Abstract
We characterized two new gene cassettes in an Acinetobacter isolate: one harbored the metallo-β-lactamase (IMP-4) gene blaIMP-4, the other harbored the rifampin ADP-ribosyltransferase (ARR-2) gene arr-2, and both arrayed with the aminoglycoside acetyltransferase [AAC(6′)-Ib7] gene cassette aacA4 in two separate class 1 integrons. The epidemiology of these gene cassettes in isolates from blood cultures obtained from 1997 to 2000 was studied. Isolates bearing either the blaIMP-4 or the arr-2 gene cassette or both represented 17.5% (10 of 57) of isolates in 1997, 16.1% (10 of 62) in 1998, 2.5% (1 of 40) in 1999, and 0% (0 of 58) in 2000. These two gene cassettes, probably borne on two separate integrons, were found in at least three genomic DNA groups, with evidence of clonal dissemination in the intensive care unit during 1997 to 1998. Seventeen of the 52 Acinetobacter baumannii (genomic DNA group 2) isolates from 1997 to 2000 harbored intI1, but only one was positive for these gene cassettes, whereas 20 of the 21 intI1-positive isolates of all other genomic DNA groups were positive for either or both of them. Reduced susceptibility to imipenem and rifampin was seen only in isolates harboring the blaIMP-4 and arr-2 cassettes, respectively. The aminoglycoside phosphotransferase [APH(3′)-VIa] gene aph(3′)-VIa was detected in all 21 isolates for which the MIC of amikacin was ≥8 μg/ml, with or without aacA4, whereas aacA4 alone was found in isolates for which the MIC of amikacin was 0.5 to 2 μg/ml. Significant differences between the 17 intI1-positive and 47 intI1-negative isolates belonging to genomic DNA group 3 from 1997 to 1998 in the MICs of amikacin, gentamicin, imipenem, sulfamethoxazole, and ceftazidime were observed (Mann-Whitney test, P < 0.001 to 0.01).
In the past decade, there has been an explosion of interest in the integron, a genetic element which facilitates gene dissemination among bacteria of different species and perhaps genera, thus establishing the possibility of a wide dispersal of antimicrobial resistance. These genetic elements are marked by the presence of two conserved regions between which gene cassettes are integrated at a recombination site known as attI. The 5′-conserved region of an integron contains an integrase gene, intI, and a promoter (Pc) which drives the expression of any integrated gene cassettes. Individual gene cassettes, existing in a free circularized form when not integrated, are comprised of an open reading frame (ORF) and a 59-base element (59-be) at the 3′ end. The 59-be is a family of recombination sites that act as substrates to integrase-mediated recombination (43). To date, over 70 resistance cassettes have been described (32, 45). According to the 3′-conserved sequence (3′-CS), integrons are divided into three classes. The majority of the resistance cassette-bearing integrons belong to class 1, and these carry, at the 3′-CS, the truncated quaternary ammonium compound resistance gene qacEΔ1, the sul1 sulfonamide resistance gene, and orf5, of unknown function (20).
Acinetobacter spp. are increasingly important nosocomial pathogens worldwide and particularly in the Hong Kong region (5, 36, 61). The therapy of Acinetobacter infections is complicated by multidrug resistance against aminoglycosides, extended-spectrum cephalosporins, and fluoroquinolones (1, 5, 9, 14, 50). The integron was found to be highly prevalent (50 to 80%) among nosocomial strains of Acinetobacter baumannii (genomic DNA group 2) (18, 49), and its association with resistance dissemination has attracted much attention. An Italian study of 36 epidemiologically unrelated A. baumannii strains isolated in six hospitals over 11 years showed that 13 of the 16 class 1 integron-positive strains carried the same array of cassettes, prompting the hypothesis of horizontal transfer of the entire integron or ancient acquisition (17).
One of the major cassette-borne resistance genes reported in recent years is the blaIMP gene encoding the metallo-β-lactamase IMP belonging to molecular class B (7). The clinical importance of IMP is highlighted by its ability to hydrolyze all penems, cephems, and carbapenems. To date, eleven blaIMP alleles have been described, and most appeared to be linked to integrons. Six of the alleles have been found in acinetobacters from various localities worldwide (9, 10, 15, 22, 23, 27, 37, 44, 58, 63). The blaIMP-4 gene in acinetobacters isolated in our hospital and encoding an IMP-4 metallo-β-lactamase that has 95.6 and 89.3% amino acid sequence homology to IMP-1 and IMP-2, respectively (9), was previously described. Further characterization here showed that blaIMP-4 was borne on a cassette arraying with three other resistance gene cassettes, namely qacG2, aacA4, and catB3, in a class 1 integron. In addition, we describe a second class 1 integron bearing the cassette array arr-2-aacA4. We examined the epidemiology of the blaIMP-4 and arr-2 cassettes in acinetobacters from blood cultures isolated during 1997 to 2000. The fact that these cassettes were apparently capable of conferring increased resistance to imipenem and rifampin (important antibacterial and antimycobacterial agents) should cause concern. However, the integrons bearing blaIMP-4 and arr-2 were readily recovered from blood culture isolates from 1997 to 1998 but apparently disappeared after 1999. This raises the question of the ability of these resistance genetic elements to persist, highlighting the need for further epidemiological studies to address such issues.
MATERIALS AND METHODS
PCR, RFLP, and DNA sequencing.
All the oligonucleotide primers used in this work are summarized in Table 1. All the PCR mixtures (50 μl) contained 25 pmol of each primer, 2 μl of genomic DNA prepared by boiling a colony in 200 μl of sterile distilled water, 0.5 U of Taq (AP Biotech, Uppsala, Sweden), and the supplied buffer. Unless otherwise stated in the reference quoted, the thermocycle employed for the PCRs was 94°C for 2 min and then 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by a final 3 min at 72°C. Amplicons were digested by the appropriate restriction endonucleases (Table 2) according to the supplier's instructions when restriction fragment length polymorphism (RFLP) analysis was performed. Restriction patterns were obtained by agarose (2%) gel electrophoresis and compared after ethidium bromide staining. When sequencing of PCR amplicons was required, amplicons were first cloned by using a TA cloning kit (Invitrogen Life Technologies, The Netherlands) and then sequenced by using a PCR sequencing kit with the ALFexpress DNA sequencer (AP Biotech) according to the manufacturers' instructions. All the DNA sequences determined were done at least twice from two PCR amplifications. Comparisons of DNA sequences and calculations of similarity were performed by the software ClustalW (56).
