Abstract
Odorant/receptor binding and initial olfactory information processing occurs in olfactory receptor neurons (ORNs) within the olfactory epithelium. Subsequent information coding involves high-frequency spike synchronization of paired mitral/tufted cell dendrites within olfactory bulb (OB) glomeruli via positive feedback between glutamate receptors and closely-associated gap junctions. With mRNA for connexins Cx36, Cx43 and Cx45 detected within ORN somata and Cx36 and Cx43 proteins reported in ORN somata and axons, abundant gap junctions were proposed to couple ORNs. We used freeze-fracture replica immunogold labeling (FRIL) and confocal immunofluorescence microscopy to examine Cx36, Cx43 and Cx45 protein in gap junctions in olfactory mucosa, olfactory nerve and OB in adult rats and mice and early postnatal rats. In olfactory mucosa, Cx43 was detected in gap junctions between virtually all intrinsic cell types except ORNs and basal cells; whereas Cx45 was restricted to gap junctions in sustentacular cells. ORN axons contained neither gap junctions nor any of the three connexins. In OB, Cx43 was detected in homologous gap junctions between almost all cell types except neurons and oligodendrocytes. Cx36 and, less abundantly, Cx45 were present in neuronal gap junctions, primarily at “mixed” glutamatergic/electrical synapses between presumptive mitral/tufted cell dendrites. Genomic analysis revealed multiple miRNA (micro interfering RNA) binding sequences in 3′-untranslated regions of Cx36, Cx43 and Cx45 genes, consistent with cell-type-specific post-transcriptional regulation of connexin synthesis. Our data confirm absence of gap junctions between ORNs, and support Cx36- and Cx45-containing gap junctions at glutamatergic mixed synapses between mitral/tufted cells as contributing to higher-order information coding within OB glomeruli.
Introduction
Major advances in understanding data encoding and initial information processing in the olfactory system resulted in awarding the Nobel Prize for Physiology or Medicine to Linda Buck and Richard Axel (reviewed by Firestein, 2005). Although each mammalian olfactory receptor neuron (ORN) is derived from a common cell lineage, each ORN expresses only one of the several hundred different odorant receptor genes (Buck & Axel, 1991; Buck, 2000), suggesting an exquisitely regulated mechanism for single-cell-specific receptor gene expression. Following odorant detection and primary information encoding in the olfactory epithelium (OE), the second level of odorant information processing occurs within individual glomeruli in the olfactory bulb (OB), where high-frequency spike synchronization occurs between receptor-specific pairs of mitral/tufted cell dendrites (Christie et al., 2005). Mitral/tufted cell-pair synchronization apparently involves direct positive feedback between glutamate receptors and closely associated Cx36-containing gap junctions at “mixed” (electrical plus chemical) synapses (Christie et al., 2005); moreover, genetic deletion of the Cx36 gene results in loss of mitral/tufted cell synchrony. Independently, neuronal gap junctions in glomeruli were shown by thin-section transmission electron microscopy (TEM) to occur almost exclusively between the dendrites of mitral/tufted cell pairs and between mitral/tufted cells and other intrinsic neurons of the OB, but none were detected between central processes of ORNs and mitral/tufted cells or between ORN terminals and periglomerular neurons in the OB (Kosaka & Kosaka, 2003, 2004, 2005).
In the OE, mRNAs for Cx36, Cx43 and Cx45 were detected in ORN somata, and Cx36 and Cx43 protein immunoreactivities were reported to be abundant in most of their somata and axons (Zhang et al., 2000; Zhang & Restrepo, 2002, 2003). Thus, Zhang and coworkers concluded that gap junctions containing these connexins were relatively abundant in most ORNs over major areas of the OE. However, numerous freeze-fracture studies did not detect gap junctions between ORNs or between their unmyelinated axons (Menco, 1980b; Miragall et al., 1996b; Blinder et al., 2003). These conflicting data have led to confusion regarding possible involvement of Cx36, Cx43 and/or Cx45 in proposed ORN/ORN gap junctions in olfactory signaling (Zufall, 2005; Copelli et al., 2005). The presence of Cx36 protein in ORNs would be consistent with widespread distribution of Cx36 in ultrastructurally-defined neuronal gap junctions elsewhere in the CNS (Rash et al., 2001). Likewise, Cx45 is reported to be a neuronal connexin based on detection of reporter gene proteins activated by the Cx45 promoter in retinal neurons (Maxeiner et al., 2003, 2005). Moreover, Cx45 protein was detected in ultrastructurally-identified gap junctions within the inner plexiform layer of the retina (Kamasawa et al., 2004; Li et al., unpublished observations). In contrast, freeze-fracture replica immunogold labeling (FRIL) has shown localization of Cx43 exclusively in CNS glial cells (Rash et al., 2001). Thus, unambiguous detection of Cx43 in ultrastructurally-identified gap junctions in ORNs would represent a novel expression pattern, but one that might be plausible. Unlike CNS neurons, ORNs are neurons of the peripheral nervous system that are derived from dermal placodes (Schlosser, 2005), which are subject to different tissue inductive factors, and therefore, might express unusual combinations of connexins.
We used confocal immunofluorescence microscopy and FRIL to visualize gap junctions and to identify their constituent connexin proteins in principal cells of the olfactory mucosa, olfactory nerve and external layers of the OB. As an aid to identifying cell types, we also labeled several replicas for aquaporin-4 (AQP4) and N-methly-d-aspartate (NMDA) glutamate receptors (NM-DAR; as NMDAR1). These combined approaches indicated complete absence of Cx43 protein in ORN somata, axons and terminals; no Cx36, Cx45 or gap junctions between ORN somata; and minimal or no Cx36 or Cx45 in ORN axons or axon terminals. As one potential explanation for possible post-transcriptional repression of connexin protein synthesis in ORNs, we examined genomic data and identified “seed matching” sequences for >25 micro interfering RNAs (miRNAs) in the 3′UTRs of each of the connexin mRNAs previously detected in ORNs.
In OB glomeruli, immunoreactivities for Cx36 protein and, less abundantly, for Cx45 are demonstrated in gap junctions on large dendrites, usually at identified glutamatergic “mixed” (chemical plus electrical) synapses. These data for co-localization of Cx36 and glutamate receptors in glomeruli of OB support current models for high-frequency direct feedback and neuronal spike synchronization as an essential component in olfactory information coding.
Materials and methods
ANIMALS AND ANTIBODIES
Young adult (6–12 weeks old) C57BL/6 and CD1 mice and adult and 4-day postnatal Sprague-Dawley rats used in this study were obtained under protocols approved by the Institutional Animal Care and Use Committees of Colorado State University, the University of Colorado Health Sciences Center, and the University of Manitoba. Antibodies against Cx36 included polyclonal 36–4600, 51–6200 and 51–6300, and monoclonal 37–4600 (Zymed Laboratories Inc., South San Francisco, CA; now Invitrogen/Zymed Laboratories), as well as previously characterized polyclonal Ab298 (Rash et al., 2000). Antibodies against Cx43 included monoclonal MAB3068 and MAB3067S and polyclonal AB1728 (Chemicon International Inc., Temecula, CA), polyclonal antibody 71–0700, monoclonal antibody 35–5000 (Invitrogen/Zymed) and polyclonal Ab18A (generously provided by Dr. E. L. Hertzberg, Albert Einstein College of Medicine, New York, NY). Monoclonal anti-Cx45 antibodies included MAB3100 and MAB3101 (Chemicon) and a newly available polyclonal anti-Cx45 (40–7000; Invitrogen/Zymed). Polyclonal antibody to AQP4 was from Chemicon (AB3068). Antibodies to NMDAR1 included Chemicon MAB363 and Pharmingen 60021A (now 556308; BDBiosciences, San Jose, CA). Antibodies to alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) glutamate receptors included “panAMPA” (anti-GluR1–4; courtesy of Elek Molnar, University of Bristol, Bristol, UK). We also investigated antibodies to the cyclic nucleotide-gated alpha-2 (CNGA2) channel subunit (APC-045; Alomone Labs Ltd, Jerusalem, Israel) as a potential cell-specific FRIL marker for ORNs in OE and OB (Dhallan et al., 1992), but FRIL labeling was not obtained, suggesting that the CNGA2 protein did not survive the required SDS washing steps. Consequently, the CNGA2 antibody currently is not useful for FRIL.
