable, or by yet unidentified nuclear components. The effects on SOS observed in this study may thus be attributed to disruption of the recognition of the first AUG sequence by the SOS mechanism when such AUGs are mutated, and/or to alterations in a specific RNA fold near this sequence caused by such mutations. The recognition of a starting point for SOS may also be hampered by drugs that, like pactamycin, adopt an RNA fold and compete, presumably by molecular mimicry, for elements of the surveillance machine that define the starting point.
Is SOS a general RNA surveillance mechanism?
A large body of literature has made it clear that 5′ splice site selection, which is essential for productive splicing events, is controlled by a number of RNA and protein elements. Not only that the final number of such elements has not yet been determined, a rule that assigns their relative weights in defining a given 5′ splice site has not yet been formulated. An apparently important factor that affects 5′ splice site selection is the compliance of the 5′ splice site with the consensus sequence, or its relative ‘strength’ as defined by a numerical score (48). Evidently however, not always a splice site that scores higher is preferred when it can potentially compete with an alternative weaker one. A striking example is the CAD gene reported here, because the Shapiro and Senepathy (48) score of the authentic 5′ splice site (AG/GTGACA) is 76.8, whereas that of the latent one (AG/GTGGGT), which resides 125 nt downstream within the intron, is 86. Yet, only the former is selected for splicing, whereas splicing involving the latent one is fully suppressed. We have therefore proposed that the occurrence or absence of stop codons, which are in the reading frame of the upstream exon, may constitute a factor that determines whether a downstream legitimate 5′ splice site would remain silent (latent) or be used for splicing (11,12).
The occurrence of this factor invoked SOS as a general surveillance mechanism that should operate on all pre-mRNAs. Thus far we have demonstrated the occurrence of SOS in two constitutive gene products—CAD and IDUA (12,19). We analyzed a large number of mutants of these genes and found that the splicing patterns of all of them were consistent with the SOS notion. The generality of the SOS mechanism was also evaluated in a computerized survey for latent 5′ splice sites. The dataset contained 2311 introns, in which we found 10 490 latent 5′ splice sites. The utilization of 10 045 (95.8%) of these sites for splicing would have led to the inclusion of an in-frame stop codon within the resultant mRNA. Statistical analysis further showed that the abundance of in-frame stop codons upstream of latent 5′ splice sites is significantly higher than that in introns that do not harbor latent 5′ splice sites, and that the probability of the occurrence of at least one in-frame stop codon upstream of a latent 5′ splice site is higher than expected for a random distribution (8,9). Finally, SOS has been shown to operate on endogenous genes. For instance, latent splicing was elicited in the endogenous CAD gene by heat treatment (11). Similarly, we show here that treatment of cells with pactamycin abrogates SOS and elicits latent splicing from CAD and IDUA endogenous genes. These findings indicate that SOS is indeed a general mechanism.
This view, however, has been challenged from both the statistical and experimental aspects. In a different statistical analysis, Zhang et al. (49) and Zhang and Chasin (50) claim to have found no significant enrichment of in-frame stop codons between authentic 5′ SS and downstream latent 5′ SS. A key feature in their analysis is an estimate of the contribution of the TRA (R = G or A) triplet within the 5′ SS to the overall frequency of in-frame stop codons in the downstream introns. In doing so they assumed that the probability of the TRA triplet to be in the reading frame of the upstream exon is one third (0.333). The real frequency however, as determined by bioinformatic analysis, is 0.182 for mammals (51) and 0.2 (52) or 0.21 (53) for human databases. When even the high value (0.21) is inserted in the equation used by Zhang and Chasin (50) the resulting curve shows that the probability of the occurrence of at least one in-frame stop codon upstream of a latent 5′ splice site is higher than expected for random sequences. It seems therefore that both different statistical analyses indicate the generality of SOS (9) (for details see http://www.weizmann.ac.il/~cosper/suppinfo/Miriami.pdf).
