Abstract
Real-time quantitative PCR (qPCR) is a powerful tool for quantifying specific DNA target sequences. Although determination of relative quantity is widely accepted as a reliable means of measuring differences between samples, there are advantages to being able to determine the absolute copy numbers of a given target. One approach to absolute quantification relies on construction of an accurate standard curve using appropriate external standards of known concentration. We have validated the use of tissue genomic DNA as a universal external standard to facilitate quantification of any target sequence contained in the genome of a given species, addressing several key technical issues regarding its use. This approach was applied to validate mRNA expression of gene candidates identified from microarray data and to determine gene copies in transgenic mice. A simple method that can assist achieving absolute quantification of gene expression would broadly enhance the uses of real-time qPCR and in particular, augment the evaluation of global gene expression studies.
INTRODUCTION
The continued improvement of methodologies for examining gene expression is essential to further our understanding of how the information contained within a genome is utilized by the cellular machinery. The focus in recent years has been on the development of techniques that allow the analysis of the expression of a large number of genes simultaneously, with the goal being to profile the global expression under a particular set of conditions. Equally important though is the refinement of techniques to more accurately quantify the level of gene expression, with the goal being to determine the absolute copy number of an expressed sequence.
The most traditional method for characterizing mRNA transcripts is northern blot analysis. While this technique is effective for sizing mature mRNA transcripts and identifying alternatively spliced forms, quantification is always relative to a reference gene. RNase protection and S1 nuclease mapping assays are potentially more amenable to modifications that would allow determination of absolute quantity but they are not typically applied in this way. Furthermore, these techniques are not easily scaleable for assessing the expression of a large number of genes in many samples.
Approaches to global expression profiling include hybridization to various DNA microarray platforms and serial analysis of gene expression (SAGE). In practice, these methods rely on analysis of differential gene expression by measurement of fold changes over a suitable control or baseline. There has not been an extensive focus on using these global mRNA profiling techniques for determining absolute quantities of expressed genes nor has it been determined whether the current technologies are capable of measuring the concentration or absolute copy number of a given expressed gene sequence in an unknown sample with reasonable accuracy. Cross-platform validation and confirmation of ratiometric differences in mRNA expression for selected genes are often sufficient for the biological objectives of many studies. However, the lack of robust and scaleable methods of absolute gene copy quantification not only represents an obvious deficiency in a most fundamental aspect of scientific measurement, but also hinders the ability to study basic questions concerning the accuracy and sensitivity of current genome-scale mRNA profiling techniques.
Real-time PCR represents an innovative evolution of conventional PCR that has emerged as a major analytical platform in molecular biology. Incorporation of fluorescence-based detection systems into real-time PCR instruments not only allows kinetic detection of the accumulation of PCR products over the cycling period, but also provides greater sensitivity for amplicon detection as compared to conventional gel-based detection. Major uses of real-time PCR include analysis of differential mRNA expression (1–3) and SNP detection (4) although additional applications have been described, including splice variant discrimination (5,6), pathogen load diagnosis (7–9), cancer marker quantification (10–12) and genotyping (13,14).
In real-time PCR, the cycle at which the amount of detectable PCR product reaches a preset threshold level is assessed (Ct). Relative quantities can be determined by normalizing against the Ct value for an internal reference gene prior to calculation of ratiometric differences between unknown samples. Several mathematical formulas that calculate relative fold changes have been proposed: (15), (16) and (17), in which E represents amplification efficiency. In contrast, the determination of absolute quantity of gene copy using real-time PCR requires the generation of a standard curve from known quantities of the PCR target sequence (18–21). Absolute quantification to measure copy number of a particular mRNA target originally present in live cells or tissue remains technically difficult due to the lack of methodology that can comprehensively account for all the variation that can occur during cDNA sample preparation. However, the quantity of target sequence in cDNA samples can be assessed with reasonable accuracy using a proper external standard. A variety of materials have been used as external standards, including plasmid DNA containing the target sequence (2,5,7,8,11,21), reverse-transcribed cRNAs (3,6,22), PCR-amplified target sequences (1,12,23) and commercially prepared DNA (9). Each has advantages and disadvantages in terms of complexity of the material, cost, accessibility, long-term stability and gene copy accuracy.
In this study, we present a simple method using standard curves generated from tissue genomic DNA (gDNA) that enable quantification of any naturally occurring gene sequence in cDNA samples. We have characterized parameters with which gDNA can be used effectively as a universal standard curve. This methodology was applied to validation of microarray data and genotyping of transgenic mice to demonstrate quantification of cDNA and gDNA, respectively.
MATERIALS AND METHODS
Plasmid dilution and calculation of plasmid copy numbers
Unless the copy numbers are specified, all green fluorescent protein (GFP) plasmid (4700 bp) standards were constructed with a 3-fold dilution series ranging from 8100 to 100 copies. The copy numbers of the GFP plasmid and PRKR plasmid (5547 bp) are calculated based on the following formula (http://www.appliedbiosystems.com/support/tutorials/pdf/quant_pcr.pdf), in which n is the number of base pairs, m is the mass of the DNA, NA is Avogadro's number (6.02 × 1023 bp/mol) and M is the average molecular weight of a base pair (610 g/mol).