LIGHT MICROSCOPE IMMUNOFLUORESCENCE
Mice were deeply anesthetized with equithesin (3 ml/kg) and then sequentially perfused transcardially with 3 ml of cold (4°C) 50 mM sodium phosphate buffer (PB), pH 7.4, containing 0.9% NaCl, 0.1% sodium nitrite and heparin (1 unit/ml), followed by perfusion with 40 ml of cold 0.16 M sodium phosphate buffer, pH 7.1, containing 1% formaldehyde and 0.2% picric acid, followed by perfusion with 10 ml of PB containing 10% sucrose. The midline nasal septum together with adjoining olfactory epithelium was removed and stored at 4°C for 48 to 72 h in cryoprotectant consisting of PB containing 10% sucrose. Olfactory epithelium supported by blocks of liver tissue was sectioned at 10 μm thickness on a cryostat, collected on gelatinized glass slides and stored at −34°C until use. Sections were thawed, washed for 20 min in 50 mM Tris-HCl, pH 7.4, containing 1.5% sodium chloride (TBS) and 0.3% Triton X-100 (TBSTr), and incubated for 24 h at 4°C with primary monoclonal or polyclonal antibodies diluted in TBSTr containing 5% normal donkey serum. All primary antibodies for immunofluorescence were used at a concentration of 2–3 μg/ml, except anti-Cx43 Ab18A, which was used at a dilution of 1:5000. Sections were then washed for 1 h in TBSTr and incubated with secondary antibody for 1.5 h at room temperature. Secondary antibodies used were FITC-conjugated horse anti-mouse IgG (Vector Laboratories, Burlingame, CA, USA) diluted 1:100, or Alexa Fluor 488-conjugated donkey anti-rabbit IgG (Vector Laboratories) diluted 1:1000 in TBSTr. After secondary antibody incubations, all sections were washed in TBSTr for 20 min, followed by two 15 min washes in 50 mM Tris-HCl buffer, pH 7.4, and cover-slipped after application of antifade medium. Fluorescence was examined on a Zeiss Axioskop2 fluorescence microscope with image capture using Axiovision 3.0 software (Carl Zeiss Canada, Toronto, Canada), and an Olympus Fluoview IX70 confocal microscope with image capture using Olympus Fluoview software (Markham, Ontario, Canada). The final images were assembled using Photoshop 6.0 (Adobe Systems, San Jose, CA, USA) and Corel Draw 8 software.
TISSUE PREPARATION FOR ELECTRON MICROSCOPY AND FRIL
Adult male (6 weeks old) C57BL/6 and CD1 mice and adult and 4-day postnatal Sprague-Dawley rats were deeply anesthetized with a mixture of ketamine/xylazine (80 mg/kg; 8 mg/kg) or sodium pentobarbital (40 mg/kg), perfused transcardially for 2–5 min with 0.15 M phosphate buffer (PB, pH 7.4) containing 10 IU/ml of heparin, and fixed by perfusion for 5–10 min with either 0%, 0.1%, 1.2% or 2% formaldehyde in PB (for FRIL) or 2.5% glutaraldehyde in PB (for thin-section transmission electron microscopy, TEM). The septum, olfactory turbinates, portions of cribriform plates, and olfactory bulbs were dissected at 4°C, cut into 100 μm-thick slices using a refrigerated Lancer 1000 Vibratome (Technical Products International, St. Louis, MO), and prepared for FRIL TEM or thin-section TEM.
PREPARATION FOR THIN-SECTION ELECTRON MICROSCOPY
Glutaraldehyde-fixed tissue slices were post-fixed with 1% OsO4 in PB, rinsed in dH2O, and either stained en bloc for 16 h with 0.5% aqueous unbuffered uranyl acetate (UAc, pH 4.5), or dehydrated in methanol series and stained for 1 h with 4% UAc in absolute methanol, a procedure that preserves glycogen and other acid-soluble polysaccharides and mucopolysaccharides (Hsu et al., 1974). Samples stained with aqueous UAc were dehydrated in graded ethanol series. Both sets of dehydrated samples were rinsed in 100% acetone, embedded in plastic resin (10% Epon 812, 20% Araldite 502, 70% dodecenyl acetic anhydride) with 1.25–1.5% DMP-30 (dimethyl amino phenol) added as catalyst, and polymerized at 70°C for 24 h (Rash et al., 1969). Silver and pale gold sections (60–100 nm thick) were cut using a Dupont Sorval Porter-Blum MT-2B ultramicrotome (no longer manufactured; successor available from RMC Products, Tucson, AZ), picked up on 200 mesh copper grids, post stained with lead citrate (Venable & Coggeshall, 1965), air dried, and examined by TEM.
FREEZE FRACTURE
For FRIL, tissue slices were cryoprotected by gradual infiltration with glycerol to a final concentration of 30% and frozen by contact with a −185°C “copper mirror” (Phillips & Boyne, 1984). Frozen samples of olfactory epithelium and olfactory bulb were fractured in a JEOL/RFD 9010 C freeze-fracture device (RMC Products; no longer manufactured), pre-coated with 1 nm of vaporized carbon (Winkler et al., 2002), replicated with 1–1.5 nm of vaporized platinum, and coated with 5–20 nm of vaporized carbon. A 200 mesh gold “index” grid (Electron Microscopy Sciences, Fort Washington, PA) was bonded to each replicated but still frozen sample using a droplet of 1.5–2% Lexan (GE Plastics, Pittsfield, MA) dissolved in dichloroethane (Rash et al., 1993). After evaporation of the dichloroethane solvent at −35°C, the resulting Lexan-replica-tissue-grid “sandwich” was thawed and photomapped by confocal microscopy (Zeiss LSM510 Meta or Molecular Dynamics Multiport 2001) prior to SDS washing and immunogold labeling.
IMMUNOGOLD LABELING
Tissue remnants were removed from the Lexan-stabilized replicas by washing in 2.5% SDS detergent for 29 h at 48.5°C. SDS washing leaves a thin film of lipid and protein molecules adhering to the highly absorptive platinum/carbon replica (Fujimoto, 1995, 1997; Rash & Yasumura, 1999; Rash et al., 2004b). However, unlike labeling in thin-section images, where sources are immobilized in plastic, the partially solubilized proteins and lipids continuously detach during SDS washing, and labeling is often detected 50–80 nanometers away from the source, or several times the 30 nanometers length attributable to the double antibody bridge (Rash & Yasumura, 1999). Moreover, specificity of labeling to the immediate vicinity of gap junctions, for example, is maintained because completely solubilized proteins from each gap junction are immediately diluted into the comparatively “infinite reservoir” of the labeling dish.
