Abstract
DNA ligation is an essential step in DNA replication, repair and recombination. Mammalian cells contain three DNA Ligases that are not interchangeable although they use the same catalytic reaction mechanism. To compare the recruitment of the three eukaryotic DNA Ligases to repair sites in vivo we introduced DNA lesions in human cells by laser microirradiation. Time lapse microscopy of fluorescently tagged proteins showed that DNA Ligase III accumulated at microirradiated sites before DNA Ligase I, whereas we could detect only a faint accumulation of DNA Ligase IV. Recruitment of DNA Ligase I and III to repair sites was cell cycle independent. Mutational analysis and binding studies revealed that DNA Ligase I was recruited to DNA repair sites by interaction with PCNA while DNA Ligase III was recruited via its BRCT domain mediated interaction with XRCC1. Selective recruitment of specialized DNA Ligases may have evolved to accommodate the particular requirements of different repair pathways and may thus enhance efficiency of DNA repair.
INTRODUCTION
Higher eukaryotes are challenged with various types of DNA damage caused by replication errors, radiation, environmental agents and by-products of cellular metabolism. Numerous repair pathways re-establishing the genetic information are known (1,2). An increasing number of proteins have been identified and assigned to these repair pathways, but the recruitment mechanisms and the spatio-temporal coordination of these repair factors at DNA damage sites remains largely unknown. One of the late steps in DNA repair is the joining of breaks in the phosphodiester backbone of duplex DNA, which is catalyzed by members of the DNA Ligase family. The ATP-dependent DNA Ligase family comprises three enzymes termed DNA Ligase I, III and IV, which all contain a highly conserved catalytic domain responsible for the ligation reaction. Although all three DNA Ligases use the same basic reaction mechanism, they have distinct functions and are not interchangeable (3,4).
DNA Ligase I is required for the joining of Okazaki fragments during lagging strand synthesis and is implicated in long-patch or replicative base-excision repair (BER) and nucleotide excision repair (NER). The end-joining activity of DNA Ligase I is directed to DNA replication sites by its interaction with PCNA, a central component of the replication machinery. This interaction and localization is mediated by the N-terminal PCNA-binding domain (PBD) of DNA Ligase I (5,6). It has been shown that loss of DNA Ligase I function, leads to abnormal joining of Okazaki fragments during S-phase (7), defective long-patch BER (8) and reduced repair of double strand breaks (DSBs) by homologous recombination (9).
DNA Ligase III is implicated in short-patch BER and single strand break (SSB) repair (SSBR) and forms a complex with XRCC1 (10–12). XRCC1 and DNA Ligase III normally exist as a preformed complex in vivo interacting via the C-terminal BRCT (BRCA1 C-terminal) domain of DNA Ligase III (10,13–15). XRCC1 also interacts with PARP-1, PARP-2, DNA polymerase β and PCNA (16) and appears to act as a scaffold protein during BER. The unique zinc finger near the N-terminus of DNA Ligase III was shown to bind DNA SSBs (17). Interestingly, this DNA Ligase III zinc finger shows homology with the two zinc finger motifs of poly(ADP-ribose) polymerase (PARP) which also bind DNA strand breaks (11). Therefore, it was suggested that binding of DNA Ligase III via its zinc finger may displace PARP from the DNA break allowing access of DNA Ligase III and other repair proteins to the DNA lesion (17). Recently DNA Ligase III was also identified as a candidate component of the non-homologous end joining (NHEJ) backup pathway (B-NHEJ) (18) and might thus be implicated in double strand break repair.
DNA Ligase IV is implicated in the NHEJ pathway and forms a complex with XRCC4 (19,20). Cultured cells that lack DNA Ligase IV are defective in V(D)J recombination and show increased sensitivity to ionizing radiation (21). Inactivation of DNA Ligase IV in mice leads to embryonic lethality, implying that DNA Ligase IV may have essential functions during early mammalian development (21,22).
We investigated the recruitment of DNA Ligases to repair sites in HeLa cells using a combination of microirradiation, live cell microscopy and binding studies. We could detect only a faint accumulation of DNA Ligase IV at laser-induced DNA damage sites. Kinetic studies and deletion analysis indicated that selective recruitment of DNA Ligase I and III to specific repair pathways is mediated through interaction with PCNA and XRCC1, respectively. These results suggest that PCNA and XRCC1 play a central role in the spatio-temporal coordination of repair factors and thereby enhance the specificity and efficiency of DNA repair in eukaryotic cells.
MATERIALS AND METHODS
Cell culture and transfection
Human HeLa or HEK 293T cells and mouse C2C12 myoblasts were cultured in DMEM containing 50 μg/ml gentamicin supplemented with 10 and 20% FCS, respectively. For transfection, cells grown on μ-slides (Ibidi) or on gridded coverslips were cotransfected with jetPEI (PolyPlus Transfection) or TransFectin transfection reagent (Bio-Rad) according to the manufacturer's instructions. Cells were subsequently incubated for 24–48 h before performing microirradiation and live cell analyses or immunostainings. 293T cells were transfected with plasmid DNA using TransFectin reagent (Bio-Rad) and incubated for 48 h before immunoprecipitations.