Primer design and selection
All primers were designed with Primer Express (PE Applied Biosystems, Perkin Elmer, Foster City, CA) based on target sequences obtained from the Affymetrix database (NetAffx™ Analysis Center, http://www.affymetrix.com/analysis) for each gene of interest. Primers were designed to target the 3′-untranslated region (3′-UTR) of transcript sequences except GFP, HsPRKR P1, HsPRKR P2 and HsPRKR P3, all of which target upstream exons. We ensured that the primer pair will specifically amplify the target sequence by searching for the nucleotide sequences that contain both primer sequences on opposing strands in the NCBI Genbank database using BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Primer pairs were discarded if the alignment of both primer sequences could result in the amplification of non-specific sequences. All new primers were tested by real-time PCR using a 3-fold serial dilution (9–0.111 ng/reaction) of gDNA and water as non-template control followed by gel electrophoresis, in order to determine amplification efficiency, specificity and the presence/absence of primer dimers. The amplification efficiency for each primer was determined from the linear slope of standard curve; only primers with a standard curve slope between −2.92 and −3.92 were used for further quantification. Inefficient and/or non-specific primers were excluded. Lastly, Verification of Amplicon Specificity using Restriction Enzymes (VASRE, see below) was performed to validate the sequence specificity of the expected amplicons. A supplementary table contains all the sequences of primers used in this paper (Online Supplementary Table).
Extraction of RNA and gDNA
RNA extraction was performed according to the manufacturer's manual for the RNeasy Kit (Qiagen). Both human and mouse gDNA from various tissues (human tonsil, liver, subcarinal lymph nodes and lung; mouse liver) was isolated according to the manufacturer's manual for the DNeasy Kit (Qiagen). Mouse-tail gDNA was isolated by phenol extraction after overnight proteinase K digestion, and then DNA is subsequently precipitated with ethanol and resuspended in 10 mM Tris, 1 mM EDTA, pH 7.4. For the generation of external standards, gDNA was stored as a 9 ng/μl stock at −20°C, and a fresh 3-fold serial dilution was prepared for each real-time PCR experiment (9–0.111 ng/reaction).
RT reaction and real-time PCR
DNase treatment was performed on RNA samples using DNA-free (Ambion Inc.) according to the manufacturer's manual in order to remove gDNA carry-over. In a single RT reaction, 5 μg of RNA was mixed with RNase-free water (Qiagen) and 1 μl of oligo(dT23) (Sigma–Aldrich Co.) to a final volume of 12 μl, then incubated at 90°C for 10 min. To this, 4 μl of 5× First-Strand buffer, 2 μl of 0.1 M DTT, 1 μl of 10 mM dNTP and 1 μl (200 U) of Superscript II (Invitrogen Co.) were added. The mixture was incubated at 42°C for 90 min followed by an extension period for 15 min at 70°C.
Real-time PCRs were performed using the ABI Prism 7900HT (PE Applied Biosystems) in 384 micro-well plates. All samples, including the external standards and non-template control, were run in triplicate. The reaction conditions had been established through a series of preliminary optimization experiments. Each 10 μl reaction contained 1× PCR buffer (Sigma–Aldrich Co.), 3 mM MgCl2, 0.2 mM dNTP, 1 nM forward and reverse primers, 1/50 dilution of ROX reference dye (Sigma–Aldrich Co.), 3/100 000 dilution of SYBR Green I (Sigma–Aldrich Co.), 0.05 U of JumpStart Taq polymerase (Sigma–Aldrich Co.) and template DNA. Template was either a 3-fold serial dilution (9–0.111 ng/reaction) of gDNA for generation of a standard curve, water for a non-template control which was included to confirm the absence of DNA contamination in the reaction mixture, 10 ng of cDNA generated from total RNA or 1 ng of cDNA generated from mRNA. The reaction was initiated by activation of Taq polymerase at 95°C for 3 min, followed by 40 three-step amplification cycles consisting of 10 s denaturation at 95°C, 15 s annealing at 65°C and 20 s extension at 72°C. A final dissociation stage was run to generate a melting curve for verification of amplification product specificity. Real-time PCR was monitored and analyzed by the Sequence Detection System version 2.0 (PE Applied Biosystems).
Verification of amplicon specificity using restriction enzymes
Our VASRE system used a panel of six restriction enzymes (HpyCH4V, HaeIII, HinP1I, HpyCH4IV, MspI and RsaI, all from New England Biolabs) to validate the sequence specificity of the amplicons produced by real-time PCR primer pairs. The PCR products produced during real-time PCR screening of new primer pairs were digested with each enzyme in a separate reaction. Each VASRE reaction contained 0.5 U of restriction enzyme, 1× of appropriate digestion buffer, 0.8 μl of 25 mM MgCl2, 0.3 μl of SYBR Green I 1/1000 dilution (Sigma–Aldrich Co.), 0.1 μl of ROX reference dye (Sigma–Aldrich Co.) and 1 μl of cDNA template taken from a 9 ng gDNA standard originally screened in real-time PCR to a final volume of 10 μl, and then incubated at 37°C for 1 h. The reaction mixture was returned to a clean PCR plate, and a dissociation curve was produced by measuring the peak fluorescence change as the temperature was increased. The dissociation curve after digestion with each restriction enzyme was compared to the dissociation curve generated after incubation of the PCR product in the absence of enzyme.
Microarray
RT reactions for microarray were carried out as described above, except that T7 oligo(dT) primer (Affymetrix, Inc.) was used instead of oligo(dT23) (Sigma–Aldrich Co.). Microarray hybridizations were performed using the Affymetrix HU133A chip (Affymetrix, Inc.) according to the user manual. The array data were analyzed with Microarray Suite ver. 5.0 (Affymetrix, Inc.).