Another attempt was made to test the generality and mechanism of SOS by using a β-globin construct with an artificial duplication of a 5′ splice site (54). Assuming that the two duplicated 5′ splice sites in this wild-type construct are equivalent, the insertion of a stop codon between them was supposed to suppress splicing from the proximal one in the mutant construct. However, splicing from the proximal 5′ splice site was not observed in both the wild-type and mutant constructs. Only upon overexpression of SF2/ASF, proximal splicing was observed for the wild-type construct and hardly for the stop codon-containing mutants. From these experiments the authors inferred that stop codons may not have a general role in regulating pre-mRNA splicing. They admit, however, that their artificial model may lack features that may be required for SOS in native systems. For one reason, this model may not be appropriate to test the generality of SOS because proximal splicing was not observed in the wild-type construct, although both its distal and proximal 5′ splice sites are identical and are flanked by identical sequences. In addition, we report here that start codon sequences, not necessarily the translation initiation codon sequence but also other AUGs even in other reading frames, are important elements in regulating SOS. It turns out that the artificial β-globin construct used by Zhang and Krainer (54) contains another open reading frame (ORF 1) that starts with an AUG, which could allow proximal splicing even when stop codons were inserted in ORF 0.
Based on the above arguments we can conclude that the SOS mechanism appears to be general rather than a gene-specific one. Our combined statistical and experimental data strongly indicate that stop codons and AUG sequences should be counted among the elements that regulate splicing through SOS, although their relative weight is not known yet and may vary among genes. These findings also imply the occurrence of a nuclear scanning mechanism that is associated with SOS. In this sense, the observation that the introduction of PTCs into exons have no effect on the rate of removal of flanking introns (55) may be interpreted as being consistent with an SOS-associated nuclear scanning, because it appears that all nuclear pre-mRNAs should be subject to scanning by the SOS mechanism. We have shown here that, despite its apparent independence on translation, SOS requires a start AUG sequence as a register to establish the reading frame. It seems therefore that AUG sequences can have additional role(s) to their known role in translation, where the translation machinery reads and translates the ORF defined by the AUG. Our results suggest the SOS mechanism as an additional machinery that is capable of recognizing the ORF for the selection of the correct 5′ splice site.
SUPPLEMENTARY DATA
Supplemenary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Drs Joshua Mendell, Kazuko Nishikura, Nahum Sonenberg, Lynne Maquat, Iain Mattaj and Paul Anderson for plasmids and antibodies. This work was supported in part by a grant from the Israel Science Foundation (R.S.) and a grant from the Israel Ministry of Commerce and Compugen Ltd (R.S. and J.S.). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 4.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 5.Burge C.B., Tuschl T.H., Sharp P.A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 6.Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 7.Sugnet C.W., Kent W.J., Ares M., Jr, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 8.Miriami E., Motro U., Sperling J., Sperling R. Conservation of an open-reading frame as an element affecting 5′ splice site selection. J. Struct. Biol. 2002;140:116–122. doi: 10.1016/s1047-8477(02)00539-7. [DOI] [PubMed] [Google Scholar]
- 9.Miriami E., Sperling R., Sperling J., Motro U. Regulation of splicing: the importance of being translatable. RNA. 2004;10:1–4. doi: 10.1261/rna.5112704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim L.P., Burge C.B. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl Acad. Sci. USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miriami E., Sperling J., Sperling R. Heat shock affects 5′ splice site selection, cleavage and ligation of CAD pre-mRNA in hamster cells, but not its packaging in lnRNP particles. Nucleic Acids Res. 1994;22:3084–3091. doi: 10.1093/nar/22.15.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B., Wachtel C., Miriami E., Yahalom G., Friedlander G., Sharon G., Sperling R., Sperling J. Stop codons affect 5′ splice site selection by surveillance of splicing. Proc. Natl Acad. Sci. USA. 2002;99:5277–5282. doi: 10.1073/pnas.082095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 14.Maquat L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 15.Lejeune F., Maquat L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Li S., Wilkinson M.F. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 17.Hentze M.W., Kulozik A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 18.Frischmeyer P.A., Dietz H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 19.Wachtel C., Li B., Sperling J., Sperling R. Stop codon-mediated suppression of splicing is a novel nuclear scanning mechanism not affected by elements of protein synthesis and NMD. RNA. 2004;10:1740–1750. doi: 10.1261/rna.7480804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maquat L.E. NASty effects on fibrillin pre-mRNA splicing: another case of ESE does it, but proposals for translation-dependent splice site choice live on. Genes Dev. 2002;16:1743–1753. doi: 10.1101/gad.1014502. [DOI] [PubMed] [Google Scholar]