TABLE 1.
PCR primers used in this work
| Forward and reverse primers | Sequence 5′-3′ | Amplicon (size in bp) | Position | Remark | Reference |
|---|---|---|---|---|---|
| int1f imp12 | CAG AAG ACG GCT GCA CTG AA AGT GTG TCC TGG GCC TGG | blaIMP-4 upstream (1,833) | −1359 to +474 (blaIMP-4 ATG) | Characterization of the upstream region of blaIMP-4 in strain 74510 | This work |
| imp11 int3c | ATG AGC AAA GTT ATC TGT ATT CT AAA GCA GTC TTG ACC TGA | blaIMP-4 downstream (2,812) | +741 (blaIMP-4 ATG) to 2071 downstream of blaIMP-4 | Characterization of the downstream region of blaIMP-4 in strain 74510 | This work |
| int2a int6b | AAC CGA GGA TGC GAA CCA CT CCG AGC CGC TCG TAT AG | intI1-arr-2-aacA4-3′CS (3,602) | +382 (intI1 ATG) to +482 (orf5 ATG) | Characterization of the regions flanking arr-2 in strain 74510 | This work |
| int2a imp12 | AAC CGA GGA TGC GAA CCA CT AGT GTG TCC TGG GCC TGG | intI1-blaIMP-4 (1,008) | +382 (intI1 ATG) to + 474 (blaIMP-4 ATG) | Combining with RFLP for detection of the intI1-blaIMP-4 sequencea | This work |
| int1b aac61bB | CAT CCA AGC AGC AAG CGC GTT A ACC CCG GIT TCT CGT AGC AT | 5′ CS-arr-2-aacA4 (1,182) | −131 (arr-2 ATG) to +460 (aacA4 ATG) | Combining with RFLP for the detection of the 5′ CS-arr-2-aacA4 sequencea | This work |
| int1e int1c | TCG TAG AGA CGT CGG AAT GG CCG AGG CAT AGA CTG TAC AA | Class 1 integrase gene intI1 (965) | −40 to +925 (intI1 ATG) | For detection of the intI1 gene | 27 |
| 5cs 3cs | GGC ATC CAA GCA GCA AG AAA GCA GAC TTG ACC TGA | Entire integron (variable) | Variable | For confirmation of the presence of class 1 integron | 27 |
| ardra1 ardra2 | TGG CTC AGA TTG AAC GCT TAC CTG TTA CGA CTT CA | 16S rRNA gene (1,500) | Variable | ARDRA of acinetobacter for genomic DNA group determination | 11 |
| BL BR | TAT GAG TGG CTA AAT CGA T CCC GCT TTC TCG TAG CA | aacA4 (395) | +115 to +509 (aacA4 ATG) | For detection of the aacA4 gene | 39 |
| aph1 aph2 | ATA CAG AGA CCA CCA TAC AGT GGA CAA TCA ATA ATA GCA AT | aph(3′)-VIa (234) | +140 to +374 [aph(3′)-VIa ATG] | For detection of the aph(3′)-VIa gene | 60 |
See Table 2 for the expected sizes of the restriction fragments.
TABLE 2.
Expected fragments in the PCR-RFLP screening for the intI1-blaIMP-4 and 5′ CS-arr-2-aacA4 sequences
| Restriction enzyme | Expected fragments (bp) |
|---|---|
| For detection of the intI1-blaIMP-4 (1,008-bp) sequence | |
| ApaLI | 96, 912 |
| BsmAI | 157, 851 |
| HinCII | 57, 268, 288, 395 |
| HindIII | 292, 716 |
| For detection of the 5′CS-arr-2-aacA4 (1,182-bp) sequence | |
| BsoBI | 73, 350, 759 |
| BspHI | 160, 599, 423 |
Isolates from blood cultures.
Table 3 shows the number of acinetobacters obtained during the 4-year period 1997 to 2000 from blood cultures. Isolates from the same patient were included if the positive blood cultures were taken at least 7 days apart or yielded acinetobacters either of different genomic DNA groups or with different novel patterns produced by amplified ribosomal DNA restriction analysis (ARDRA [11]) (Table 1). They were stored on nutrient agar slopes at room temperature until further examination. Isolates obtained in 1997 and the first 8 months of 1998 had been used in another study (21). No outbreak of Acinetobacter infection was noted during the study years. Methods of genus confirmation by the transformation assay of Juni and genomic DNA group determination by ARDRA have been described elsewhere (11, 25).
TABLE 3.
Acinetobacters in blood cultures from 1997 to 2000a
| Genomic DNA group | No. of acinetobacters in blood cultures
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1997
|
1998
|
1999
|
2000
|
||||||||||||
| Total | PCR positive for:
|
Total | PCR positive for:
|
Total | PCR positive for:
|
Total | PCR positive for:
|
||||||||
| intI1 | A | B | C | intI1 | A | B | C | intI1 | C | intI1 | |||||
| 2 | 8 | 3 | 16 | 8 | 1 | 12 | 1 | 16 | 5 | ||||||
| 3 | 22 | 8 | 3 | 5 | 23 | 9 | 2 | 6 | 1 | 8 | 12 | 1 | |||
| 5 | 2 | 4 | |||||||||||||
| 8/9 | 1 | 2 | |||||||||||||
| 10 | 1 | 1 | |||||||||||||
| 12 | 2 | ||||||||||||||
| 13TU | 10 | 12 | 5 | 15 | |||||||||||
| 15 | 1 | 3 | |||||||||||||
| 16 | 1 | ||||||||||||||
| 17 | 4 | 1 | 1 | 1 | 1 | ||||||||||
| U | 11 | 1 | 1 | 9 | 7 | 1 | 1 | 6 | |||||||
| Totalb | 56 (48) | 13 (12) | 3 (3) | 5 (5) | 2 (2) | 63 (53) | 17 (16) | 3 (3) | 6 (6) | 1 | 40 (30) | 2 (2) | 1 | 58 (41)c | 6 (5) |
Abbreviations: A, both the intI1 -blaIMP-4 and 5′ CS-arr-2-aacA4 sequences; B, the intI1-blaIMP-4 sequence; C, the 5′ CS-arr-2-aacA4 sequence; U, unclassifiable by ARDRA.