Expression plasmids
Mammalian expression constructs encoding translational fusions of human DNA Ligase I, human FEN1 and human PCNA with either green (GFP) or red (RFP) fluorescent protein were previously described (23). Red variants of the previously described GFP-Ligase III (24) and GFP-XRCC1 (25) were generated by replacing GFP with RFP (26) and termed RFP-Ligase III and RFP-XRCC1, respectively. Deletion expression constructs were amplified by PCR with primers containing a SalI and BamHI restriction site and subcloned into the SalI and BamHI site of the peGFP-C1 vector (Clontech) downstream of GFP. The Ligase I PBD-GFP construct was made by subcloning oligonucleotides corresponding to the first 20 amino acids of DNA Ligase I into the EcoRI and XmaI site of peGFP-N2 (Clontech). The GFP-Ligase IV construct was generated by cloning the human DNA Ligase IV cDNA (11) into the peGFP-C1 (Clontech) vector. DNA Ligase IV was amplified using the following oligonucleotides as primers for the PCR: forward 5′-gggg gtc gac gct gcc tca caa ac-3′; reverse 5′-cccc gga tcc aat caa ata ctg gtt ttc-3′. The residues in bold indicate a SalI and a BamHI site encoded in the forward and reverse primer, respectively, for subcloning the PCR fragment into the SalI and BamHI sites of the peGFP-C1 vector downstream of GFP. PCR amplified sequences were verified by DNA sequencing. The Ku70-GFP construct (27) was provided by William Rodgers. In all cases expression was under the control of the CMV promoter. All fusion proteins were tested by expression in HEK 293T cells followed by western blot analysis.
Immunofluorescence and chemicals
Cells were fixed in 3.7% formaldehyde for 10 min and permeabilized with ice-cold methanol for 5 min. The following primary antibodies (diluted in PBS containing 2% BSA) were used: anti-γ H2AX (Ser139) rabbit antibodies (Upstate), anti-PAR mouse monoclonal antibody (Trevigen), anti-DNA Ligase III mouse monoclonal antibody (Gene-Tex), rabbit affinity purified DNA Ligase I antibodies (5), anti-XRCC1 mouse monoclonal antibody (Abcam) and anti-PCNA rat monoclonal antibody. Primary antibodies were detected using secondary antibodies (diluted 1:400 in PBS containing 2% BSA) conjugated to Alexa Fluor 488, 555 or 647 (Molecular Probes). Cells were counterstained with DAPI and mounted in Vectashield (Vector Laboratories).
Microscopy
Time series were taken with a Leica TCS SP2/AOBS confocal laser scanning microscope equipped with a HCX PL 63x/1.4 oil objective using a 488 nm Ar laser line and a 561 nm DPSS laser line. Before and after microirradiation confocal image series of one mid z-section were recorded at 2 s time intervals (typically 6 pre-irradiation and 150 post-irradiation frames) with a pixel size of 90 nm.
Images of fixed cells were taken with a Zeiss Axiophot 2 widefield epifluorescence microscope using a Zeiss Plan-Apochromat 63x/1.40 oil objective and a cooled CCD camera (Visitron Systems).
UVA laser microirradiation
Cells were either seeded on μ-slides (ibidi) or on 40 mm round coverslips and sensitized for microirradiation by incubation in a medium containing BrdU (10 μg/ml) for 24–48 h. For live cell microscopy and irradiation round coverslips were mounted in a POC live-cell chamber (Visitron Systems). Microirradiation was carried out with a 405 nm diode laser coupled into a Leica TCS SP2/AOBS confocal laser scanning microscope. The 405 nm laser was set to maximum power at 100% transmission and was focused through a UV transmitting Leica HCX PL APO 63x/1.40–0.60 oil objective to locally irradiate preselected spots of ∼1 μm in diameter within the nucleus for 1 s. For evaluation of the recruitment kinetics, fluorescence intensities of the irradiated region were corrected for background and for total nuclear loss of fluorescence over the time course and normalized to the pre-irradiation value.
Immunoprecipitations
HEK 293T cells were transiently cotransfected with expression plasmids as described. After 48 h the transfection rate was checked by fluorescence microscopy. About 70–90% of the cells were coexpressing the GFP and RFP fusion constructs. For immunoprecipitations ∼2 × 107 cells were harvested in ice cold 1× PBS, washed twice and subsequently homogenized in 200 μl lysis buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 2 mM PMSF and 0.5% NP40). After a centrifugation step (10 min, 20 000× g, 4°C) the supernatant was adjusted with dilution buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 2 mM PMSF) to 500 μl. Totally 50 μl were added to SDS-containing sample buffer (referred to as input). For immunoprecipitation 1 μg of a purified α-GFP antibody was added and incubated for 2 h rotating at 4°C. For pull down of immunocomplexes 25 μl of equilibrated protein A agarose beads (Amersham Pharmacia, NJ, USA) were added and incubated for 1 h. After centrifugation (2 min, 5000× g, 4°C) the supernatant was removed and 50 μl were collected (referred to as non-bound). The beads were washed two times with 1 ml dilution buffer containing 300 mM NaCl. After the last washing step 100 μl of the supernatant was collected (referred to as wash) and the beads were resuspended in 2× SDS-containing sample buffer and boiled for 10 min at 95°C.
Western blot analysis
For immunoblot analysis 1% of the input, the non-bound and the wash fractions as well as 20% of the soluble supernatants were separated on 12 or 10% SDS–PAGE and then electrophoretically transferred to a nitrocellulose membrane (Bio-Rad Laboratories, CA, USA). The membrane was blocked with 3% milk in PBS and incubated overnight at 4°C with either a mouse monoclonal α-GFP antibody or an α-mRFP rabbit polyclonal antibody. After washing with PBS containing 0.1% Tween-20, the blots were incubated with the appropriate secondary antibody conjugated with horseradish peroxidase. Immunoreactive bands were visualized with ECL plus Western Blot Detection Kit (Amersham Biosciences, NJ, USA).