RESULTS
Calculation of gene copy number in gDNA
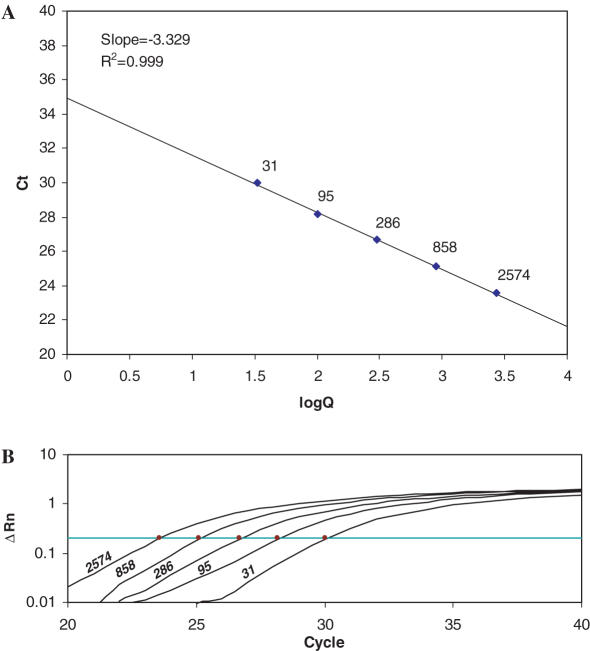
Absolute quantification by real-time PCR requires amplification of the target sequence from a series of external standards of known quantities to generate a standard curve from which the quantity of the target sequence in the unknown sample can be calculated (18–21). Since all gene sequences for a given species are represented, gDNA could serve as a universal standard for the absolute quantification of any expressed gene. The mass of the haploid human genome (C-value) is ∼3.5 pg (http://www.genomesize.com), and therefore 1 ng of gDNA contains ∼286 copies of a single-copy gene. Generation of standard curves ranging up to 10 000 copies would therefore require <35 ng of gDNA. We typically generate a standard curve using a 3-fold dilution series to produce five quantities ranging from 9 ng (∼2574 copies) to 0.111 ng (∼31 copies) of gDNA isolated from tonsils as a source of normal diploid cells. A typical standard curve is shown in Figure 1 generated from primers designed against the human CCL2 gene. The Ct values range between cycles 23 and 30 (threshold = 0.20), producing a standard curve with a slope of −3.33 (R2 = 0.999).
Figure 1.
(A) A typical standard curve of Ct versus log copy number. The points comprising the line are labeled with the copy number. (B) Ct values are obtained from amplification plots which indicate the change in normalized signal for the five standards (indicated with copy numbers) between cycles 20 and 40 of the PCR. Ct is the cycle at which fluorescence crosses a threshold value.
Considerations for the design and testing of gene-specific primers to generate a gDNA-based standard curve
An essential consideration for using gDNA as an external standard in real-time PCR is designing target-specific primer pairs that do not span introns. Although it is common to design intron-spanning primer pairs to mitigate signals arising from contamination of RNA samples with gDNA, we routinely treat samples with DNase to avoid this non-specific amplification.
Primer pairs designed against a gene of interest were experimentally tested by generating a standard curve with tissue-derived gDNA. Acceptable primers meet the following criteria for efficiency and specificity.
- High amplification efficiency: Efficiency of amplification can be determined by
using the slope from a standard curve plot (Ct versus log copy number). Efficiency (100%) is therefore defined by a slope of −3.32. We typically utilize primer pairs that produce standard curves with slopes between −2.92 and −3.92 (Efficiency = 100 ± 20%). - Specificity
- Dissociation plots are generated at the end of the qPCR by slowly increasing the reaction temperature and measuring the rate of decrease in fluorescence as the strands of the final product separate. The dissociation plot should consist of a single peak to indicate a single product, and the peak of the curve should occur at the expected melting temperature of the amplicon.
- Specific amplification can be further verified by assessing the size of the PCR product by agarose gel electrophoresis.
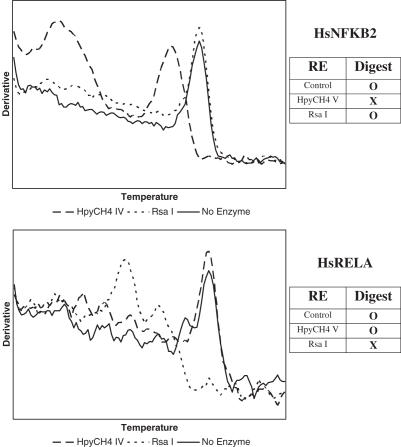
- Verification of the Amplicon Specificity using Restriction Enzymes (VASRE). We have developed the VASRE assay to examine the dissociation curve after digestion of the PCR products with a panel of restriction enzymes. By identifying restriction sites in the expected amplification product, we can predict which digests will result in shifts in the dissociation peak. VASRE analysis of the products amplified from the genes RELA and NFKB2 is shown in Figure 2. Shifts in the peak of the dissociation curve were observed only after digestion with enzymes having a restriction site in the expected product.
Figure 2.
Verification of Amplicon Specificity using Restriction Enzymes (VASRE) on two real-time PCR amplicons. Expected digestion patterns are shown in the panels on the right. Dissociation curves after digestion with HpyCH4V and RsaI confirm the specificity of two amplicons when compared to expected results.
Effect of sample complexity on amplification efficiency
While gDNA has the advantage of acting as a source for any gene target, it also contains a large excess of non-target sequence. In contrast, plasmids or cDNA standards contain a relatively small amount of non-target sequence. We postulated that amplification efficiency may be affected by both the inherent properties of the primer pair and the complexity and nature of the DNA sample containing the target to be amplified. As gDNA is to be used as an external standard for the quantification of gene expression in cDNA, it is necessary to ensure that the differences in complexity do not confound these measurements.