Numbers in parentheses indicate the numbers of patients.
One isolate from 2000 was not done.
PFGE.
Pulsed-field gel electrophoresis (PFGE) fingerprints were generated by using a contour-clamped homogeneous electric field electrophoresis apparatus (CHEF-MAPPER; Bio-Rad, Richmond, Calif.) as described in a previous study (21). The restriction endonuclease ApaI was used for the in situ digestion of intact Acinetobacter genomic DNA embedded in 1% agarose gel blocks prepared according to previously described methods (21). Samples were loaded into 1% certified PFGE grade (Bio-Rad) agarose gels and electrophoresed with 0.5× Tris-borate-EDTA buffer with an electric field of 6 V/cm, an included angle of 120°, and a pulse time of 5 to 35 s over 32 h at 14°C. Images of ethidium bromide-stained gels were digitized by use of a gel documentation system (Imagemaster; AP Biotech). Clusters of possibly related isolates were identified and interpreted by using the Dice coefficient of similarity and the unweighted pair group method (21, 55).
MICs of antimicrobials.
The agar dilution method was used to determine the MICs of the following major classes of agents: amikacin, gentamicin, ceftazidime, nalidixic acid, tetracycline, sulfamethoxazole, and rifampin (all from Sigma, St. Louis, Mo.), ciprofloxacin (Bayer, Elberfeld, Germany), imipenem (Merck Sharp and Dohme, Hoddesdon, United Kingdom), netilmicin (Schering-Plough Corporation, Kenilworth, N.J.), and sulbactam (Pfizer, Sandwich, United Kingdom). Inocula of 104 CFU/spot were inoculated onto Mueller-Hinton agar plates with a multipoint inoculator (Dynatech Laboratories, Alexandria, Va.), and the plates were incubated at 35°C for 18 h. The MIC was defined as the lowest concentration which inhibited visible growth. Control strains Escherichia coli ATCC 35218 and Pseudomonas aeruginosa ATCC 27853 were included (35). Nonparametric tests (SPSS version 10) were used for statistical analysis.
Southern hybridization.
In order to identify whether the integrons were plasmid borne, the extraction method described by Kado and Liu (26) was employed to obtain plasmids of over 50 Kb from acinetobacters. The plasmid extractions were then electrophoresed by using an agarose gel (0.7%), blotted onto nylon membranes following standard protocols (46), and hybridized with two probes. One probe was a blaIMP-4 fragment (1 to 474 bp), and the other was an arr-2 fragment (36 to 417 bp). A DIG system was used for signal detection, and the manufacturer's instructions were followed (Boehringer Mannheim, Mannheim, Germany).
Nucleotide sequence accession numbers.
The sequences described in this article are available from the EMBL and GenBank databases under the accession numbers AF445082 and AY038837.
RESULTS
Class 1 integron bearing the blaIMP-4-qacG2-aacA4-catB3 cassette array.
By use of a PCR method based on the published metallo-β-lactamase IMP-1 gene sequence (27), a variant of the gene (738 bp) from an acinetobacter (strain 74510) in a culture collection had previously been identified (9). Strain 74510 belonging to genomic DNA group 13TU was isolated from a wound in 1995. The metallo-β-lactamase was subsequently designated IMP-4, and the gene was designated blaIMP-4 (9). From the presence of adjacent GTTRRRY integrase-dependent recombination motifs, blaIMP-4 was speculated to be gene cassette borne (9).
In this work, a series of PCR primers were designed to amplify the flanking regions of blaIMP-4, based also on the class 1 integron bearing the IMP-1 sequence (27). Two sets of primers produced positive results: (i) int1f and imp12 produced an amplicon of 1,833 bp, including 1,359 bp upstream of the IMP-4 gene; (ii) imp11 and int3c produced an amplicon of 2,812 bp, including 2,071 bp downstream of the IMP-4 gene (Table 1). DNA sequencing of the amplicons confirmed that blaIMP-4 was borne on a gene cassette in a class 1 integron. The 59-be of the blaIMP-4 cassette has the typical inverted repeat sequence related to a 59-bp consensus sequence (54).
Only one plasmid (estimated size, 275 Kb) was extracted from the isolate by using a method specialized for large plasmids (26). The presence of the IMP-4 sequence on this large plasmid was demonstrated by Southern hybridization of the extracted plasmid, with a blaIMP-4 fragment used as the probe (result not shown).
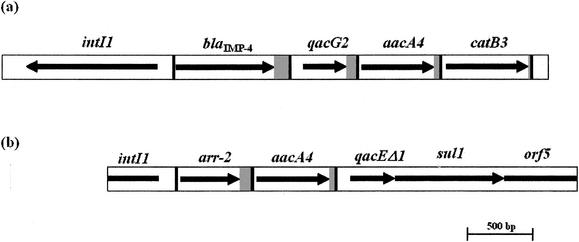
The sequence of the integron bearing the blaIMP-4 gene cassette identified in this work is schematically represented in Fig. 1. A typical class 1 integrase gene, intI1, was found upstream of the blaIMP-4 gene cassette, and downstream were three more gene cassettes: (i) a variant of qacG encoding a small protein that confers resistance to quaternary ammonium compounds (27), (ii) aacA4 encoding an aminoglycoside acetyltransferase [aac(6′)-Ib7] (28), and (iii) catB3 encoding a chloramphenicol acetyltransferase (6). The intI1 gene was identical to that of In1 except for the two strength-related hexameric motifs TTGACA(−35) and TAAGCT(−10) in the promoter Pc. While TTGACA(−35) was found to constitute a strong promoter, TAAGCT(−10) had previously been seen in a weak promoter (29). This hexamer combination has also been reported to exist in an integron in Enterobacter aerogenes before, but its exact strength is yet to be determined (38).