After SDS washing, replicas were rinsed in “blocking buffer” (10% goat serum, 1.5% fish gelatin in Sorensen’s phosphate buffer, pH 7.4; Dinchuk et al., 1987) and labeled for 90–180 min using appropriate combinations of mouse monoclonal and rabbit polyclonal antibodies. Labeled samples were rinsed and counter-labeled using goat anti-rabbit IgG and goat anti-mouse IgG, each coupled to a separate size of uniform-diameter gold beads (6 nm, 12 nm, and 18 nm from Jackson; 10 and 20 nm from Chemicon; and 5-, 10-, 20-and 30-nm from BBInternational (Cardiff, UK). Because both electron opacity and cross-sectional area increase with the square of the diameter of the gold beads used for labeling, the electron contrast/electron density (“detectability”) of gold beads increases with the fourth power of the diameter. Although the smaller gold beads are difficult to detect, even at 100,000× magnification, the 5 nm and 6 nm gold beads were used because their labeling efficiency is four- to eight-fold greater than that of 18 nm or 20 nm gold beads (Rash et al., 2004b; Kamasawa et al., 2005), and they are easily counted in high-magnification, reverse stereoscopic electron micrographs (see below).
FREEZE-FRACTURE TERMINOLOGY; MEMBRANE SPLITTING AT GAP JUNCTIONS
This report uses the internationally-recognized freeze-fracture nomenclature (Branton et al., 1975) in which the “P-face” represents the replicated fracture face of the membrane leaflet remaining in contact with the “plasm” (cytoplasm, nucleoplasm, mitochondrial plasm); whereas the “E-face” represents the replicated membrane leaflet remaining in contact with the “extraplasmic” space (extracellular space, extranuclear space, extramitochondrial space). In addition, the abbreviation “IMP” is used synonymously to designate either “intramembrane particle” or “intramembrane protein”. Because gap junctions represent paired cell appositions, and because connexins of two coupled cells are always separated at the point of connexon contact in the extracellular space (Staehelin, 1974), both the P- and E-fracture faces of gap junctions always overlie a complete plaque of connexons. Thus, hemiplaques label equally well beneath both E- and P-faces (Fujimoto, 1995; Yasumura & Rash, 1997).
FRIL ANALYSIS OF CONNEXINS AND AQUAPORINS IN OLFACTORY MUCOSA AND OLFACTORY BULB
To detect gap junctions, identify the connexin(s), and identify the several cell types present in the olfactory mucosa and olfactory bulb, 24 samples were prepared. Of these, 12 were examined by FRIL after labeling with various combinations of monoclonal and polyclonal antibodies to Cx36, Cx43, Cx45, AQP4 and NMDAR1 and 12 were discarded due to undissolved bone, high “background”, excessive replica fragmentation, or other technical faults. In most replicas, we labeled multiple connexins in multiple cell types because failure to detect a connexin in one cell type does not constitute strong evidence for its absence unless the same connexin is simultaneously demonstrated in ultrastructurally-defined gap junctions in other cell types in the same replica or in companion FRIL replicas prepared simultaneously under identical conditions. Consequently, replicas of OE were labeled for Cx36 at the same time as replicas of OB, or contained both OE and OB in the same replica (see next).
Of the six replicas from OE that were examined (Table 1), one was double-labeled for Cx43 (one polyclonal and two monoclonal antibodies; two sizes of gold), four were double-labeled for Cx36 and Cx43, and one was double-labeled for Cx43 and Cx45. Of five replicas of OB, one was single-labeled for Cx36, two were double-labeled for Cx36 and Cx45, one was triple-labeled for Cx36, Cx43, and NMDAR1, and one was “double-double”labeled (quadruple-labeled) for Cx45+ AQP4 and Cx36+ NMDAR1. In addition, one sample of mouse containing OE, cribriform plate, and OB was triple-labeled for Cx43, Cx45, and AQP4. The quadruple-labeled sample of OB was labeled in pairs, with AQP4 + Cx45 labeled with both 6 nm and 18 nm gold and Cx36+ NMDAR1 labeled with 12 nm gold. Just as the same color stain is often used to label recognizably-different structures in LM immunohistochemistry (e.g., nucleus vs. collagen fibers), paired sizes of immunogold labels were occasionally used to identify two or more recognizably different structures in FRIL. In the absence of appropriate primary antibodies from four different species, the use of the same size labels for both NM-DAR1 and Cx36 is acceptable because connexons are always P-face IMPs, whereas glutamate receptors are always E-face IMPs, which cannot be confused with connexon IMPs. Likewise, the use of the same sizes of gold (6 plus 18 nm goat anti-rabbit IgG) for both AQP4 and Cx45 did not lead to confusion in samples of OB because AQP4 in astrocyte square arrays could not be confused with labeling for Cx45 in neuronal gap junctions, and Cx45 does not occur in astrocyte gap junctions. However, one potential for ambiguity arose in OE, where “cryptic” labeling for AQP4 in the cell membrane immediately beneath the replica and immediately adjacent to the gap junction could have been mistaken for Cx45 in the gap junction. Special notice for this potential ambiguity is made in the text.
Table 1.
Number of samples examined, their labeling combinations, and the figures documenting the listed combinations of antibody labels.
|
Tissue |
||||
|---|---|---|---|---|
| Labels | OE | OE, CP, OB | OB | Figures |
| Cx43 | 1 | INS | ||
| Cx43 | ||||
| Cx43 | 4 | 3E, G | ||
| Cx36 | 2C–E; 4A–D; 8A–B* | |||
| Cx43 | 1 | INS | ||
| Cx45 | ||||
| Cx43 | 3F, H–I; | |||
| Cx45 | 1 | 5C; 7A–C | ||
| AQP4 | 10A–E; 11A | |||
| Cx36 | 1 | INS | ||
| Cx36 | 2 | 9A–E; | ||
| Cx45 | 12B–D | |||
| Cx36 | ||||
| Cx43 | 1 | INS | ||
| NMDAR1 | ||||
| Cx36 + NMDAR1 | 1 | 11B; 12A | ||
| Cx45 + AQP4 | 14A–E | |||
OE = olfactory epithelium
CP = cribriform plate
OB = olfactory bulb
INS = images not shown
All figures were from adult mouse, except for Figs. 2C–E, 4A–D, 8A–B, which were from 4-day postnatal rat. No differences were detected in cell-specificity of connexin labeling in adult mouse vs. 4 day postnatal rat.
After rinsing in distilled H2O, labeled replicas were air dried and coated on the labeled side with 10–20 nm of evaporated carbon. The second carbon coat not only surrounds and further stabilizes the immunogold beads, it also stabilizes the entire replica by annealing the myriad cracks caused by thermal expansion when the replicated samples were warmed from −180°C to 22°C. To permit viewing by TEM, the Lexan support film was removed by immersing the grids in dichloroethane solvent for 1–2 h, followed by air drying.