RESULTS
DNA Ligase I and III accumulate at DNA repair sites
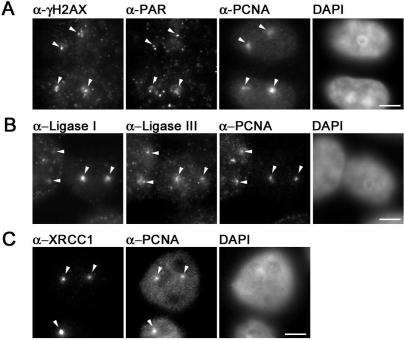
Ligation of DNA is the ultimate step in DNA repair to restore genome integrity. To investigate the involvement of DNA Ligase I and III in DNA repair we performed immunostainings of microirradiated HeLa cells. We employed a confocal laser scanning microscope to locally generate DNA damage at preselected subnuclear sites in BrdU-sensitized cells. In contrast to previous studies we used a long wavelength ultraviolet 405 nm diode laser for microirradiation. Using specific antibodies for different types of DNA damage we could detect phosphorylated histone variant H2AX (γ−H2AX), a marker for DSBs, at microirradiated sites. In addition, we detected poly(ADP-Ribose) which is generated by PARP at SSBs (Figure 1A). These results show that microirradiation with a 405 nm laser generates a mixture of different types of DNA damage that are substrates for different DNA repair pathways involving either DNA Ligase I or III. Immunofluorescence stainings with specific antibodies revealed that endogenous DNA Ligase I and III as well as their binding partners PCNA and XRCC1 are present at DNA damage sites as early as 5 min after irradiation (Figure 1B and C). These results demonstrate that this microirradiation technique allows the direct comparison of factors from different repair pathways within the same cell.
Figure 1.
Immunochemical detection of DNA repair intermediates after laser microirradiation. Widefield fluorescence images of HeLa cells are shown. Cells were fixed ∼5 min after laser microirradiation. Arrows mark sites of irradiation. (A) Laser microirradiation with 405 nm results in local generation of DSBs and SSBs detected by antibodies against γ-H2AX and PAR, respectively. Accumulation of endogenous PCNA can be observed at these sites. (B) Both, DNA Ligase I and III accumulate at sites of DNA damage and colocalize with PCNA. (C) XRCC1 and PCNA accumulate at laser-induced DNA damage sites. Scale bars, 5 μm.
Recruitment kinetics of DNA Ligases at repair sites
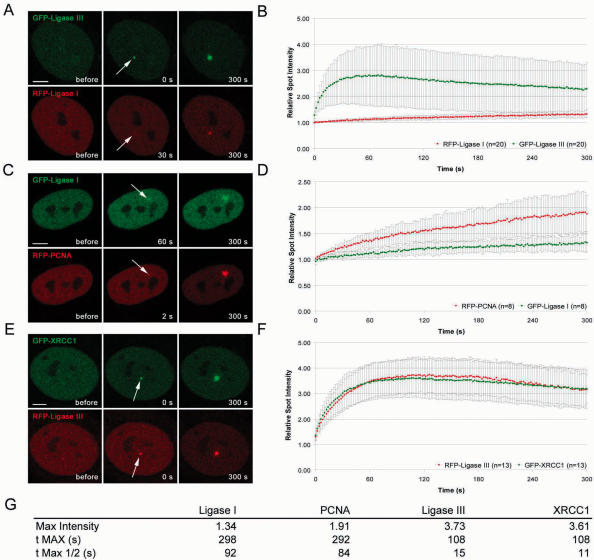
The fact that DNA Ligase I and III although being involved in different repair pathways are both present at microirradiated sites raised the question whether they are recruited by similar mechanisms. First hints for kinetic differences in the recruitment of repair factors involved in SSB repair and BER came from single measurements with GFP-fusion proteins (28). To directly compare the recruitment kinetics of DNA Ligase I and III we microirradiated BrdU-sensitized cells and quantified the accumulation of different GFP- and RFP-tagged proteins at DNA damage site (Figure 2). The intensity values were corrected for background and for total nuclear loss of fluorescence over the time course and normalized to the pre-irradiation value. Mean values of at least eight different cells are shown. A direct comparison of GFP- and RFP-tagged Ligases showed a significantly slower recruitment of DNA Ligase I in contrast to the very fast accumulation of DNA Ligase III at microirradiated sites (Figure 3A and B and Supplementary Fig3video1). Recruitment kinetics of DNA Ligase I and III were independent of the fluorescence tag as swapping of GFP and RFP gave similar results (Supplementary Figure 1). To gain further insights into the mechanisms underlying the different recruitment kinetics of DNA Ligases to repair sites we extended our analysis to their reported interaction partners. Since DNA Ligase I is targeted to replication sites by PCNA in S phase we compared the accumulation of DNA Ligase I with that of PCNA and found similar recruitment kinetics (Figure 3C and D and Supplementary Fig3video2). Both proteins showed a slow but constant accumulation at DNA repair sites and fluorescence intensities reached a maximum after ∼5 min. This suggests that PCNA might be responsible for recruitment of DNA Ligase I to sites of DNA repair.
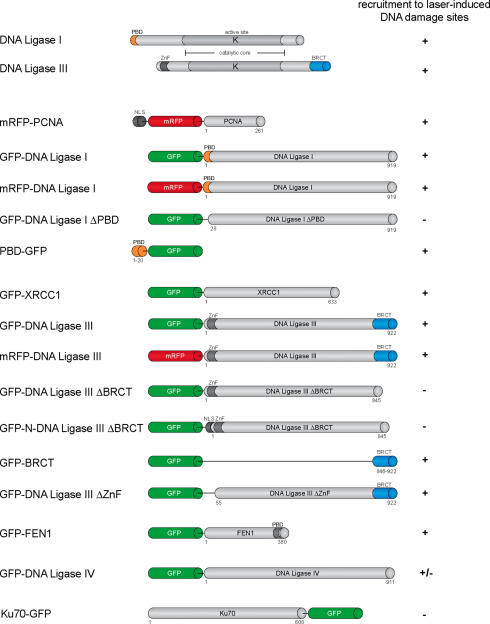
Figure 2.
Schematic outline of the structure of DNA Ligase I and III and of fusion proteins used in this study. The catalytic core (highlighted in grey shading) is highly similar in both Ligases. The relative position of the conserved lysine residue (K) in the catalytic center is indicated.