FIG. 1.
Schematic representation of the sequences of the integrons bearing the blaIMP-4 cassette (a) and the arr-2 cassette (b) determined in this work. The thick vertical lines show the borders of the gene cassettes. The arrows represent the ORFs, and the grey areas show the 59-be of individual cassettes.
The blaIMP-4 gene had 95.6% nucleotide sequence identity to the blaIMP-1 gene initially found in Japan (37). Examination of all the 59-be linked to the published IMP alleles (blaIMP-1 in a class 1 integron [GenBank accession no. AJ223604] [27], blaIMP-1 in a class 3 integron [accession no. AF419627] [3], blaIMP-2 in a class 1 integron [accession no. AJ243491] [44], blaIMP-7 in a class 1 integron [accession no. AF318077] [15], blaIMP-8 in a class 1 integron [accession no. AF322577] [63]) revealed that the blaIMP-4 cassette 59-be is more closely related to that of blaIMP-7 (sharing 93% nucleotide sequence identity) than to that of blaIMP (86% sequence identity), blaIMP-2 (44% sequence identity), and blaIMP-8 (27% sequence identity). However, the ORFs of blaIMP-4 and blaIMP-7 share a lower identity (89%) than their 59-be comparison.
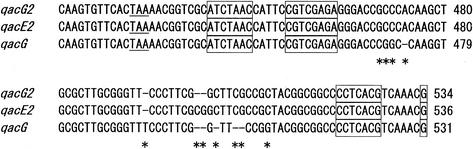
The qacG2 cassette downstream of blaIMP-4 including a long untranslated leading sequence is identical to the qacG cassette described for In31 (27) except for the 59-be (88% nucleotide sequence identity) (Fig. 2). Interestingly, the 59-be of qacG2 is almost identical to the qacE2 cassette found in a class 1 integron in Aeromonas salmonicida subsp. Salmonicida except for a 2-bp (CG) insertion between the 2L and 2R core sites (Fig. 2). The qacE2 cassette has also a long untranslated leading sequence identical to qacG2 in length and >99% identical in sequence, but the ORF of qacE2 has two nucleotide differences translating into two amino acid changes (Phe13→Ser and Gly57→Ala). The long untranslated leading sequence is believed to be a secondary promoter containing two putative strength-related hexameric motifs reported also as existing in qacG in In31 (27).
FIG. 2.
Comparison of the 59-be of the gene cassettes qacG2 (accession no. AF445082), qacG (AJ223604), and qacE2 (AF327731). Underlining indicates stop codons of the ORFs, boxes show imperfect inverted repeat core sites, and asterisks indicate base differences. The base numbers shown correspond to the individual gene cassettes.
The aacA4 gene cassette is identical to the aacA4 cassette previously found in a class 1 integron in Campylobacter jejuni (28). It contains the aminoglycoside acetyltransferase AAC(6′)-Ib7 gene. The substrate-determining residue 102 is occupied by a serine; hence, it should confer resistance to tobramycin and slightly increased resistance to gentamicin but no resistance to amikacin (42). Compared to the first correctly described aacA4, the cassette has 2 extra C bases in a 2C-rich region in the 59-be (43). Also, the ORFs differ by one T→C accounting for the substrate-decisive Leu102→Ser and by one T→A changing the Val183 to Asp (43).
The ORF of the catB3 cassette in this work differs from the catB3 previously described for Enterobacter by one amino acid residue (Tyr81→Cys), and the two cassettes share 99% nucleotide sequence identity (6). Major differences were in the 59-be. The catB3 in this work has a four-base deletion between the 2L and 2R core sites and a T→C base change between the 2R and 1R core sites.
The presence of 3′-CS typical of sul1-associated integrons following the catB3 gene cassette suggested that this integron contained only the four described cassettes (19).
Class 1 integron bearing the arr-2-aacA4 cassette array.
During the process of identifying sequences flanking the IMP-4 gene by using PCR mapping, a set of primers, int2a and int6b, resulted in an amplicon of 3,602 bp that did not contain blaIMP-4 (Table 1). Upon sequencing, the amplicon was found to be a fragment of a class 1 integron that contained two gene cassettes: (i) rifampin ADP-ribosyltransferase (arr-2) and (ii) the aminoglycoside acetyltransferase AAC(6′)-Ib7 (aacA4). The sequence is schematically represented in Fig. 1. Three hundred eighty-two base pairs of a class 1 integrase gene intI1 was found at the 5′ end of the PCR amplicon originally targeting the blaIMP-4-aacA4 integron. The Pc promoter has the hexameric motifs TCGACA(−35) and TAAACT(−10). TAAACT(−10) has previously been found to constitute a strong Pc promoter (29), but TCGACA(−35) has not been described before. However, this combination was previously reported as occurring in a sequence upstream to an aminoglycoside 3′-N-acetyltransferase gene, aacC1, described in Serratia marcescens; but the strength of it as a promoter is not known (GenBank accession no. S68049 [24]).
The arr-2 cassette was found also in a class 1 integron in Klebsiella pneumoniae previously (GenBank accession no. AJ277027 [4]). It differs from another arr-2 cassette reported to occur in P. aeruginosa and E. coli by one base (A→G), translating to one amino acid change (Lys98→Arg) (34, 57). The ribosyltransferase ARR-2 described in this work has 55% amino acid sequence identity to ARR-1 found in the naturally resistant Mycobacterium smegmatis (41). Related arr-2 gene cassettes have also been reported from two other studies, both in the same geographical area as ours (16, 34).