ELECTRON MICROSCOPY AND PHOTOGRAPHIC REPRODUCTION
Thin sections and FRIL replicas were examined at 100 kV in JEOL 2000 EX-II and JEOL 1200 EX transmission electron microscopes (TEMs; JEOL USA Inc., Peabody, MA). Stereoscopic images (8° included angle; tilt ranges of ±60° and ±45°) were obtained at TEM magnifications from 50×–100× (for correlation with confocal photomaps) to 100,000×. Measured differences in magnifications of the two TEMs were corrected during photographic enlargement. TEM negatives were digitized at 2500 dpi using an ArtixScan 2500f digital scanner (Microtek; Carson, CA), and processed using Photoshop 7.0.1 and CS2 (Adobe Systems, San Jose, CA). Stereoscopic images were used to assess complex three-dimensional membrane topography, as well as to confirm that each label is on the tissue side of the replica. In contrast, relatively rare non-specific binding of gold beads was primarily on the Lexan-coated side of the replica (Rash & Yasumura, 1999). Selected regions are presented as stereoscopic “triplets” consisting of a stereo pair (with left pair of three images viewed using a stereopticon-type viewer) and a reverse or “intaglio” pair (right pair of images). (Note: Stereoscopy is reversed from the above description when viewed by the “crossed-eyes” method.) The reverse stereo images are required for discriminating the 5 nm and 6 nm gold beads from platinum-coated 6–10 nm IMPs (Rash & Yasumura, 1999).
TWO INDEPENDENT SEARCH STRATEGIES
The primary search strategy was to examine FRIL replicas at low magnification for electron-dense gold beads (10,000× image viewed with 10× binoculars), then identify gap junctions and cells at higher magnification (typically, 30,000×–100,000×). The secondary search strategy was to scan for specific cell types, examine the cells at high magnification for the presence of gap junctions, either labeled or unlabeled, then determine if any immunogold beads were present. More than 250,000 μm2 of replicated plasma membranes were searched at high magnification for immunogold beads, including >1000 μm2 from >100 identified ORN somata.
LIMIT OF DETECTABILITY OF LABELS BY FLUORESCENCE MICROSCOPY VS. FRIL
The limit of photon production and detectability for fluorescence labeling of connexins is not established for confocal microscopy, but gap junctions with <15–30 connexons were not reliably detected at high LM magnifications (Meier et al., 2004), whereas FRIL often revealed gap junctions with as few as two connexons (Meier et al., 2004), reliably detected junctions with >10 connexons, and detected virtually all gap junctions with >50 connexons when they had been labeled with the appropriate anti-connexin antibodies (Rash & Yasumura, 1999). Consequently, we compared detection limits for Cx43 and Cx36 in OE using both confocal immunofluorescence microscopy and FRIL.
IDENTIFICATION OF miRNA “SEED MATCHING” SEQUENCES IN mRNAS FOR Cx36, Cx43, AND Cx45
The annotated 3′ UTRs for mCx36, mCx43, and mCx45 were retrieved from Ensembl genome database using MartView (http://www.ensembl.org/Multi/martview) and extended to 2000 nucleotide bases by including downstream flanking sequences. Mouse miRNA sequences were retrieved from miRBase; Release 7.0 (http://microrna.sanger.ac.uk/sequences/). To predict miRNAs for Cx36, Cx43, Cx45 and Cx43, consistent with the possibility that the synthesis of these three connexins may be down-regulated by miRNA mechanisms, we compared data from TargetScan (http://genes.mit.edu/targetscan/) and Miranda (http://www.microrna.org/).
Results
LM IMMUNOFLUORESCENCE FOR Cx43 IN OLFACTORY EPITHELIUM AND UNDERLYING CONNECTIVE TISSUE
A low magnification LM immunofluorescence image of labeling for Cx43 in mouse olfactory epithelium and a corresponding LM image of a plastic embedded semi-thin section are shown in Fig. 1(A–C), respectively. The fluorescence image is from a horizontal, midline section taken at a dorsal level, including the posterior extreme of the epithelium where it abuts the cribriform plate. Epithelia bilaterally line the midline nasal septum. Tissue adjacent to the nasal lumen on the upper and lower left two-thirds of the immunofluorescent image are sections of liver blocks that were used to support the septum and its epithelia in a vertical position during cryostat sectioning. Intense labeling for Cx43 is evident in the lamina propria, which is of variable thickness along its course beneath the sensory epithelium, often displaying large voids in Cx43 immunoreactivity. Labeling for Cx43 was undetectable in most areas of sensory epithelium examined at low magnification. The relatively evenly-spaced olfactory knobs (Fig. 1(C), arrows) of the ORNs extend above the surrounding sustentacular cells (SCs) and into the weakly-stained layer of cilia and microvilli. Within the lamina propria (Fig. 1(C), LP) are large bundles of olfactory axons, secretory Bowman’s glands, and the septal bone, which is ensheathed by epithelioid cells of the periosteum (Figs. 1(C), 3(A)). Within the septum and turbinate bones, osteocytes are present within clear “lakes” or lacunae (Fig. 1(C); higher magnification in Fig. 3(A)).
Fig. 1.
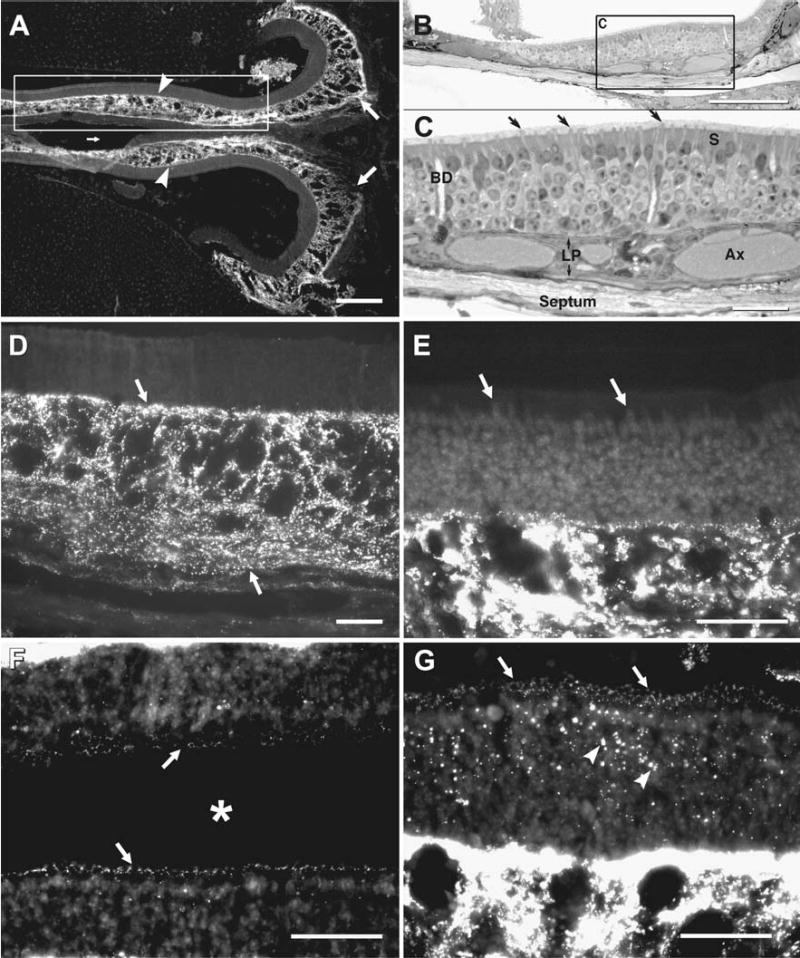
Immunofluorescence labeling of Cx43 in adult mouse OE. (A) Low magnification micrograph of a midline horizontal section through the posterior pole of the OE abutting the osseous nasal septum medially (small arrow) and the cribriform plate posteriorly (right, large arrows). On both left and right sides, dense immunolabeling for Cx43 is seen within the lamina propria (arrowheads). (B) Semi-thick plastic-embedded section showing a similar view to that in boxed area in (A). (C) Higher magnification view of inscribed area in “B”, showing olfactory knobs (arrows), sustentacular cells (S), and Bowman’s gland ducts (BD) traversing the sensory epithelium. Within the lamina propria (LP) are axon bundles (Ax). The supporting septal bone (bottom) is lined by epithelioid cells of the periosteum. (D–E) Higher magnifications of OE showing a high concentration of punctate labeling for Cx43 in the lamina propria (D, arrows), and little detectable labeling in most regions of sensory epithelium (E, arrows). (F, G) Areas of sensory epithelium bordering the nasal lumen (asterisk) from a different area than in (A, D) showing a band of sparse punctate labeling for Cx43 along the base of the olfactory cilium (F, arrows), and a section obliquely through this band showing annular rings of puncta (G, arrows), with larger puncta in the sensory epithelium (G, arrowheads) and comparison of the much more dense labeling for Cx43 in the lamina propria (bottom half of image). Scale bars: A–B, 200 μm; C–G, 50 μm.