Standard curves were generated by amplifying a dilution series of plasmid containing the gene for GFP by real-time PCR and plotting the Ct values against the concentration of plasmid. To examine the effect of sample complexity on amplification efficiency, increasing amounts of gDNA or cDNA were spiked into the GFP dilution series. The quantities of gDNA used were identical to quantities we typically use for generating standard curves (9–0.111 ng), and the quantities of cDNA represent typical amounts used in our experiments. The standard curves were assessed for two properties, the coefficient of correlation (R2), and the amplification efficiency (E) determined from the slope of the standard curve. As shown in Table 1, high-quality standard curves were produced in the absence or presence of background DNA (R2 > 0.989). Furthermore, neither the presence of background gDNA or cDNA significantly affected amplification efficiency.
Table 1.
Effect of sample complexity on amplification efficiency: standard curves were constructed from a plasmid containing varying copies of the green fluorescent protein gene. The slope, R2 value and efficiency of standard curves are shown in the absence of background material and in the presence of increasing quantities of gDNA or CDNA
| BDC | Water | gDNA | cDNA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.111 | 0.333 | 1 | 3 | 9 | 1 | 3 | 9 | 27 | |
| Slope | −3.82 | −3.39 | −3.26 | −3.20 | −3.56 | −3.31 | −3.43 | −3.48 | −3.57 | −3.36 |
| R2 | 0.993 | 0.993 | 0.989 | 0.994 | 0.997 | 0.992 | 0.995 | 0.995 | 0.995 | 0.993 |
| Efficiency | 83 | 97 | 103 | 105 | 91 | 101 | 96 | 94 | 91 | 98 |
BDC, background DNA concentration (ng/reaction).
We extended this analysis by assessing GFP amplification in the presence of even higher concentrations of DNA ranging between 30.8 and 60 ng. GFP amplification was inhibited in the presence of higher gDNA concentrations (Table 2) whereas cDNA quantities over this range was not inhibitory (data not shown), suggesting that the bulk amount of background nucleic acid may not necessarily be the sole determinant in inhibiting PCR amplification and that the structural nature of the gDNA may also contribute. Mechanical shearing of gDNA effectively removed much of the inhibition of GFP amplification (Table 2).
Table 2.
Effect of sample complexity on amplification efficiency: GFP plasmid amplification in presence of varying concentrations of unsheared or sheared gDNA
| BDC | 0 | 30.8 | 38.6 | 48 | 60 | |
|---|---|---|---|---|---|---|
| Unsheared | Slope | −3.30 | −1.74 | −1.41 | −0.96 | −0.48 |
| R2 | 0.990 | 0.865 | 0.660 | 0.643 | 0.176 | |
| Efficiency | 101 | 275 | N/A | N/A | N/A | |
| Sheared | Slope | −3.30 | −2.97 | −3.17 | −3.15 | −2.78 |
| R2 | 0.990 | 0.994 | 0.986 | 0.995 | 0.980 | |
| Efficiency | 101 | 117 | 107 | 108 | 129 |
N/A, not available due to poor correlation between replicates.
Although the range of gDNA that we typically use in our standard curves does not impair amplification from GFP plasmid, we also wanted to ensure that amplification directly from the gDNA is not affected by the mechanical shearing that may result in loss of target sequences. To examine this, we generated standard curves using sheared or unsheared gDNA in the range of 9–0.111 ng with eight different primer pairs. Shearing the DNA affected neither the slope nor the correlation coefficient for any of the primer pairs tested (Table 3).
Table 3.
Effect of sample complexity on amplification efficiency: effect of shearing on amplification of eight target genes
| Gene Primer Pair | Slope | R2 | Efficiency | |||
|---|---|---|---|---|---|---|
| Unsheared | Sheared | Unsheared | Sheared | Unsheared | Sheared | |
| HsIFNB1 | −3.36 | −3.26 | 0.978 | 0.984 | 99 | 102 |
| HsTLR3 | −3.28 | −3.31 | 0.985 | 0.979 | 102 | 101 |
| HsIRF7 | −2.83 | −3.06 | 0.925 | 0.967 | 125 | 112 |
| HsSCYA2 | −3.09 | −3.35 | 0.988 | 0.990 | 111 | 99 |
| HsPRKR P2 | −3.27 | −3.43 | 0.993 | 0.996 | 102 | 96 |
| HsOAS2 | −3.29 | −3.25 | 0.994 | 0.993 | 102 | 103 |
| HsIFIT1 | −3.36 | −3.36 | 0.918 | 0.982 | 98 | 99 |
| HsGAPD | −3.43 | −3.42 | 0.981 | 0.991 | 96 | 96 |
R2 indicates the degree of correlation between replicates.
Amplification efficiency (%) = [10(−1/slope) − 1] × 100.
Comparison between external standards
Plasmid DNA is often used as an external standard for absolute quantification in real-time PCR. We compared amplification from ∼14.5 fg (∼2574 copies) of plasmid containing the human PRKR gene to amplification from the same target in 9 ng (∼2574 copies) of gDNA extracted from three different tissues (HT, Human tonsil; HS, Human subcarinal lymph node; and HL, Human lung tissue). The Ct score measured in each sample (n = 12) was not significantly different between the groups (Supplementary Figure A).
HT gDNA was used to construct a standard curve from which the copy numbers of eight different target genes were determined in human gDNA from both subcarinal lymph node and lung. Each genomic sample was assessed at serial dilutions that covered the range of the standard curve. At each input level, the number of copies was not significantly different for each of the eight targets. Furthermore, the number of copies determined from each source was in agreement with each other at each dilution (Supplementary Figure B).
The three gDNA sources were compared by constructing standard curves from each and assessing gene expression in various cDNA samples. The number of copies of the PRKR target was determined in 10 ng of cDNA from U937 cell line treated with and without interferon for 4 h and in 1 ng of cDNA from peripheral CD4+ and CD8+ T cells. No significant differences were seen between the values obtained from each standard curve (Supplementary Figure C).