The aacA4 cassette encoding the aminoglycoside acetyltransferase AAC(6′)-Ib7 is identical to that of the blaIMP-4-bearing integron. Southern hybridization of the plasmid extract with an arr-2 fragment used as a probe demonstrated the presence of this integron on the large plasmid (estimated size, 275 Kb), where the blaIMP-4-bearing integron also resided (result not shown).
Distribution of blaIMP-4, arr-2, and intI1.
A PCR-based method targeting the intI1-blaIMP-4 fragment (1,008 bp) was designed to detect the intI1-blaIMP-4 sequence in our acinetobacters isolated from blood cultures in 1997 to 2000. The identity of the amplicons was confirmed by digestion by four restriction endonucleases (Table 2). Also, a similar approach with an amplicon of 1,182 bp digested by two restriction endonucleases was employed to detect the 5′-CS-arr-2-aacA4 sequence (Table 2). Another PCR, previously described, was used to detect the intI1 sequence (27).
Table 3 shows the numbers and genomic DNA groups of acinetobacters obtained from blood cultures from 1997 to 2000 and their distribution according to genomic DNA groups. Results of PCR to the class I integrase gene intI1, the intI1-blaIMP-4, and/or 5′-CS-arr-2-aacA4 sequences are also shown.
The blaIMP-4 cassette linked to intI1 was found only in genomic DNA groups 3 and 2 (A. baumannii), and the 5′-CS-arr-2-aacA4 was found in genomic DNA groups 3 and 17 as well as in an unclassifiable isolate. Isolates bearing either the intI1-blaIMP-4 sequence or the arr-aacA4 sequence or both were found mainly in 1997 and 1998, representing 17.5% of the isolates in 1997, 16.1% in 1998, and 2.5% in 1999. Of the 52 A. baumannii isolates from 1997 to 2000, 17 (33%) harbored intI1 but only 1 was PCR positive for intI1-blaIMP-4 and 5′-CS-arr-2-aacA4; whereas 20 of the 21 intI1-positive isolates of all other genomic DNA groups were associated with the presence of intI1-blaIMP-4 and/or 5′-CS-arr-2-aacA4 sequences.
Figure 3 shows the relatedness of all the genomic DNA group 3 isolates from 1997 to 1998. The five isolates harboring both blaIMP-4 and arr-2 cassettes were grouped into two clusters of four isolates and one isolate, linked together at a Dice coefficient of similarity of ≤65% (Fig. 3). Four of these five isolates were obtained from the ICU. The 11 isolates harboring the blaIMP-4cassette only were divided into four clusters linked together at a Dice coefficient of similarity of ≤65%, indicating that they were possibly not related (Fig. 3). Six of these 11 isolates came from the ICU. PFGE of all genomic DNA group 2 isolates from 1997 to 1998 showed that the single isolate containing both cassettes was linked to others with a Dice coefficient of similarity of <65% (data not shown).
FIG. 3.
Dendrogram based on Dice coefficients of similarity for 45 genomic DNA group 3 isolates from blood cultures obtained in 1997 and 1998. The dendrogram was constructed by the unweighted pair group method with arithmetic averages UPGMA (BioNumerics; Applied Maths, Sint-Martens-Latem, Belgium). A, isolates bearing both the intI1-blaIMP-4 and 5′-CS-arr-2-aacA4 sequences; B, isolates bearing the intI1-blaIMP4 sequence (one isolate was not studied); C, isolates bearing the 5′-CS-arr-2-aacA4 sequence; 3C, 8A, 8B, 4E, CBMT, CCC, etc., designations of wards.
MIC studies.
With the exception of the unclassifiable isolate from 1999 (Table 4), reduced susceptibility to imipenem and rifampin was seen only in isolates harboring the intI1-blaIMP-4 and 5′-CS-arr-2-aacA4 sequences, respectively. The MIC of imipenem for the 17 isolates bearing intI1-blaIMP-4 was 1 to 16 μg/ml. For three of the four isolates bearing 5′-CS-arr-2-aacA4 only, the imipenem MIC was 0.03 to 0.25 μg/ml. The MIC of imipenem for the remaining isolate (1999, unclassifiable) was >32 μg/ml. The MIC of rifampin for the 10 isolates bearing 5′-CS-arr-2-aacA4 was ≥32 μg/ml, and that for the 11 nonbearers was 1 to 2 μg/ml. The MICs of 11 antimicrobial agents for genomic DNA group 3 isolates positive for both or either of the intI1-blaIMP-4 and 5′-CS-arr-2-aacA4 sequences were compared with those for isolates from 1997 to 2000 negative for those sequences (Table 4). There is a significant difference between the 17 intI1-positive and the 48 intI1-negative isolates (1997 to 2000) in the MICs of amikacin, gentamicin, imipenem, sulfamethoxazole, and ceftazidime (Mann-Whitney U test, P < 0.001 to 0.01) (Table 4). Similar statistical results were obtained when the 17 intI1-positive isolates were compared with 27 intI1-negative isolates (1 isolate was not done) from 1997 to 1998. The numbers of isolates in other genomic DNA groups were too small for statistical analysis (data not shown).
TABLE 4.