Figure 3.
Recruitment kinetics of DNA Ligase I and III at DNA damage sites in living cells. Live cell imaging of microirradiated HeLa cells coexpressing various combinations of GFP- and RFP-tagged DNA Ligase I, DNA Ligase III, PCNA and XRCC1. For determination of the recruitment kinetics the relative fluorescence intensity at the irradiated spot was calculated and plotted as a function of time. The displayed curves were generated after integrating data from at least eight independent experiments. Error bars represent the SD. (A and B) Accumulation of DNA Ligase III at DNA repair sites precedes accumulation of DNA Ligase I (Supplementary Fig3video1). (C and D) Accumulation of RFP-PCNA can be observed as early as 2 s after irradiation, while DNA Ligase I accumulates with a delay of ∼30–60 s (Supplementary Fig3video2). (E and F) Immediate and fast recruitment of GFP-XRCC1 and DNA Ligase III to DNA damage sites (Supplementary Fig3video3). Scale bars, 5 μm. The table in (G) summarizes the kinetic parameters of PCNA, DNA Ligase I, XRCC1 and DNA Ligase III recruitment. Mean values of at least eight different cells are shown.
Since biochemical studies had suggested an interaction of DNA Ligase III with XRCC1 during short-patch BER (10,13–15) we directly compared their recruitment to microirradiated sites. Remarkably, we found that DNA Ligase III and XRCC1 redistributed to sites of DNA damage with similar kinetics. After a very fast initial accumulation, intensities of both proteins reached a maximum within 100–120 s after irradiation and then began to decline. These observations support the idea that DNA Ligase III is targeted to DNA repair sites through interaction with XRCC1.
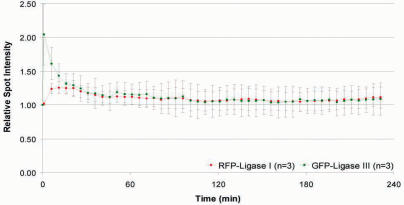
To test whether DNA Ligase I and III not only differ in their recruitment but also in their release kinetics we performed long-term observations of irradiated HeLa cells with 5 min time intervals to compare their release from repair sites. After reaching a maximum the mean fluorescence intensities of PCNA, XRCC1, DNA Ligase I and III decreased gradually at the irradiated sites (Figure 4 and data not shown). Both, DNA Ligase I and III reached basal levels ∼120 min after irradiation (Figure 4). We found similar release kinetics for their respective binding partners PCNA and XRCC1 (data not shown). This indicates that although DNA Ligase I and III differ in their recruitment to repair sites they show similar release kinetics.
Figure 4.
Release kinetics of DNA Ligase I and III at DNA repair sites. Microirradiated HeLa cells were followed up several hours with a time interval of 5 min. Maximum projections of 10–12 z-Stacks were collected and the fluorescence intensities at the irradiated sites calculated and plotted as a function of time.
Next we tested whether the recruitment of GFP-Ligase I and GFP-Ligase III is cell cycle dependent. The characteristic focal distribution of RFP-PCNA and GFP-Ligase I allowed identification of S phase in living cells (29,30). Accumulations of GFP-Ligase I and RFP-PCNA at DNA damage sites could be observed in all S phase stages (Supplementary Figure 2). We could also detect recruitment of RFP-PCNA and GFP-DNA Ligase I to sites of DNA damage in mouse C2C12 cells in S and non-S phase (Supplementary Figure 3). These results indicate that DNA Ligase I and PCNA are recruited with similar kinetics in both human and mouse cells. We also observed accumulation of DNA Ligase III in S phase cells (Supplementary Figure 4).
As laser microirradiation generates DSBs (Figure 1), we tested whether DNA Ligase IV, which is involved in NHEJ, gets recruited to laser-induced DNA damage sites.
We found only a very faint accumulation at repair sites that was barely detectable and in some cases not distinguishable from the nuclear background of unbound GFP-DNA Ligase IV (Supplementary Figure 5A). This is consistent with earlier reports failing to detect recruitment of factors involved in DSB repair like Ku70, DNA-PK and Smc (31). In agreement with this report, we found that Ku70-GFP was not recruited to DNA damage sites after 405 nm irradiation (Supplementary Figure 5B). These results suggest that the number of DSBs generated by microirradiation and the stoichiometry of the repair complex yield only faint signals that are barely above background and hard to analyse with this experimental system.
Taken together, these results show that DNA Ligase I and III are recruited to microirradiated sites with distinct kinetics which match similar differences of their respective binding partners. The recruitment of both DNA Ligases occurred independently of the cell cycle stage in S and in non-S phase cells.
Recruitment of DNA Ligase I to sites of DNA damage depends on its interaction with PCNA
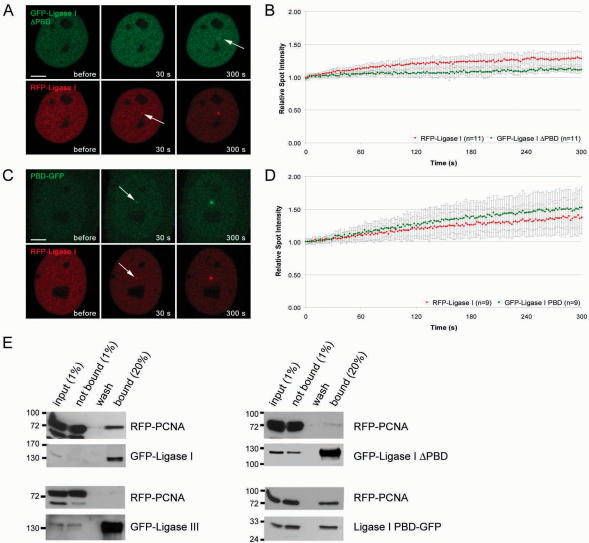
During S phase, DNA Ligase I is targeted to sites of DNA replication via its PCNA-binding domain (PBD) (5,6). To test whether PCNA also mediates recruitment of DNA Ligase I during DNA repair, we deleted the PBD of DNA Ligase I (GFP-Ligase I ΔPBD) and expressed the deletion construct in HeLa cells together with full-length RFP-Ligase I. After laser microirradiation GFP-Ligase I ΔPBD showed only minor accumulation at irradiated sites compared to the wild type fusion construct (Figure 5A and B). This suggests that the PBD plays a critical role in the recruitment of DNA Ligase I to repair sites. The observed residual accumulation of the deletion construct is likely owing to recruitment via trimerization (32) with the endogenous or the full length DNA Ligase I construct. To directly study the function of the PBD we fused the PBD alone with GFP (PBD-GFP). Besides association with replication sites, the PBD fusion protein was recruited to sites of DNA damage with kinetics similar to the full-length DNA Ligase I construct (Figure 5C and D and Supplementary Figure 6).