- 21.Aoufouchi S., Yelamos J., Milstein C. Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell. 1996;85:415–422. doi: 10.1016/s0092-8674(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 22.Naeger L.K., Schoborg R.V., Zhao Q., Tullis G.E., Pintel D.J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of final spliced product. Genes Dev. 1992;6:1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- 23.Lozano F., Maertzdorf B., Pannell R., Milstein C. Low cytoplasmic mRNA levels of immunoglobulin kappa light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 1994;13:4617–4622. doi: 10.1002/j.1460-2075.1994.tb06783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gersappe A., Burger L., Pintel D.J. A premature termination codon in either exon of minute virus of mice P4 promoter-generated pre-mRNA can inhibit nuclear splicing of the intervening intron in an open reading frame-dependent manner. J. Biol. Chem. 1999;274:22452–22458. doi: 10.1074/jbc.274.32.22452. [DOI] [PubMed] [Google Scholar]
- 25.Wang J., Hamilton J.I., Carter M.S., Li S., Wilkinson M.F. Alternatively spliced TCR mRNA induced by disruption of reading frame. Science. 2002;297:108–110. doi: 10.1126/science.1069757. [DOI] [PubMed] [Google Scholar]
- 26.Mendell J.T., ap Rhys C.M.J., Dietz H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 27.Gingras A.C., Gygi S.P., Raught B., Polakiewicz R.D., Abraham R.T., Hoekstra M.F., Aebersold R., Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gingras A.C., Raught B., Gygi S.P., Niedzwiecka A., Miron M., Burley S.K., Polakiewicz R.D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava S.P., Kumar K.U., Kaufman R.J. phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J., Sun X., Qian Y., Maquat L.E. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramakers C., Ruijter J.M., Deprez R.H.L., Moorman A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 32.Peirson S.N., Butler J.N., Foster R.G. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter M.S., Doskow J., Morris P., Li S., Nhim R.P., Sandstedt S., Wilkinson M.F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 35.Rajavel K.S., Neufeld E.F. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 2001;21:5512–5519. doi: 10.1128/MCB.21.16.5512-5519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodersen D.E., Clemons W.M., Jr, Carter A.P., Wimberly B.T., Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 38.Richter J.D., Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 39.Chiu S.Y., Lejeune F., Ranganathan A.C., Maquat L.E. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishigaki Y., Li X., Serin G., Maquat L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 41.Bedwell D.M., Kaenjak A., Benos D.J., Bebok Z., Bubien J.K., Hong J., Tousson A., Clancy J.P., Sorscher E.J. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nature Med. 1997;3:1280–1284. doi: 10.1038/nm1197-1280. [DOI] [PubMed] [Google Scholar]
- 42.Li S., Leonard D., Wilkinson M.F. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun X., Perlick H.A., Dietz H.C., Maquat L.E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lykke-Andersen J., Shu M.D., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 45.Mendell J.T., Medghalchi S.M., Lake R.G., Noensie E.N., Dietz H.C. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iborra F.J., Jackson D.A., Cook P.R. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 47.Dahlberg J.E., Lund E., Goodwin E.B. Nuclear translation: what is the evidence? RNA. 2003;9:1–8. doi: 10.1261/rna.2121703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro M.B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X., Lee J., Chasin L.A. The effect of nonsense codons on splicing: A genomic analysis. RNA. 2003;9:637–639. doi: 10.1261/rna.5060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X.H., Chasin L.A. Latent splice sites and stop codons revisited. RNA. 2004;10:5–6. doi: 10.1261/rna.5211704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomita M., Shimizu N., Brutlag D.L. Introns and reading frames: correlation between splicing sites and their codon positions. Mol. Biol. Evol. 1996;13:1219–1223. doi: 10.1093/oxfordjournals.molbev.a025687. [DOI] [PubMed] [Google Scholar]
- 52.Long M., de Souza S.J., Rosenberg C., Gilbert W. Relationship between ‘proto-splice sites’ and intron phases: evidence from dicodon analysis. Proc. Natl. Acad. Sci. USA. 1998;95:219–223. doi: 10.1073/pnas.95.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long M., Deutsch M. Association of intron phases with conservation at splice site sequences and evolution of spliceosomal introns. Mol. Biol. Evol. 1999;16:1528–1534. doi: 10.1093/oxfordjournals.molbev.a026065. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z., Krainer A.R. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Lytle J.R., Steitz J.A. Premature termination codons do not affect the rate of splicing of neighboring introns. RNA. 2004;10:657–668. doi: 10.1261/rna.5241404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.