Comparison of MICs for genomic DNA group 3 isolates from blood cultures 1997-2000a
| Antibiotic | MIC (μg/ml) for isolates:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bearing intI1-blaIMP-4 and/or 5′CS-arr-2-aacA4 (n = 17)b
|
Not bearing either intI1-blaIMP-4 or 5′CS-arr-2-aacA4 (n = 41)c
|
|||||||||
| Geomean | MIC50 | MIC90 | Min MIC | Max MIC | Geomean | MIC50 | MIC90 | Min MIC | Max MIC | |
| AMK | 18.08 | 32 | 64 | 0.50 | 64 | 1.33 | 2 | 4 | 0.06 | 64 |
| GEN | 1.44 | 1 | 8 | 0.06 | 64 | 0.61 | 1 | 2 | 0.06 | 128 |
| NET | 1.45 | 1 | 8 | 0.25 | 32 | 0.71 | 1 | 2 | 0.06 | 8 |
| IPM | 3.69 | 4 | 8 | 0.25 | 16 | 0.15 | 0.13 | 0.25 | 0.03 | 1 |
| CIP | 0.42 | 0.50 | 2 | 0.03 | 32 | 0.33 | 0.25 | 1 | 0.02 | 128 |
| SUL | 1.08 | 1 | 2 | 0.50 | 4 | 0.84 | 1 | 2 | 0.06 | 4 |
| TET | 1.57 | 2 | 4 | 0.13 | 128 | 1.66 | 2 | 4 | 0.13 | 256 |
| SMX | 128 | 128 | 128 | 128 | 128 | 16.83 | 8 | 1,024 | 0.25 | 1,024 |
| RIF | 6.80 | 2 | 128 | 1 | 128 | 1.93 | 2 | 4 | 0.25 | 32 |
| CAZ | 96.22 | 128 | 128 | 16 | 256 | 8.56 | 8 | 32 | 0.50 | 128 |
| CHL | 128 | 256 | 256 | 16 | 256 | 47.55 | 64 | 256 | 0.50 | 512 |
Geomean, geometric mean of MIC; MIC50 and MIC90, MICs at which 50 and 90% of isolates are inhibited, respectively; Min MIC, minimum MIC; Max MIC, maximum MIC; AMK, amikacin; GEN, gentamicin; NET, netilmicin; IPM, imipenem; CIP, ciprofloxacin; SUL, sulbactam; TET, tetracycline; SMX, sulphamethoxazole; RIF, rifampin; CAZ, ceftazidime; CHL, chloramphenicol. By the Mann-Whitney test, for isolates bearing the intI1-blaIMP-4 and/or 5′CS-arr-2-aacA4 sequences versus isolates not bearing those sequences, P < 0.001 to 0.01 for AMK, GEN, IMP, SMX, CAZ; for other agents, P > 0.05.
Fifteen isolates were tested for NET, and 5 were tested for CHL.
Forty-one isolates were tested for all agents except CHL, for which 21 were tested.
Detection of the aph(3′)-VIa and aacA4 genes in isolates from 1997 to 1998.
There were 21 isolates for which the MIC of amikacin was ≥8 μg/ml; 15 of them were positive by PCR for both aph(3′)-VIa (60) and aacA4, and 6 were positive only for aph(3′)-VIa. Seventy-six isolates for which the MIC of amikacin was ≤0.12 to 4 μg/ml were examined in a similar manner; all were negative for aph(3′)-VIa, but five isolates (MIC of amikacin, 0.5 to 2 μg/ml) were positive for aacA4 PCR and all of them were carriers of either or both cassettes on the two integrons described. None of the PCR products was sequenced to determine the exact aacA4 type.
DISCUSSION
To date, over 70 cassettes capable of conferring antimicrobial resistance have been characterized (45), including the largest class 1 integron found so far, In53, in an E. coli isolate, containing nine different antibiotic resistance genes of different classes (34). In comparison, there are only a few epidemiological studies of specific resistance integrons. The carriage of integron may vary among different types of clinical specimens and different genera (13, 30, 47, 48, 59, 62). In our study, 33% (17 of 52) of A. baumannii (genomic DNA group 2) isolates from 1997 to 2000 carried class 1 integron but only 1 isolate carried both the blaIMP-4 and arr-2 cassettes (Table 3); whereas, for other genomic DNA groups, intI1 carriage appeared to be closely related to the presence of blaIMP-4 and arr-2-aacA4 (Table 3). No isolate belonging to genomic DNA group 13TU, another member of the Acinetobacter calcoaceticus-A. baumannii complex, was PCR positive for intI1, or intI1-blaIMP-4, or 5′-CS-arr-2-aacA4 sequences (5). In studies of resistant nosocomial isolates of A. baumannii, 70 to 80% were found to be PCR positive for the intI1 gene (14, 40). As reported by others, we found that integron carriage was significantly associated with reduced susceptibility to the aminoglycosides, β-lactams, ciprofloxacin, and cotrimoxazole tested (30).
In a previous study, it was reported that blaIMP-4 was present in the Prince of Wales Hospital culture collections of acinetobacters as early as 1994 (9). Cassettes of other blaIMP alleles (IMP-2, IMP-7, and IMP-8) have since been described for other members of Enterobacteriaeae and as integron borne (15, 44, 63). Further characterization of blaIMP-4 here showed that it was also integron cassette borne. Acinetobacters that were PCR positive for either or both of the intI1-blaIMP-4 and 5′-CS-arr-2-aacA4 sequences were nearly all from patients who had stayed in an ICU in which there had not been an overt change in the practice of antibiotic prescription over the study period. The two integrons bearing these gene cassettes were found in at least three genomic DNA groups (Table 3), indicating the presence of horizontal dissemination. There was also evidence of longitudinal dissemination of possibly clonally related isolates during 1997 to 1998 (Fig. 3). The four isolates that were PCR positive for both intI1-blaIMP-4 and 5′-CS-arr-2-aacA4 formed a cluster with a Dice coefficient of similarity of >80% (Fig. 3) (21). Another cluster with a similar Dice coefficient of similarity was formed by the three isolates that were PCR positive for intI1-blaIMP-4 only (Fig. 3).
It is not clear why the carriage of both integrons bearing the intI1-blaIMP-4 and/or 5′-CS-arr-2-aacA4 sequences had declined from blood culture isolates by 1999. There was also a corresponding fall in the carriage of class 1 integrons (Table 3) by isolates from 1999 to 2000. No specific containment measures for infection control were implemented in the ICU during this period, as these isolates were not clustered temporally. It is worth noting that the imipenem MIC for intI1-blaIMP-4-positive isolates was only modestly raised (1 to 16 μg/ml). Integron-borne resistance to older antibiotics such as trimethoprim and the early aminoglycosides, e.g., dfrIa-aadA1a and dfr12-orfF-aadA2, is frequently conserved, suggesting that these gene cassettes have become preserved and stably integrated over a long period of time (8, 31). Should there be a genetic linkage of the two resistance cassettes discussed here to other resistance determinants, removal of selective pressure may not bring about a reduction of resistance within a useful time (12). To devise effective preventive programs, there is therefore a need to understand fully the appearance, maintenance, and decline of antibiotic resistance.