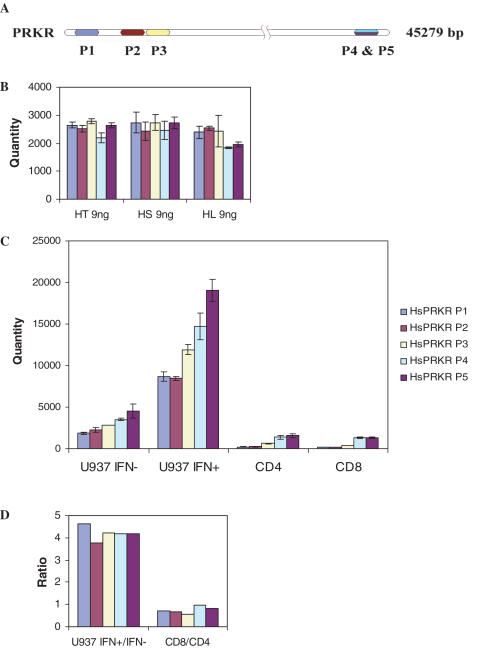
Reproducibility of quantification between multiple primer pairs and the effect of primer sequence position
We have assessed copy numbers using five different primer pairs targeting different regions of the PRKR gene (Figure 3A) in 9 ng of human tonsil, subcarinal lymph node and lung gDNA. No significant differences were observed in the assessed quantity between the three sources of gDNA regardless of the target position (Figure 3B). In contrast, the copy number determined in cDNA samples obtained from the U937 cell line treated with and without interferon for 4 h or from peripheral CD4+ and CD8+ T cells is affected by the target site (Figure 3C). The assessed quantity of transcript copy decreases as the target region moves upstream from the 3′ end of the transcript. Primers closest to each other in the 3′-UTR (HsPRKR P4 and HsPRKR P5 in Figure 3A) are generally in agreement although less so when assessing expression in interferon treated cells due to the high copy numbers which exceed the standard curve. In spite of the difference in copy numbers between each primer pair, the ratios of target copy numbers between IFN treated and untreated samples and between CD8+ and CD4+ samples were comparable for each primer pair (Figure 3D).
Figure 3.
Effect of target position in assessment of PRKR copy number in gDNA and cDNA. (A) Target regions of five primers in PRKR transcript. (B) Absolute quantities in 9 ng of three gDNA (HT, human tonsil; HS, human subcarinal lymph node; HL, human lung) assessed using five primer pairs targeting different regions of the transcript. (C) Absolute quantities in 10 ng of cDNA from IFN treated or untreated U937 human monocytic cell line and 1 ng of cDNA from human peripheral CD4 and CD8 cells assessed using five primer pairs targeting different regions of the transcript. (D) Ratio of PRKR copy numbers between IFN treated and untreated U937 cells, and CD8+ and CD4+ T cells.
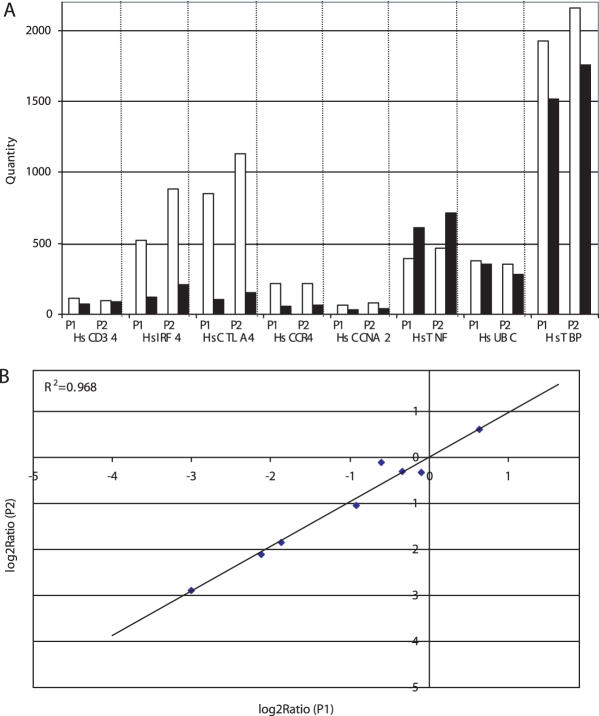
This position-dependent effect is likely due to the efficiency of the reverse transcription reaction, which has long been appreciated, and it highlights the importance of designing primers near the 3′ end to help mitigate this effect. We examined this further by assessing the transcript copy number for eight different genes in CD4+ and CD8+ T cell cDNA using two sets of primer pairs designed against the 3′ region of each gene (Figure 4A). The assessed quantity was highly correlated (R2 = 0.968) between the two sets of primers (Figure 4B).
Figure 4.
Assessment of gene transcript copy number using different real-time PCR targets. Expression level of eight genes was determined in CD4+ (open) and CD8+ (closed) T cells by amplification of two target sites in each gene. Absolute quantity was assessed in 1 ng of cDNA from each type of T cell. (A) The graph shows the quantity determined with each primer pair (P1 versus P2). (B) The correlation (0.968) between Log2 Ratio (CD8+/CD4+) for the two sets of primers is shown.