Fig. 3.
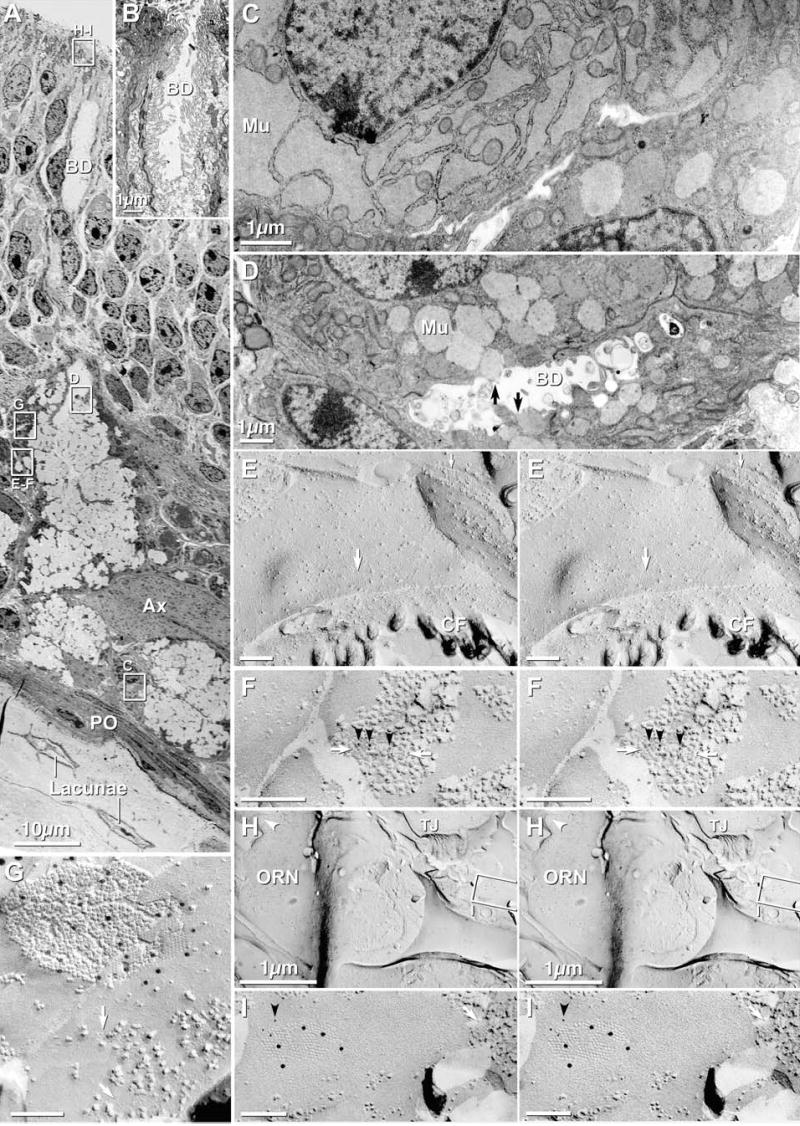
TEM and FRIL images of Bowman’s gland cells in olfactory mucosa of adult mouse. (A) Low-magnification overview of olfactory mucosa. Clear areas in Bowman’s gland are globules of condensed/incompletely hydrated mucous. Images similar to the inscribed areas “C–I” are shown at higher magnification in succeeding panels, either as thin sections C–D or FRIL images. BD = Bowman’s gland duct; Ax = axon bundle; PO = periosteum; Lacunae contain osteocytes. (B) Opening of Bowman’s gland duct onto OE surface. Note the abundant microvilli within the duct (BD) and extending onto the OE surface. Duct cells are linked by abundant tight junctions, gap junctions and belt desmosomes. (C) Distended rough endoplasmic reticulum contain mucus (Mu) destined for packaging in spherical mucous granules. (D) Spherical mucous granules, some apparently being released (arrows) into the lumen of the duct (BD). (E) Stereoscopic E-face image of square arrays (large arrow = E-face; small arrow = P-face) in basal plasma membrane of Bowman’s gland duct cell. The abundance of AQP4 arrays implies high capacity for water transport, CF = Collagen Fibers. (F) FRIL image of square arrays (white arrows) after immunogold-labeling for AQP4 (6 nm gold, black arrowheads). IMP clumping in this sample was due to poor fixation with formaldehyde. (G) FRIL image of gap junction between Bowman’s gland duct cells; immunogold-labeled for Cx43 (12 nm gold). E-face image of square array is indicated (white arrow). (H) Low magnification FRIL image of Bowman’s gland duct cell opening onto surface of OE, adjacent to ORN. Inscribed area is shown at higher magnification in “I”. TJ = tight junctions. Arrowhead = cilium of ORN. (I) FRIL image of Bowman’s gland gap junction labeled for Cx43 (12 nm gold) and Cx45 (6 nm gold, black arrowhead). The E-face imprint of an AQP4 array is indicated (white arrow).
At higher magnification (Fig. 1D), labeling for Cx43 in the lamina propria appears exclusively as intense immunofluorescent puncta, with no clear localization to any particular cells, which in any case are not separately discernable in these non-counterstained sections. Cx43-puncta are concentrated around voids of labeling and were frequently present as linear arrays of puncta, particularly within the lower half of the lamina propria. Still higher magnifications were used to compare labeling intensity of Cx43 in the lamina propria to that observed in most regions of the sensory epithelium (Fig. 1(E)). Despite heavy labeling in the lamina propria, Cx43 was not visible as punctate labeling at low magnification in the vast majority of sensory epithelial areas examined. However, after compensation for relative fluorescence intensity, more ventral and posterior regions of sensory epithelium displayed clear labeling for Cx43 that was localized as a very narrow band of extremely fine puncta just below the ciliary knobs at the approximate location of the “terminal web junctional complex” (Hull & Staehelin, 1979). The fine puncta are visible on epithelia adjacent to each side of the nasal lumen (Fig. 1(F), arrows). In a tangential section through this band, allowing a somewhat en face view, these fine puncta appear as annular profiles of dots (Fig. 1G, arrows), possibly surrounding individual SCs. In addition, sparsely scattered, larger puncta are evident in the sensory epithelium (Fig. 1(G), arrowheads), but in these LM views of unstained sections, it was not possible to identify the epithelial cell types having Cx43 puncta. From a comparison of puncta located at the epithelial surface (Fig. 1(F), (G)) with those located beneath and in the lamina propria (Figs. 1(E), (G)), surface puncta were on average much fainter and several fold smaller/less intensely labeled than those in deeper regions.