Gene cassettes bearing resistance to rifampin have been reported from two other studies, both in the same geographical area as ours (16, 34). In our hospital, the arr-2 gene cassette appeared to be a major contributor to rifampin resistance in acinetobacters at the time of study, since for all arr-2 positive isolates the MIC of rifampin was ≥32 μg/ml whereas for all negative isolates it was ≤8 μg/ml.
In clinical isolates of Acinetobacter spp. the production of aminoglycoside-modifying enzymes is thought to account for most of the resistance (2, 33, 50, 51). PCR and hybridization data showed that some strains contained more than one aminoglycoside resistance gene (50, 52, 53). Of the aminoglycoside-modifying enzymes detected in Acinetobacter so far, only the APH(3′)-VI and some versions of AAC(6′)-I enzymes confer resistance to amikacin (39, 60). The aph(3′)-VIa gene encoding the APH(3′)-VIa phosphotransferase was detected in all of our 21 isolates for which the MIC of amikacin was ≥8 μg/ml with or without aacA4 gene cassettes. The exact identification of the aacA4 cassettes encountered in the blood culture isolates is uncertain because the amplicons were not sequenced.
The results of our study on blood culture collections of acinetobacters over a 4-year period show that the blaIMP-4-bearing and arr-2-bearing integrons, apparently residing on a large plasmid(s), were disseminated horizontally in three different genomic DNA groups and longitudinally mainly in the ICU during 1997 to 1998. They were associated with increased resistance to imipenem and rifampin. Their carriage became undetectable in 1999 to 2000, suggesting that they were yet to be established firmly as resistance determinants in Acinetobacter and judicial use of antimicrobials may delay such an outcome. Nevertheless, further studies similar to ours should be carried out for a better understanding of the impact of integrons on the persistence of antimicrobial resistance in clinical practice.
Acknowledgments
This study was supported by a grant (4290/99 M) from the Research Grants Council, Hong Kong, SAR, People's Republic of China.
REFERENCES
- 1.Afzal-Shah, M., and D. M. Livermore. 1998. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J. Antimicrob. Chemother. 41:576-577. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, M. H., A. Rechenmacher, and A. Bock. 1980. Interaction between aminoglycoside uptake and ribosomal resistance mutations. Antimicrob. Agents Chemother. 18:798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arakawa, Y., M. Murakami, K. Susuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlet, G., D. Nadjar, J. L. Herrmann, J. L. Donay, M. Rouveau, P. H. Lagrange, and A. Philippon. 2001. Plasmid-mediated rifampin resistance encoded by an arr-2-like gene cassette in Klebsiella pneumoniae producing an ACC-1 class C β-lactamase. Antimicrob. Agents Chemother. 45:2971-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunny, K. L., R. M. Hall, and H. W. Stokes. 1995. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob. Agents Chemother. 39:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, C. Y., L. L. Chang, Y. H. Chang, T. M. Lee, and S. F. Chang. 2000. Characterization of drug resistance gene cassettes associated with class 1 integrons in clinical isolates of Escherichia coli from Taiwan, ROC. J. Med. Microbiol. 49:1097-1102. [DOI] [PubMed] [Google Scholar]
- 9.Chu, Y.-W., M. Afzal-Shah, E. T. S. Houang, M.-F. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Da Silva, G. J., M. Correia, C. Vital, G. Ribeiro, J. C. Sousa, R. Leitao, L. Peixe, and A. Duarte. 2002. Molecular characterization of blaIMP-5, a new integron-borne metallo-beta-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 215:33-39. [DOI] [PubMed] [Google Scholar]
- 11.Dijkshoorn, L., B. Van Harsselaar, I. Tjernberg, P. J. Bouvet, and M. Vaneechoutte. 1998. Evaluation of amplified ribosomal DNA restriction analysis for identification of Acinetobacter genomic species. Syst. Appl. Microbiol. 21:33-39. [DOI] [PubMed] [Google Scholar]
- 12.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. (Erratum, 357: 1890.) [DOI] [PubMed] [Google Scholar]
- 13.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 14.Gallego, L., and K. J. Towner. 2001. Carriage of class 1 integrons and antibiotic resistance in clinical isolates of Acinetobacter baumannii from northern Spain. J. Med. Microbiol. 50:71-77. [DOI] [PubMed] [Google Scholar]
- 15.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M.-F. I. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girlich, D., L. Poirel, A. L. A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum-β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gombac, F., M. L. Riccio, G. M. Rossolini, C. Lagatolla, E. Tonin, C. Monti-Bragadin, A. Lavenia, and L. Dolzani. 2002. Molecular characterization of integrons in epidemiologically unrelated clinical isolates of Acinetobacter baumannii from Italian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob. Agents Chemother. 46:3665-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez, G., K. Sossa, H. Bello, M. Dominguez, S. Mella, and R. Zemelman. 1998. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol. Lett. 161:125-128. [DOI] [PubMed] [Google Scholar]
- 19.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 21.Houang, E. T. S., Y.-W. Chu, C. M. Leung, K. Y. Chu, J. Berlau, K. C. Ng, and A. F. B. Cheng. 2001. Epidemiology and infection control implications of Acinetobacter spp. in Hong Kong. J. Clin. Microbiol. 39:228-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyobe, S., H. Kusadokoro, J. Ozaki, N. Matsumura, S. Minami, S. Haruta, T. Sawai, and K. O'Hara. 2000. Amino acid substitutions in a variant of IMP-1 metallo-beta-lactamase. Antimicrob. Agents Chemother. 44:2023-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyobe, S., H. Kusadokoro, A. Takahashi, S. Yomoda, T. Okubo, A. Nakamura, and K. O'Hara. 2002. Detection of a variant metallo-beta-lactamase, IMP-10, from two unrelated strains of Pseudomonas aeruginosa and an Alcaligenes xylosoxidans strain. Antimicrob. Agents Chemother. 46:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javier Teran, F., M. Alvarez, J. E. Suarez, and M. C. Mendoza. 1991. Characterization of two aminoglycoside-(3)-N-acetyltransferase genes and assay as epidemiological probes. J. Antimicrob. Chemother. 28:333-346. [DOI] [PubMed] [Google Scholar]