Figure 5.
Recruitment of DNA Ligase I to DNA damage sites is mediated by PCNA. Recruitment kinetics were determined as described in Figure 3. (A) Live cell imaging of a microirradiated HeLa cell coexpressing GFP-DNA Ligase I ΔPBD and RFP-Ligase I. Deletion of the PBD in GFP-DNA Ligase I ΔPBD almost completely abolishes recruitment to sites of DNA damage, whereas RFP-Ligase I accumulates at these sites as seen in Figure 3 (arrow). (C) A HeLa cell coexpressing DNA Ligase I PBD-GFP and RFP-Ligase I shows accumulation of both, RFP-Ligase I and DNA Ligase I PBD-GFP, at sites of microirradiation (arrows). Times after microirradiation are indicated. Scale bars, 5 μm. (B and D) Recruitment kinetics of fluorescence-tagged proteins at microirradiated sites. (E) Coimmunoprecipitations were performed in 293T cells coexpressing different combinations of RFP and GFP fusion constructs. For interaction mapping the same deletion constructs as in A–D were used. Immunoprecipitations were performed with an antibody against GFP. Precipitated fusion proteins were then detected with specific antibodies against RFP and GFP on western blots. RFP-PCNA coprecipitated with GFP-Ligase I but not with GFP-Ligase III (left panel). RFP-PCNA was also coprecipitated with Ligase I PBD-GFP but not with GFP-Ligase I ΔPBD (right panel).
These kinetic measurements of deletion constructs indicated that recruitment to repair sites is mediated by an interaction of the PBD of DNA Ligase I with PCNA. To test this interaction we performed coimmunoprecipitation assays with the same constructs as used for live cell microscopy. We found that deletion of the PBD abolished the interaction of DNA Ligase I with PCNA, while the PBD alone was sufficient to coprecipitate PCNA (Figure 5E). These lines of evidence strongly suggest that the PBD-mediated interaction of DNA Ligase I with PCNA is necessary and sufficient for targeting of DNA Ligase I to repair sites.
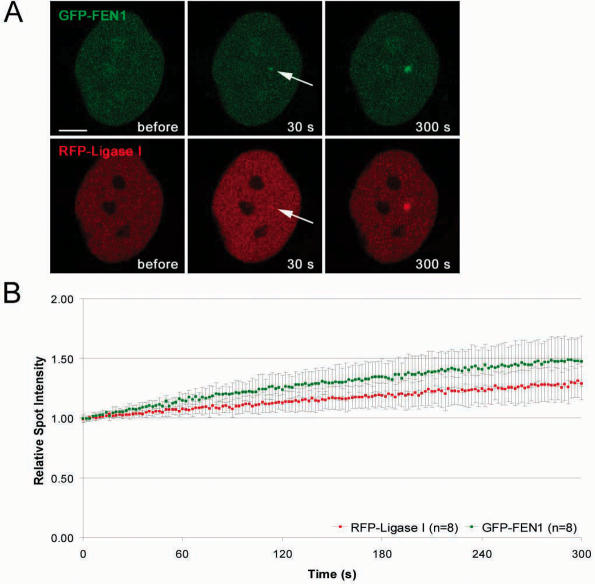
To test whether this mechanism applies also to other PCNA binding proteins we compared the recruitment kinetics of DNA Ligase I with FEN-1, another PBD-containing protein involved in DNA replication and repair. After microirradiation FEN-1 accumulated at DNA repair sites with similar recruitment kinetics as DNA Ligase I (Figure 6). These results point to a common mechanism for the recruitment of PBD containing enzymes like DNA Ligase I and FEN-1 in DNA replication and repair.
Figure 6.
Recruitment kinetics of the PCNA interacting proteins DNA Ligase I and FEN-1 are similar. The structure of the fusion proteins is depicted in Figure 2. (A) Live cell imaging of a microirradiated HeLa cell coexpressing GFP-FEN-1 and RFP-Ligase I. Both FEN-1 and DNA Ligase I accumulate at DNA repair sites (arrow) with similar kinetics. Times after microirradiation are indicated. Scale bars, 5 μm. (B) Recruitment kinetics of fluorescence-tagged proteins at microirradiated sites.