TEM OVERVIEW OF OLFACTORY MUCOSA: OLFACTORY EPITHELIUM AND SUPPORTING CONNECTIVE TISSUE
In thin-section TEM images of the olfactory epithelium and supporting lamina propria in adult mice and early postnatal rats (Fig. 2), ciliated ORNs were surrounded by sustentacular cells (SCs), which are characterized by abundant microvilli on their apical surfaces (Fig. 2(A), “S”). Deeper in the olfactory mucosa are loosely-connected fibroblasts of the lamina propria (Figs. 2(A), 3(A)), as well as cuboidal epithelial cells comprising Bowman’s glands (Fig. 3), which produce most of the aqueous nasal secretions that bathe the sensory cilia. Consistent with previous ultrastructural studies (Miragall et al., 1992, 1996a; Blinder et al., 2003), areas of close membrane apposition identified as gap junctions linked fibroblasts of the lamina propria (Fig. 2(B)), Bowman’s gland cells (Fig. 3), and axon bundle ensheathing cells (described below).
Fig. 2.
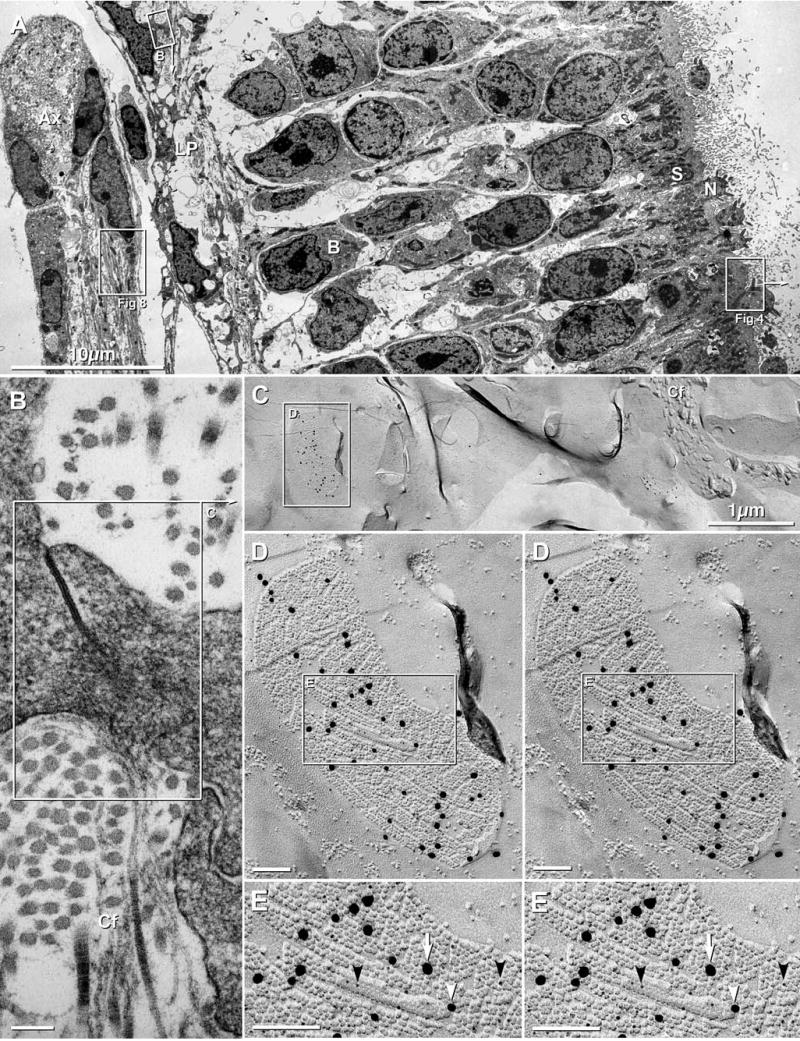
Lamina propria and olfactory epithelium in adult mouse and 4 day postnatal rat. (A) Overview of olfactory mucosa in adult mouse, including olfactory epithelium with olfactory receptor neurons (N), sustentacular cells (S; box labeled “Fig. 4”), basal cells (B), and axons (Ax) surrounded by ensheathing cells (box labeled “Fig. 8”). In the lamina propria (LP), fibroblasts are linked by gap junctions (box B; enlarged as 2B). (B) Higher magnification image of gap junction between two fibroblasts of the lamina propria. Fibroblasts secrete and are surrounded by collagen fibers (Cf). The inscribed area “C” corresponds to the FRIL image in Fig. 2(C). (C) Low-magnification FRIL image of fibroblasts in lamina propria of postnatal day 4 rat. Collagen fibers (Cf) are adjacent to fibroblasts. Even at low magnification, immunogold beads call attention to gap junctions of various sizes, from small (center of image) to very large (inscribed box labeled “D”; enlarged as 2D). (D) Higher magnification stereoscopic image of inscribed gap junction in “C”. Of the three sizes of gold beads (5 nm, 18 nm and 20 nm) labeling this gap junction, only the 18 nm and 20 nm gold beads are easily discerned at this magnification. However, high-magnification analysis revealed 89 5-nm gold beads, 17 18-nm gold beads, and 23 20-nm gold beads; for a labeling ratio of 4.5:1:1. Inscribed area is shown in higher magnification reverse stereoscopic perspective as “E” (below). Gap junctions in fibroblasts consist of smaller subdomains of connexins, separated by connexin-free voids. (E) In high-magnification reverse stereoscopic images, 33 5-nm gold beads (black arrowheads) are more readily evident than in conventional stereoscopic images. There are also 7 18-nm gold beads and 6 20-nm gold beads. Note the high labeling efficiency and very low non-specific “background” labeling. In FRIL and TEM images, calibration bars are 0.1μm, unless otherwise indicated.
Lamina propria by TEM and FRIL
Between the olfactory epithelium and the periosteum (PO) that surrounds the turbinate bones and septum (Figs. 1(A–C), 3(A)) is a variable thickness layer of connective tissue, containing one or more of the following: connective tissue proper, called lamina propria (Fig. 2); secretory epithelial cells of Bowman’s glands (Fig. 3); bundles of unmyelinated olfactory axons (Figs. 2(A), 8(A); detailed descriptions below) surrounded by epithelioid “ensheathing cells” (Figs. 2(A), 8(B), below); plus capillaries, arteries, veins, venous sinuses, and a variety of pressure-sensitive, multilayered nerve terminals, primarily Pacinian corpuscles (the latter five cell types not shown). At low magnification, fibroblasts of the lamina propria (Fig. 2(C)) were irregularly distributed but were always adjacent to or surrounded by bundles of collagen fibers (Figs. 2(B), (C)). Adjacent fibroblasts were linked by irregular cytoplasmic processes that frequently had gap junctions at the point of contact (Fig. 2(B)). These structures were identified as gap junctions based on their extended areas of close parallel membrane apposition and the increased electron density at the area of contact. (Criteria for identifying gap junctions in thin-section images are presented in Rash et al., 1998a).
Fig. 8.