- 25.Juni, E. 1972. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J. Bacteriol. 112:917-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J. M. Frère, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, M. D., S. Sanchez, M. Zimmer, U. Idris, M. E. Berrang, and P. F. McDermott. 2002. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob. Agents Chemother. 46:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Freijo, P., A. C. Fluit, F. J. Schmitz, V. S. Greks, J. Verhoef, and M. E. Jones. 1998. Class 1 integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42:689-696. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Freijo, P., A. C. Fluit, F. J. Schmitz, J. Verhoef, and M. E. Jones. 1999. Many class 1 integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob. Agents Chemother. 43:686-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, G. H., F. J. Sabatelli, L. Naples, R. S. Hare, and K. J. Shaw. 1995. The most frequently occurring aminoglycoside resistance mechanisms—combined results of surveys in eight regions of the world. J. Chemother. 7(Suppl. 2):17-30. [PubMed] [Google Scholar]
- 34.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing; 9th informational supplement. M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 36.Ng, T. K. C., J. M. Ling, A. F. B. Cheng, and S. R. Norrby. 1996. A retrospective study of clinical characteristics of Acinetobacter bacteremia. Scan. J. Infect. Dis. 101(Suppl.):26-32. [PubMed] [Google Scholar]
- 37.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters, E. D. J., M. A. Leverstein-van Hall, A. T. A. Box, J. Verhoef, and A. C. Fluit. 2001. Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 45:2961-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ploy, M. C., H. Giamarellou, P. Bourlioux, P. Courvalin, and T. Lambert. 1994. Detection of aac(6′)-I genes in amikacin-resistant Acinetobacter spp. by PCR. Antimicrob. Agents Chemother. 38:2925-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan, S., H. Venter, and E. R. Dabbs. 1997. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob. Agents Chemother. 41:2456-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rather, P. N., H. Munayyer, P. A. Mann, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174:3196-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 44.Riccio, M., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe-Magnus, D. A., and D. Mazel. 1999. Resistance gene capture. Curr. Opin. Microbiol. 2:483-488. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schmitz, F. J., D. Hafner, R. Geisel, P. Follmann, C. Kirschke, J. Verhoef, K. Kohrer, and A. C. Fluit. 2001. Increased prevalence of class 1 integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in a German university hospital. J. Clin. Microbiol. 39:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitz, F. J., P. Martinez-Freijo, S. Theis, A. C. Fluit, J. Verloef, H. P. Heinz, and M. E. Jones. 1998. Class 1 integrons: prevalence and impact on antibiotic susceptibility in 278 consecutive unrelated Gram negative blood isolates. Clin. Microbiol. Infect. 5:496-498. [DOI] [PubMed] [Google Scholar]
- 49.Seward, R. J. 1999. Detection of integrons in worldwide nosocomial isolates of Acinetobacter spp. Clin. Microbiol. Infect. 5:308-318. [DOI] [PubMed] [Google Scholar]
- 50.Seward, R. J., T. Lambert, and K. J. Tower. 1998. Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J. Med. Microbiol. 47:455-462. [DOI] [PubMed] [Google Scholar]
- 51.Shannon, K., and I. Phillips. 1982. Mechanisms of resistance to aminoglycoside in clinical isolates. J. Antimicrob. Chemother. 9:91-102. [DOI] [PubMed] [Google Scholar]
- 52.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw, K. J., R. S. Hare, F. J. Sabatelli, M. Rizzo, C. A. Cramer, L. Naples, S. Kocal, H. Munayyer, P. Mann, G. H. Miller, L. Verbist, H. V. Landuyt, Y. Glupcaynski, M. Catalno, and M. Woloj. 1991. Correlation between aminoglycoside resistance profiles and DNA hybridization of clinical isolates. Antimicrob. Agents Chemother. 35:2253-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 55.Tenover, F. C., R. D. Arbeit, R. V. Goerling, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tribuddharat, C., and M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tysall, L., M. W. Stockdale, P. R. Chadwick, M. F. Palepou, K. J. Towner, D. M. Livermore, and N. Woodford. 2002. IMP-1 carbapenemase detected in an Acinetobacter clinical isolate from the UK. J. Antimicrob. Chemother. 49:217-218. [DOI] [PubMed] [Google Scholar]
- 59.van Belkum, A., W. Goessens, C. van der Schee, N. Lemmens-den Toom, M. C. Vos, J. Cornelissen, E. Lugtenburg, S. de Marie, H. Verbrugh, B. Lowenberg, and H. Endtz. 2001. Rapid emergence of ciprofloxacin-resistant Enterobacteriaceae containing multiple gentamicin resistance-associated integrons in a Dutch hospital. Emerg. Infect. Dis. 7:862-871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vila, J., J. Ruiz, M. Navia, B. Becerril, I. Garcia, S. Perea, I. Lopez-Hernandez, I. Alamo, F. Ballester, A. M. Planes, J. Martinez-Beltran, and T. Jimenez De Anta. 1999. Spread of amikacin resistance in Acinetobacter baumannii strains isolated in Spain due to an epidemic strain. J. Clin. Microbiol. 37:758-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, J. T., L. C. McDonald, S. C. Chang, and M. Ho. 2002. Community-acquired Acinetobacter baumannii bacteremia in adult patients in Taiwan. J. Clin. Microbiol. 40:1526-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan, J. J., W. C. Ko, and J. J. Wu. 2001. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-β-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:2368-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]