Recruitment of DNA Ligase III to sites of DNA damage depends on its interaction with XRCC1
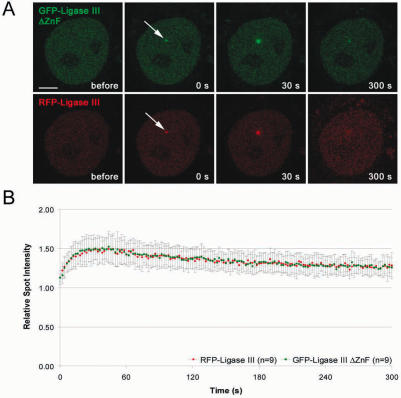
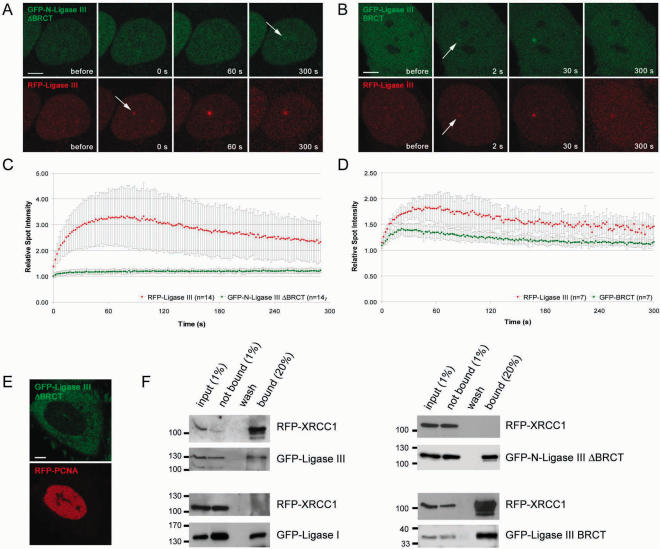
The unique Zn-Finger motif at the N-terminus of DNA Ligase III binds to unusual DNA secondary structures and it was suggested that this domain could recruit DNA Ligase III to damaged DNA (17,33,34). We generated a deletion construct of DNA Ligase III lacking the N-terminal ZnF motif (GFP-Ligase III ΔZnF) and expressed this construct together with the full length RFP-Ligase III in HeLa cells. After microirradiation, no difference could be observed and both fusion proteins showed similar recruitment kinetics (Figure 7). We then tested whether the BRCT domain of DNA Ligase III, which was described to be essential for the interaction with XRCC1 (10,13–15) is required for recruitment to repair sites in vivo. We generated a deletion construct lacking the C-terminal BRCT domain of DNA Ligase III (GFP-Ligase III ΔBRCT). This fusion protein did not enter the nucleus but remained in the cytoplasm indicating that the BRCT domain is responsible for nuclear localization of DNA Ligase III (Figure 8E). After addition of an SV40-NLS (GFP-N-Ligase III ΔBRCT) the fusion protein entered the nucleus but showed, in comparison with the full-length construct, only a minor accumulation at DNA repair sites (Figure 8A and C). Having shown that deletion of the BRCT domain abolishes recruitment of DNA Ligase III, we next fused the BRCT domain alone to GFP (GFP-BRCT). The BRCT fusion protein redistributed to DNA repair sites and showed a similar although weaker accumulation at these sites as the full-length DNA Ligase III (Figure 8C and D). The slightly reduced accumulation of the isolated BRCT domain could be explained by additional protein sequences of DNA Ligase III enhancing recruitment or proper protein folding.
Figure 7.
Deletion of the ZnF motif does not abolish recruitment of DNA Ligase III. Recruitment kinetics were determined as described in Figure 3. (A) Live cell imaging of a microirradiated HeLa cell coexpressing GFP-DNA Ligase III ΔZnF and RFP-Ligase III which accumulate at DNA repair sites (arrow). Times after microirradiation are indicated. Scale bars, 5 μm. (B) Recruitment kinetics of GFP-DNA Ligase III ΔZnF and RFP-Ligase III at microirradiated sites.
Figure 8.
Recruitment of DNA Ligase III to DNA damage sites is mediated by XRCC1. Recruitment kinetics were determined as described in Figure 3. (A) Live cell imaging of a microirradiated HeLa cell coexpressing GFP-N-DNA Ligase III ΔBRCT, containing an additional SV40 NLS and RFP-Ligase III. Deletion of the BRCT domain in GFP-N-DNA Ligase III ΔBRCT abolishes recruitment to sites of DNA damage, whereas RFP-Ligase III accumulates at these sites as seen in Figure 3 (arrow). (B) A HeLa cell coexpressing GFP-Ligase III BRCT and RFP-Ligase III which both accumulate at sites of microirradiation (arrows). Times after microirradiation are indicated. Scale bars, 5 μm. (C and D) Recruitment kinetics of fluorescence-tagged proteins at microirradiated sites. (E) Deletion of the BRCT domain results in cytoplasmic localization of the fusion protein. (F) Coimmunoprecipitations were performed with 293T cells coexpressing RFP-XRCC1 and GFP-Ligase I or GFP-Ligase III, respectively. For interaction mapping the same deletion constructs as in A–D were used. Immunoprecipitations were performed with an antibody against GFP. Precipitated fusion proteins were then detected with specific antibodies against RFP and GFP on western blots. RFP-XRCC1 was coprecipitated with GFP-Ligase III but not with GFP-Ligase I (left panel). RFP-XRCC1 was also coprecipitated with GFP-Ligase III ΔBRCT but not with GFP-N-Ligase III ΔBRCT (right panel).
To investigate the role of the BRCT domain in the interaction of DNA Ligase III with XRCC1 we performed coimmunoprecipitation assays. We found that deletion of the BRCT domain abolished binding of DNA Ligase III to XRCC1, while the BRCT domain alone was sufficient for interaction (Figure 8F). These data fit well with the results obtained from kinetic measurements which altogether indicate that the BRCT domain is necessary and sufficient for recruitment of DNA Ligase III to repair sites while the ZnF domain does not seem to be involved.