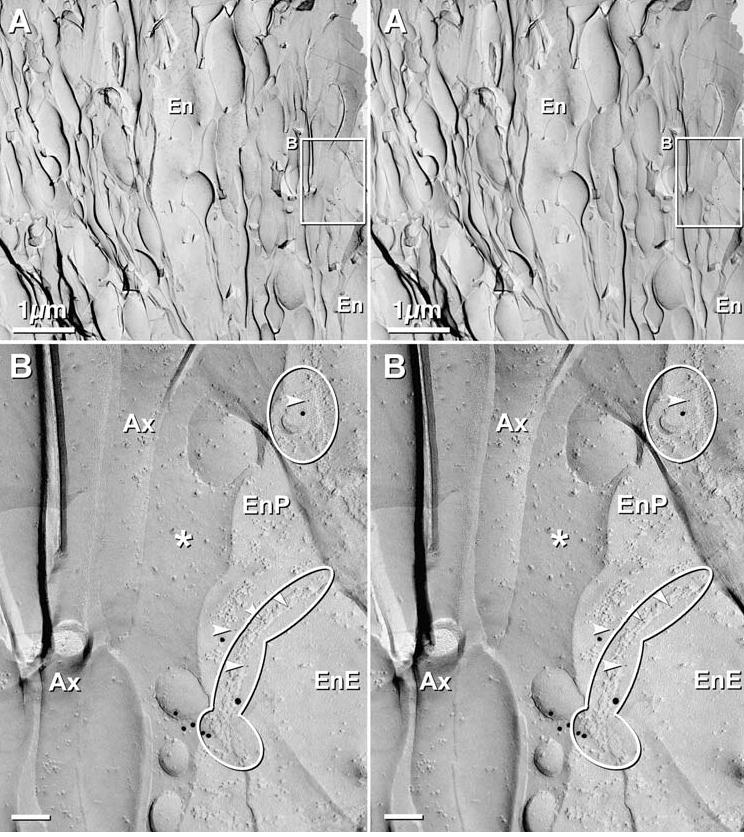
Stereoscopic images of axon bundle and surrounding ensheathing cells in OE of 4 day postnatal rat after double-labeling for Cx36 and Cx43, using 5 sizes of gold (Cx36 = 10 nm and 12 nm; Cx43 = 5 nm, 18 nm, and 20 nm). (A) Low magnification image of an axon bundle, with an intrafascicular ensheathing cell (En, top center) and an external ensheathing cell (En at lower right; and inscribed box, shown at higher magnification below). (B) Higher magnification stereoscopic image of the inscribed area “B”, above. Two ensheathing cell gap junctions are delineated by ovals. Each gap junction is labeled for Cx43, with 5 nm, 18 nm and 20–22 nm gold beads. No 10 nm or 12 nm gold beads (for Cx36) are present on gap junctions in ensheathing cells or in axons. The small clump of 18nm gold beads (bottom center) was counted as a single “hit” (Rash and Yasumura, 1999). * = extracellular space; Arrowheads = 5 nm gold beads; Ax = axon; EnE = ensheathing cell E-face; EnP = ensheathing cell P-face.
By FRIL, immunogold beads for Cx43 called attention to abundant gap junctions between fibroblasts of the lamina propria (Figs. 2(C–E)), but neither Cx45 nor Cx36 were detected in gap junctions within the lamina propria. For example, in high magnification reverse stereoscopic images (Fig. 2(E)) showing samples that were double-labeled for Cx43 and Cx36, three sizes of gold for Cx43 (5 nm, 18 nm, and 20 nm) were present on each fibroblast gap junction, whereas immunogold beads for Cx36 (10 nm and 12 nm) were not present. Reverse stereoscopy was essential for detecting most 5 nm gold beads (Fig. 2E, arrowheads) against the equally electron-opaque replica. To maximize the probability for detecting Cx36 and/or Cx43 in hypothetical ORN gap junctions, we used two sizes of gold beads for Cx36 and three sizes of gold beads for two different Cx43 antibodies. However, despite heavy labeling for Cx43 in gap junctions of fibroblasts in nearby lamina propria, Bowman’s glands, and SCs (see below), no gap junctions or connexin labeling for Cx36 or Cx43 were detected in any of the dozens of ORNs examined (Table 2).
Table 2.
Cell types and the connexins expressed in their gap junctions.
| Cell types: Olfactory mucosa | Cell types: Olfactory bulb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ← | → | |||||||||
| Connexin | Fibroblast | BG | SC | ORN | EC | Pia | Astro | PG | ORN | M/T |
| Cx43 | + + + + | + + | + + | − | + + | + + + | + + + | − | − | − |
| Cx45 | − | + ? | + | − | − | − | − | − | − | + |
| Cx36 | − | − | − | − | − | − | − | ? | − | + + |
BG = Bowman’s gland; SC = sustentacular cell; ORN = olfactory receptor neuron; EC = ensheathing cell; Pia = pia mater. Astro = astrocyte; PG = periglomerular neuron; M/T = mitral/tufted cell.
Bowman’s gland by TEM and FRIL
The secretory portions of each Bowman’s gland (BG) are composed of cuboidal and flattened epithelial cells (Figs. 3(A–D)) that are enriched in rough endoplasmic reticulum (RER) and secretory vesicles (Figs. 3(C), (D), respectively). Near the OE surface, the simple cuboidal epithelium narrows to thin epithelioid BG duct cells having short microvilli projecting into their lumens (Figs. 3(A–B)). The microvilli within BG ducts become more densely packed at the transition zone between the cuboidal secretory gland cells and the flattened duct cells (Frisch, 1967). At the opening of the ducts, densely-packed microvilli extend above the OE surface (Fig. 3(B)), thereby causing the apices of these cells to resemble the apical surfaces of SCs (see below). In their cytoplasms, BG cells exhibited either distended RER containing finely granular serous or mucous material (Fig. 3(C)) or spherical vesicles representing serous or mucous granules ready for release into the ducts (Fig. 3(D), black arrows). Cells containing a combination of distended RER and spherical vesicles were less frequently encountered, confirming previous detailed TEM observations of Bowman’s glands (Frisch, 1967).
Serous fluid is released primarily by merocrine secretion of vesicles into the lumen of BG ducts (Fig. 3(D), arrows) (Moran et al., 1982). Copious aqueous secretion to bathe the olfactory cilia requires abundant transport of water into BG cells for packaging and hydration of serous and mucous fluids. Thus, it is not surprising that the basal and lateral plasma membranes of cuboidal BG cells contained densely-packed “square arrays” (Fig. 3(E), arrows), similar to those previously identified as AQP4 arrays in astrocytes and ependymocytes (Rash et al., 1998b). In triple-labeled samples, abundant immunogold labeling for AQP4 (Fig. 3F; 6 nm gold beads, black arrowheads) was present beneath many square array P-face IMPs (Fig. 3(F), white arrows), identifying them as AQP4 arrays; whereas immunogold labels for Cx45 and Cx43 were not present in this area. [Fixation of this sample at 4°C resulted in low temperature-induced phase separation of lipids and clumping of IMPs (Maul, 1979), thereby partially obscuring the square arrays.] No other cell types in the olfactory mucosa contained densely-packed square arrays, and no other cell type was strongly labeled for AQP4. Because BG cells secrete into the lumen, the presence of AQP4 on the basal (ablumenal) plasma membrane implies that the pathway of water for hydration of mucous and serous granules is through square arrays into BG cells, rather than out of the cell via AQP4 arrays, as is commonly assumed to occur in astrocyte endfeet (Nielsen et al., 1997; Rash et al., 2004a).