Real-time PCR and microarray
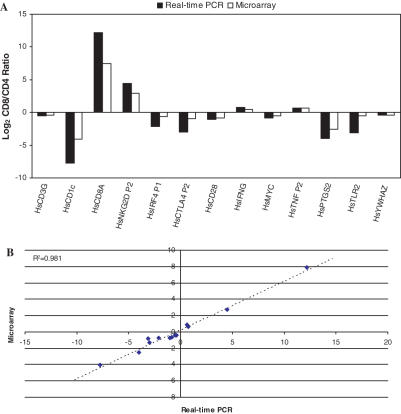
Quantitative real-time PCR is often used to validate the results obtained from microarray experiments (24–26). We examined the relative gene expression profiles of human CD4+ and CD8+ T cells obtained by microarray analysis (M. Hyrcza, M. Ostrowski and S. D. Der, unpublished data) and validated the levels of 43 single-copy genes. Of these, 29 were not different between the two cell types (data not shown). The remaining 14 were differently expressed on the microarray and the direction of change for each was confirmed by real-time PCR although the log2 CD8+/CD4+ ratios tended to be higher in real-time PCR than in microarray analysis (Figure 5A). Nevertheless, the quantities determined by qPCR were highly correlated (R2 = 0.981) with the quantities determined by microarray analysis (Figure 5B).
Figure 5.
Validation of microarray data using real-time PCR with a gDNA standard. (A) Ratio of gene expression between CD4+ and CD8+ T cells as assessed by real-time PCR (closed) or Affymetrix Microarray (open). (B) Comparison of Log2 Ratios for each gene. The slope of the regression line (R2 = 0.981) is 0.59 indicating that microarray values are ∼60% of those seen by real-time PCR.
Mouse genotyping
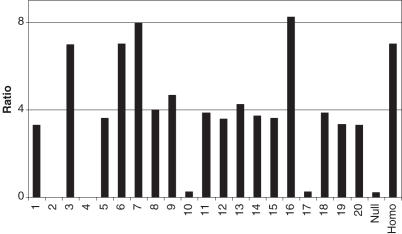
Other applications that have relied upon real-time PCR are single nucleotide polymorphism detection and genotyping (4,13,14). We have applied the gDNA external standard to genotype human-tau transgenic mice using real-time PCR. Mouse-tail gDNA samples were isolated from the F2 generation of a homozygous/null cross. Expected ratios were 1/2 heterozygous, 1/4 homozygous and 1/4 null. The gDNA was amplified with two human-tau specific primer pairs as well as two primer pairs for internal reference genes (GRO1 and IL6). Human tonsil gDNA was used as an external standard to measure human-tau gene, and mouse liver gDNA was used to quantify the internal reference genes. The quantity of each gene obtained by real-time PCR was normalized with the internal reference genes to account for multiple insertions. A clear distinction is evident between the three populations with 12/20 heterozygous, 4/20 homozygous and 4/20 null (Figure 6). Multiple transgene copies of the inserted gene are also indicated, three to four copies in the haploid genome.
Figure 6.
Genotyping of human-tau transgenic mice using mouse-tail gDNA as standard for real-time PCR. Null and homo indicate the expected ratio between absolute quantity of transgene and of internal reference gene in null (tau −/−) and homozygous (tau +/+) transgenic mice.
DISCUSSION
Quantitative real-time PCR is a well-proven and valuable tool for assessing gene expression. When determining the change in expression of a particular gene in response to a specific treatment or the difference in expression between two samples, amplification of an internal control or reference gene that is assumed to be constant between samples is generally used to normalize measurements. While assessment of changes in gene expression is often sufficient for most experimental purposes, the importance of determining the absolute copy number for an expressed sequence can be highlighted in cases where the degree of fold change may over- or under-signify the biological phenomena. For example, an increase from 10 to 100 copies (10-fold change) may carry less biological significance compared to an increase from 1000 to 5000 copies (5-fold change) even though in relative terms the former appears to have a greater induction. Furthermore, with relative quantification it is difficult to compare gene expression levels between studies that do not use the same baseline control, or where the baseline may not be constant. Determination of absolute copy numbers removes the need for a common baseline. This is particularly useful in pathogen load diagnosis (7–9) and cancer marker detection (10–12), in which normal controls are often different between studies.
Using real-time PCR to determine absolute copy number requires that a series of standards of known quantity be assessed in the same assay. The Ct values obtained from these standards can be used to generate a standard curve from which the quantity of unknown samples can be calculated. There is an absolute requirement that each standard contains the same target region that is amplified in the sample. In our laboratory, tissue gDNA is used as an external standard for calculation of copy numbers in unknown samples. There are several advantages to using gDNA over other materials such as plasmid DNA, purified PCR products or reverse-transcribed cRNA. First, the heterogeneous nature of a cDNA population is better represented by gDNA as opposed to the more homogenous pools found in these other materials. Second, the quantity of plasmid DNA necessary to achieve a useable copy number is generally in the femtogram range. Purified PCR product and reverse-transcribed cRNA require similar quantities. For example, 1 ng of plasmid DNA contains ∼100 million copies of target sequence (1.9 × 108 copies of GFP plasmid and 1.2 × 108 copies of PRKR plasmid). After dilution to concentrations at the lower range of spectrophotometric detection, generation of external standards would still require multiple-step dilutions to achieve copy numbers approaching those expected in expressed sequences. This presents an opportunity for the introduction of small errors at each step to be amplified to significant variation in standard quantity. Furthermore, dilution of DNA to such low concentrations is not suitable for long-term storage. The addition of carrier nucleic acids to plasmid DNA may improve its long-term storage, but not solve the problem of multiple-step dilutions. Lastly, the same gDNA pool can be used as an external standard for the amplification of any gene product as it contains a copy of all genes. The other external standards require unique pools that have to be designed and constructed for each gene of interest, a time-consuming process that can also be very costly for large-scale experiments.