DISCUSSION
Different DNA repair pathways have evolved to deal with various types of DNA damage caused by normal cellular metabolism or exogenous factors. The common essential step in all these different repair pathways is the joining of DNA ends by members of the DNA Ligase family. Although the catalytic core of DNA Ligase I and III is highly conserved they have no or only poorly overlapping functions and are not interchangeable (7–10,18,35). Extracts from the cell line EM9 which is deficient in DNA Ligase III (10) are defective in short-patch BER (12) while extracts from the DNA Ligase I deficient cell line 46BR.1G1 (36) are defective in long-patch BER (8). Furthermore, generation of partially DNA Ligase I defective mouse embryonic stem cells revealed that DNA Ligase III could not compensate for loss of DNA Ligase I function in cell proliferation (35).
To explore possible differences that could explain the non-redundant functions of these highly homologous enzymes we compared the recruitment kinetics of DNA Ligase I and III at local DNA lesions generated by laser microirradiation. We found that DNA Ligases I and III accumulated at DNA damage sites with distinct kinetics suggesting that they catalyze the same reaction but use different mechanisms for recruitment. With deletion and binding studies we could demonstrate that the PCNA binding domain (PBD) of DNA Ligase I mediates targeting DNA repair sites. Interestingly, the PBD is not required for enzyme activity in vitro but rescue experiments with DNA Ligase I deficient cells demonstrated that the PBD is essential in vivo (8,9,35). These results suggest that PCNA mediated recruitment of DNA Ligase I could enhance the efficiency of the ligation reaction in vivo by locally concentrating DNA Ligase I at sites of replication and repair.
In further studies, we also observed recruitment of FEN-1 to DNA repair sites, which like DNA Ligase I interacts with PCNA during DNA replication (37,38) and is implicated in long-patch BER (39,40). Remarkably, FEN-1 showed the same recruitment kinetics as DNA Ligase I although it has a completely different function in replication and repair. Likewise, the PBD of DNA methyltransferase 1 (Dnmt1) is also necessary and sufficient for accumulation of Dnmt1 at repair sites (24). Taken together our results show that various PBD-containing proteins involved in the restoration of genetic and epigenetic information are recruited to replication as well as repair sites by PCNA.
On one hand, it has been proposed that the ZnF motif of DNA Ligase III could act as a nick sensor, recruiting DNA Ligase III to DNA nicks and altered DNA structures (17,33,34). We found, however, that deletion of the ZnF did not influence the recruitment kinetics of DNA Ligase III. On the other hand, biochemical studies have suggested that the BRCT domain of DNA Ligase III is essential for its interaction with XRCC1 (10,13–15). Here, we demonstrate that the deletion of the BRCT domain of DNA Ligase III abolishes recruitment of DNA Ligase III to repair sites in vivo. Moreover, the BRCT domain alone was sufficient to mediate recruitment of the fusion protein to DNA repair sites and is essential for nuclear localization of DNA Ligase III.
These different mechanisms for the localization of DNA Ligases at repair sites are consistent with specific characteristics of the respective repair pathways. The continuous synthesis of long stretches of DNA during long patch BER resembles the process of DNA replication. Consequently, also similar recruitment mechanisms seem to be used. In both processes DNA Ligase I is recruited through interaction with the sliding clamp and processivity factor PCNA. In contrast, replacement of just a single nucleotide during short patch BER does not require a processive repair machinery sliding along the DNA but rather a stationary repair complex recruiting DNA Ligase III.
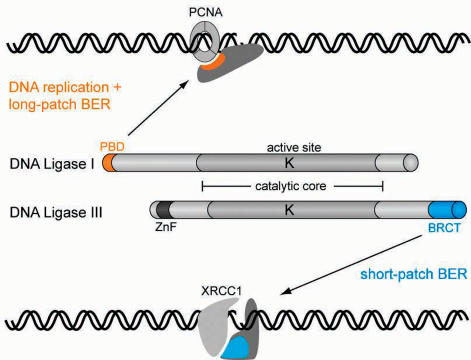
In summary, although DNA Ligase I and III share a highly similar catalytic core, they have distinct functions in DNA replication and repair and are not interchangeable. Here we identified differences in the adjacent regulatory domains of DNA Ligases which may explain their non-redundant functions in eukaryotic cells. Thus, the PBD domain of DNA Ligase I and the BRCT domain of DNA Ligase III mediate interaction with PCNA and XRCC1, respectively, and target them to different repair pathways (Figure 9). This selective recruitment may contribute to the spatio-temporal coordination of different repair factors and could thus enhance accuracy and efficiency of DNA repair in eukaryotic cells.
Figure 9.
Model for selective targeting of DNA Ligase I and III to DNA replication and different repair pathways. All DNA Ligases use the same catalytic mechanism and show high sequence similarity in the catalytic core (grey shading). The active site lysine residue (K) in the center of the catalytic domain is directly involved in the ligation reaction. However, DNA Ligases have non-overlapping functions in DNA repair and replication and are not interchangeable. We could show that DNA Ligase I and III are targeted to different repair pathways through their regulatory PBD and BRCT domains which mediate interaction with PCNA and XRCC1, respectively. This selective recruitment of specialized DNA Ligases may accommodate the specific requirements of different repair pathways and thereby enhance repair efficiency.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors are indebted to Dr R. Tsien for providing the mRFP1 expression vector, Dr G. de Murcia for providing the GFP-XRCC1 expression vector, Dr W. Rodgers for providing the Ku70-GFP expression vector and Dr T. Lindahl for providing cDNAs of DNA Ligases. The authors are grateful to A. Gahl, I. Grunewald and K. Zolghadr for their help and to Lothar Schermelleh and Fabio Spada for helpful comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Volkswagenstiftung to H.L. and M.C.C. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft (DFG).