Our results show that PCR efficiency is affected by the complexity of the material from which a target is amplified. This includes both the quantity as well as the composition of background DNA. Real-time PCR is inhibited at high background gDNA concentrations and/or in the presence of long strands of structurally complex DNA. While these inhibitory effects do not appear at the gDNA concentrations that are typically used to generate standard curves in our laboratory, it can affect quantification when using higher concentrations of gDNA standards or when assessing copy numbers in unknown gDNA samples. In such circumstances, the inhibitory effects can be overcome by moderate shearing of gDNA, which removes some of this complexity. We have additionally demonstrated that shearing of gDNA does not adversely affect the target amplification within the gDNA due to the potential loss of target sites.
gDNA can be extracted easily from a variety of sources, and it is expected that amplification of a target sequence with a given primer pair should be equally efficient regardless of this source. The standard curves constructed from the amplification of a target region in the PRKR gene from three gDNA preparations (tonsil, subcarinal lymph node and lung) or within a plasmid did not differ significantly in quality or in efficiency of amplification. Furthermore, the quantity of PRKR transcript assessed in various cDNA samples was the same regardless of the gDNA source used to construct a standard curve. The equivalency of these three gDNA standards was further verified by amplification with primers for a number of randomly selected genes.
We have highlighted in this paper the importance of choosing an optimal target region to assess gene expression. Consistent quantification was observed between amplification from target regions that were located in similar regions of a gene. Generally, the assessed copy number determined from targets in the 5′ region of the gene was lower than that determined from targets in the 3′ region. This is likely due to inefficient reverse transcription reaction resulting in incomplete cDNA representation of the 5′ ends of the transcripts. Assessment of copy number was not affected by the primers' target location in gDNA. In general, when amplifying from cDNA, it is therefore preferable to target the 3′-UTR to minimize underestimation of the transcript copy number.
In our laboratory, real-time PCR is used to verify changes in gene expression observed with DNA microarrays. This is the first step to eliminate false positives prior to pursuing further analysis on the functional characterization of identified genes. We have shown in this paper that the real-time PCR using a gDNA standard is effective at verifying the differences in gene expression between two different human T cell types (CD4+ and CD8+). The need for a higher degree of accuracy for assessing global gene expression with high-throughput platforms is increasing, and the ability to determine absolute copy numbers with real-time PCR will play a crucial role in validating the design of these platforms and the algorithms used for their analysis. Real-time PCR is also a valuable tool for other applications beyond assessing gene expression such as assessment of copy number in transgenic mice. We have shown that a litter of mice could effectively be classified into null, heterozygous and homozygous groups for the gene in question.
The use of any external standard does not preclude the need for appropriate internal references, which monitor aspects of sample preparation such as RNA isolation and RT efficiency. While we believe that the relatively simple approach described in our study for quantification of gene targets in cDNA samples provides a richer form of data than relative quantification, there has not yet been methodologies described for achieving absolute quantification of transcript copies in live cells or tissues that have been robustly tested and universally accepted. Simultaneous quantification of an internal reference can be used to further adjust measured target quantities that are independent of the specific experimental procedure. An effective combination of both external standard and internal reference provides the best means by which to most accurately quantify gene expression. Furthermore, intra-assay variability has long been an issue for microarray studies, and is being addressed by development of a set of universal RNA reference materials (27). Adoption of similar reference material for qPCR studies could further contribute toward standardization of gene expression data especially when comparing real-time PCR data to that obtained from microarray analysis.
In summary, there are many advantages to using gDNA as an external standard for quantitative real-time PCR. This includes the structural similarity of sheared gDNA to cDNA, the ability to use preparations of gDNA without excessive dilution, and the greater stability of these more concentrated solutions during long-term storage. Although there has been an effort to construct a DNA external standard containing multiple target sequences for several primer pairs (23), gDNA is a truly universal external standard applicable to all endogenous genes in the genome. The cost, time and effort required for the preparation of gDNA are minimal compared to those for construction of external standards using purified plasmid or cDNA, which contain a unique target sequence. With all these advantages, we anticipate that the use of gDNA as external standard will contribute significantly to the accurate quantification of copy number using quantitative real-time PCR.
Acknowledgments
We thank Drs Jeremy Mogridge for providing the GFP plasmid and Mingyao Liu for providing subcarinal lymph nodes gDNA. This work was supported by grants from CIHR and Genome Canada to S.D.D. Funding to pay the Open Access publication charges for this article was provided by Genome Canada and CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rose'Meyer R.B., Mellick A.S., Garnham B.G., Harrison G.J., Massa H.M., Griffiths L.R. The measurement of adenosine and estrogen receptor expression in rat brains following ovariectomy using quantitative PCR analysis. Brain Res. Brain Res. Protoc. 2003;11:9–18. doi: 10.1016/s1385-299x(02)00219-2. [DOI] [PubMed] [Google Scholar]