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg E.C. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Timson D.J., Singleton M.R., Wigley D.B. DNA ligases in the repair and replication of DNA. Mutat. Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 4.Martin I.V., MacNeill S.A. ATP-dependent DNA ligases. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-4-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardoso M.C., Joseph C., Rahn H.P., Reusch R., Nadal-Ginard B., Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell. Biol. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montecucco A., Savini E., Weighardt F., Rossi R., Ciarrocchi G., Villa A., Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. Embo J. 1995;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackenney V.J., Barnes D.E., Lindahl T. Specific function of DNA ligase I in simian virus 40 DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. J. Biol. Chem. 1997;272:11550–11556. doi: 10.1074/jbc.272.17.11550. [DOI] [PubMed] [Google Scholar]
- 8.Levin D.S., McKenna A.E., Motycka T.A., Matsumoto Y., Tomkinson A.E. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 9.Goetz J.D., Motycka T.A., Han M., Jasin M., Tomkinson A.E. Reduced repair of DNA double-strand breaks by homologous recombination in a DNA ligase I-deficient human cell line. DNA Repair (Amst) 2005;4:649–654. doi: 10.1016/j.dnarep.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Caldecott K.W., McKeown C.K., Tucker J.D., Ljungquist S., Thompson L.H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei Y.F., Robins P., Carter K., Caldecott K., Pappin D.J., Yu G.L., Wang R.P., Shell B.K., Nash R.A., Schar P., et al. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappelli E., Taylor R., Cevasco M., Abbondandolo A., Caldecott K., Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 13.Taylor R.M., Wickstead B., Cronin S., Caldecott K.W. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 14.Beernink P.T., Hwang M., Ramirez M., Murphy M.B., Doyle S.A., Thelen M.P. Specificity of protein interactions mediated by BRCT domains of the XRCC1 DNA repair protein. J. Biol. Chem. 2005;280:30206–30213. doi: 10.1074/jbc.M502155200. [DOI] [PubMed] [Google Scholar]
- 15.Dulic A., Bates P.A., Zhang X., Martin S.R., Freemont P.S., Lindahl T., Barnes D.E. BRCT domain interactions in the heterodimeric DNA repair protein XRCC1-DNA ligase III. Biochemistry. 2001;40:5906–5913. doi: 10.1021/bi002701e. [DOI] [PubMed] [Google Scholar]
- 16.Fan J., Otterlei M., Wong H.K., Tomkinson A.E., Wilson D.M., III XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackey Z.B., Niedergang C., Murcia J.M., Leppard J., Au K., Chen J., de Murcia G., Tomkinson A.E. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Rosidi B., Perrault R., Wang M., Zhang L., Windhofer F., Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 19.Critchlow S.E., Bowater R.P., Jackson S.P. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 20.Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T.E., Mann M., Lieber M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 21.Frank K.M., Sekiguchi J.M., Seidl K.J., Swat W., Rathbun G.A., Cheng H.L., Davidson L., Kangaloo L., Alt F.W. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 22.Barnes D.E., Stamp G., Rosewell I., Denzel A., Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 23.Sporbert A., Domaing P., Leonhardt H., Cardoso M.C. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 2005;33:3521–3528. doi: 10.1093/nar/gki665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortusewicz O., Schermelleh L., Walter J., Cardoso M.C., Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy N., Martz A., Bresson A., Spenlehauer C., de Murcia G., Menissier-de Murcia J. XRCC1 is phosphorylated by DNA-dependent protein kinase in response to DNA damage. Nucleic Acids Res. 2006;34:32–41. doi: 10.1093/nar/gkj409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodgers W., Jordan S.J., Capra J.D. Transient association of Ku with nuclear substrates characterized using fluorescence photobleaching. J. Immunol. 2002;168:2348–2355. doi: 10.4049/jimmunol.168.5.2348. [DOI] [PubMed] [Google Scholar]
- 28.Lan L., Nakajima S., Oohata Y., Takao M., Okano S., Masutani M., Wilson S.H., Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonhardt H., Rahn H.P., Weinzierl P., Sporbert A., Cremer T., Zink D., Cardoso M.C. Dynamics of DNA replication factories in living cells. J. Cell. Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sporbert A., Gahl A., Ankerhold R., Leonhardt H., Cardoso M.C. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 2002;10:1355–1365. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- 31.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell. Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimitriadis E.K., Prasad R., Vaske M.K., Chen L., Tomkinson A.E., Lewis M.S., Wilson S.H. Thermodynamics of human DNA ligase I trimerization and association with DNA polymerase beta. J. Biol. Chem. 1998;273:20540–20550. doi: 10.1074/jbc.273.32.20540. [DOI] [PubMed] [Google Scholar]
- 33.Kulczyk A.W., Yang J.C., Neuhaus D. Solution structure and DNA binding of the zinc-finger domain from DNA ligase IIIalpha. J. Mol. Biol. 2004;341:723–738. doi: 10.1016/j.jmb.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 34.Taylor R.M., Whitehouse J., Cappelli E., Frosina G., Caldecott K.W. Role of the DNA ligase III zinc finger in polynucleotide binding and ligation. Nucleic Acids Res. 1998;26:4804–4810. doi: 10.1093/nar/26.21.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrini J.H., Xiao Y., Weaver D.T. DNA ligase I mediates essential functions in mammalian cells. Mol. Cell Biol. 1995;15:4303–4308. doi: 10.1128/mcb.15.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prigent C., Satoh M.S., Daly G., Barnes D.E., Lindahl T. Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell Biol. 1994;14:310–317. doi: 10.1128/mcb.14.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tom S., Henricksen L.A., Bambara R.A. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 38.Frank G., Qiu J., Zheng L., Shen B. Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J. Biol. Chem. 2001;276:36295–36302. doi: 10.1074/jbc.M103397200. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto Y., Kim K., Hurwitz J., Gary R., Levin D.S., Tomkinson A.E., Park M.S. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem. 1999;274:33703–33708. doi: 10.1074/jbc.274.47.33703. [DOI] [PubMed] [Google Scholar]
- 40.Gary R., Kim K., Cornelius H.L., Park M.S., Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.