- 2.Li X., Wang X. Application of real-time polymerase chain reaction for the quantitation of interleukin-1beta mRNA upregulation in brain ischemic tolerance. Brain Res. Brain Res. Protoc. 2000;5:211–217. doi: 10.1016/s1385-299x(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 3.Tseng C.P., Cheng A.J., Chang J.T., Tseng C.H., Wang H.M., Liao C.T., Chen I.H., Tseng K.C. Quantitative analysis of multidrug-resistance mdr1 gene expression in head and neck cancer by real-time RT–PCR. Jpn. J. Cancer Res. 2002;93:1230–1236. doi: 10.1111/j.1349-7006.2002.tb01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yip S.P., Lee S.Y., To S.S., Wong M.L. Improved real-time PCR assay for homogeneous multiplex genotyping of four CYP2C9 alleles with hybridization probes. Clin. Chem. 2003;49:2109–2111. doi: 10.1373/clinchem.2003.023622. [DOI] [PubMed] [Google Scholar]
- 5.Vandenbroucke II, Vandesompele J., Paepe A.D., Messiaen L. Quantification of splice variants using real-time PCR. Nucleic Acids Res. 2001;29:E68–E68. doi: 10.1093/nar/29.13.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickert L., Steinkruger S., Abiaka M., Bolkenius U., Purps O., Schnabel C., Gressner A.M. Quantitative monitoring of the mRNA expression pattern of the TGF-beta-isoforms (beta 1, beta 2, beta 3) during transdifferentiation of hepatic stellate cells using a newly developed real-time SYBR Green PCR. Biochem. Biophys. Res. Commun. 2002;295:330–335. doi: 10.1016/s0006-291x(02)00669-1. [DOI] [PubMed] [Google Scholar]
- 7.Polanco J.C., Rodriguez J.A., Corredor V., Patarroyo M.A. Plasmodium vivax: parasitemia determination by real-time quantitative PCR in Aotus monkeys. Exp. Parasitol. 2002;100:131–134. doi: 10.1016/S0014-4894(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 8.Mengelle C., Pasquier C., Rostaing L., Sandres-Saune K., Puel J., Berges L., Righi L., Bouquies C., Izopet J. Quantitation of human cytomegalovirus in recipients of solid organ transplants by real-time quantitative PCR and pp65 antigenemia. J. Med. Virol. 2003;69:225–231. doi: 10.1002/jmv.10277. [DOI] [PubMed] [Google Scholar]
- 9.Dworkin L.L., Gibler T.M., Van Gelder R.N. Real-time quantitative polymerase chain reaction diagnosis of infectious posterior uveitis. Arch. Ophthalmol. 2002;120:1534–1539. doi: 10.1001/archopht.120.11.1534. [DOI] [PubMed] [Google Scholar]
- 10.Mocellin S., Rossi C.R., Marincola F.M. Quantitative real-time PCR in cancer research. Arch. Immunol. Ther. Exp. (Warsz) 2003;51:301–313. [PubMed] [Google Scholar]
- 11.Bolufer P., Barragan E., Verdeguer A., Cervera J., Fernandez J.M., Moreno I., Lerma E., Esquembre C., Tasso M., Fuster V., et al. Rapid quantitative detection of TEL-AML1 fusion transcripts in pediatric acute lymphoblastic leukemia by real-time reverse transcription polymerase chain reaction using fluorescently labeled probes. Haematologica. 2002;87:23–32. [PubMed] [Google Scholar]
- 12.Kreuzer K.A., Lass U., Bohn A., Landt O., Schmidt C.A. LightCycler technology for the quantitation of bcr/abl fusion transcripts. Cancer Res. 1999;59:3171–3174. [PubMed] [Google Scholar]
- 13.Aarskog N.K., Vedeler C.A. Real-time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot-Marie-Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Hum. Genet. 2000;107:494–498. doi: 10.1007/s004390000399. [DOI] [PubMed] [Google Scholar]
- 14.Ponchel F., Toomes C., Bransfield K., Leong F.T., Douglas S.H., Field S.L., Bell S.M., Combaret V., Puisieux A., Mighell A.J., et al. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak K.J. ABI Prism 7700 Sequence Detection System, User Bulletin 2. 1997. PE Applied Biosystems. Foster City, CA. [Google Scholar]
- 16.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Saint D.A. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal. Biochem. 2002;302:52–59. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- 18.Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 19.Rutledge R.G., Cote C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 2003;31:e93. doi: 10.1093/nar/gng093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peirson S.N., Butler J.N., Foster R.G. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whelan J.A., Russell N.B., Whelan M.A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods. 2003;278:261–269. doi: 10.1016/s0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- 22.Poola I. Molecular assay to generate expression profile of eight estrogen receptor alpha isoform mRNA copy numbers in picogram amounts of total RNA from breast cancer tissues. Anal. Biochem. 2003;314:217–226. doi: 10.1016/s0003-2697(02)00614-0. [DOI] [PubMed] [Google Scholar]
- 23.Dumoulin F.L., Nischalke H.D., Leifeld L., von dem Bussche A., Rockstroh J.K., Sauerbruch T., Spengler U. Semi-quantification of human C–C chemokine mRNAs with reverse transcription/real-time PCR using multi-specific standards. J. Immunol. Methods. 2000;241:109–119. doi: 10.1016/s0022-1759(00)00210-6. [DOI] [PubMed] [Google Scholar]
- 24.Vallat L., Magdelenat H., Merle-Beral H., Masdehors P., Potocki de Montalk G., Davi F., Kruhoffer M., Sabatier L., Orntoft T.F., Delic J. The resistance of B-CLL cells to DNA damage-induced apoptosis defined by DNA microarrays. Blood. 2003;101:4598–4606. doi: 10.1182/blood-2002-06-1743. [DOI] [PubMed] [Google Scholar]
- 25.de Vos S., Hofmann W.K., Grogan T.M., Krug U., Schrage M., Miller T.P., Braun J.G., Wachsman W., Koeffler H.P., Said J.W. Gene expression profile of serial samples of transformed B-cell lymphomas. Lab. Invest. 2003;83:271–285. doi: 10.1097/01.lab.0000053913.85892.e9. [DOI] [PubMed] [Google Scholar]
- 26.Rajeevan M.S., Ranamukhaarachchi D.G., Vernon S.D., Unger E.R. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods. 2001;25:443–451. doi: 10.1006/meth.2001.1266. [DOI] [PubMed] [Google Scholar]
- 27.Cronin M., Ghosh K., Sistare F., Quackenbush J., Vilker V., O'Connell C. Universal RNA reference materials for gene expression. Clin. Chem. 2004;50:1464–1471. doi: 10.1373/clinchem.2004.035675. [DOI] [PubMed] [Google Scholar]