Where BG ducts empty onto the OE surface (Fig. 3(B)), thin section images of BG cells closely resembled images of sustentacular cells (Fig. 2(A)); however, BG cells remained identifiable based on the occasional presence of AQP4 arrays, which decreased in density toward the surface of the OE. Square arrays have also been reported on sustentacular cells in rat olfactory epithelium (Miragall et al., 1984). Because sustentacular cells in OE and respiratory epithelium of lower vertebrates and in the respiratory epithelium of mammals perform some of the same secretory roles as BG in the mammalian OE (Moran et al., 1982), square arrays in sustentacular cells of rodents may reflect an evolutionary vestige of that secretory function. However, it also is possible that some BG cells in mammalian OE have been misidentified as sustentacular cells having square arrays.
On their apical and lateral surfaces, BG duct cells are linked by desmosomes (apposed dark patches in Fig. 3(B)), tight junctions (not shown) and gap junctions (Figs. 3H–I). BG cells also share tight junctions with sustentacular cells at the epithelial surface (TJ, Fig. 3(H)). In FRIL replicas that had been single- or double-labeled for Cx43 plus Cx43 (monoclonal and polyclonal antibodies), Cx43 plus Cx45, or Cx36 plus Cx43, gap junctions between BG cells were strongly labeled for Cx43 (Fig. 3(G)), and occasionally for both Cx43 and Cx45 (Figs. 3(H–I)), but never for Cx36 (Table 2). However, the simultaneous use of the same sizes of gold beads for both Cx45 and AQP4 (Fig. 3(I)) did not allow unambiguous identification of Cx45 in the gap junction because of the possibility of “cryptic” labeling of AQP4 in the adjacent cell. Because AQP4 is much reduced at the apical surface of BG, and no other AQP4 arrays were detected or labeled in this area, the labeling within the gap junction perimeters was attributed to Cx45 rather than AQP4. Clarification of this issue would require labeling using Cx45 and Cx43 as double labels.
Cx43 AND Cx45 IN SUSTENTACULAR CELLS OF THE OLFACTORY EPITHELIUM; ABSENCE OF Cx36
Sustentacular cells (SCs)
Four general classes of cells are exposed on the osmosensory surface—SCs, ORNs, microvillar cells and BG cells. SCs, which are characterized by the presence of abundant microvilli and absence of cilia on their apical surfaces, are interspersed among ORNs in the olfactory epithelium (Figs. 1(B), 2(A), 4(A)). Previous thin-section electron microscopic studies have identified gap junctions linking SCs (Kerjaschki & Hörandner, 1976; Menco, 1980a; Mack & Wolburg, 1986; Miragall et al., 1992), and intracellular recording revealed that SCs are weakly coupled to other SCs but not to ORNs (Vogalis et al., 2005a). Moreover, previous thin-section immunogold-labeling TEM studies demonstrated that Cx43 is present in SC gap junctions (Miragall et al., 1993, 1996a, Miragall et al., b).
Fig. 4.
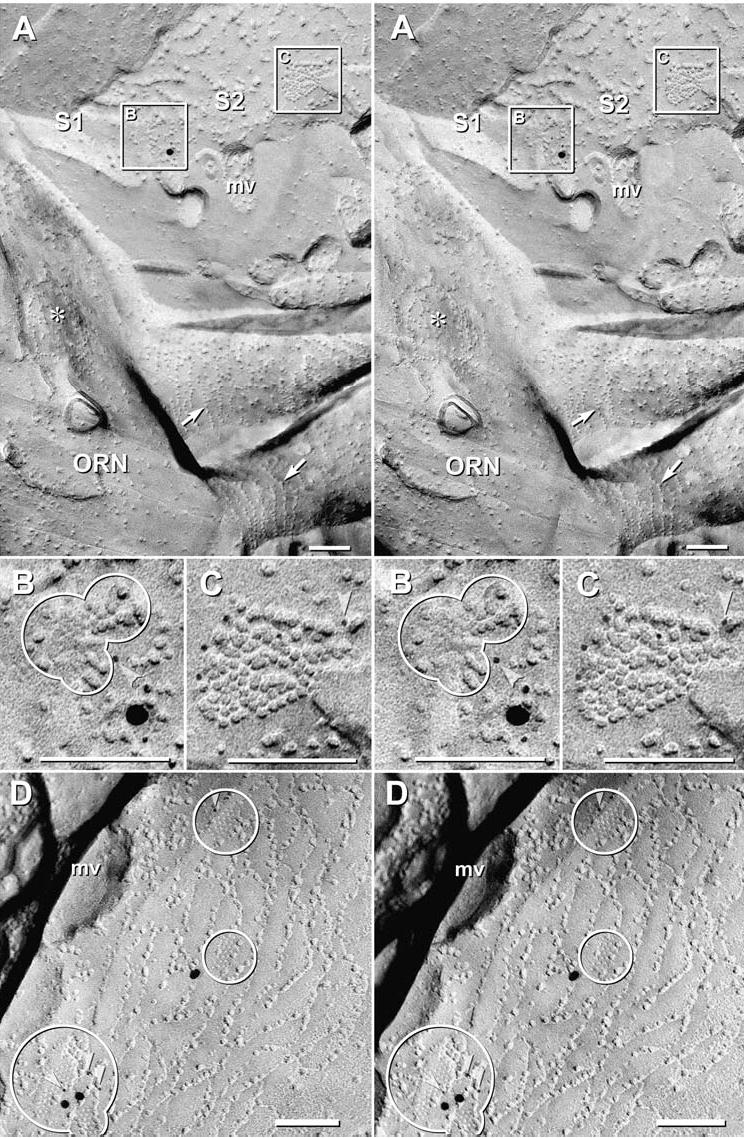
Cx43 immunogold labeling in small gap junctions between sustentacular cells in 4 day postnatal rat. (A) Stereoscopic image of tight junctions and gap junctions linking two sustentacular cells (S1, S2) that are adjacent to an olfactory receptor neuron (ORN). The bases of two ORN cilia are surrounded by strands of IMPs called “ciliary necklaces” (arrows). A circular “target array” (*) is another marker for the ORN plasma membrane (Menco, 1980c). Inscribed areas “B” and “C” are shown at higher magnification below. (B–C) Stereoscopic images of gap junction E- and P-faces that are double-immunogold labeled for Cx36 (10 nm and 12 nm; none present) and Cx43 (5 nm, 18 nm and 20 nm gold beads; arrowheads point to 5 nm gold beads). (D) Gap junctions are labeled for Cx43 (arrowheads); some were unlabeled (not shown). Note that the tight junction strands are interlocking. mv = cross fractured base of microvillus.
In FRIL replicas, SCs were identified by microvilli extending from their apical plasma membranes (Figs. 4(A), (D), “mv”) and by the presence of gap junctions and extensive tight junction complexes, which consisted of either of two morphologies: a) parallel strands of primarily E-face IMPs and P-face pits vs. b) interlocking strands of IMPs and pits on both P- and E-faces (Figs. 4 (A), (D)). Tight junctions having multiple interlocking strands were noted between adjacent SCs, between SCs and microvillar cells, and between SCs and BG duct cells, whereas tight junctions with parallel strands having few cross-links were between SCs and ORNs (Menco, 1980b; also see below). However, unlike the “particle partitioning coefficient” (Satir & Satir, 1979) for tight junctions in cells fixed with glutaraldehyde (CP = 1.0; or 100% of IMPs on P-faces and 100% of pits on E-faces), tight junctions fixed with formaldehyde (a requirement for FRIL) have CP = 0.3–0.5. Consequently, tight junctions in FRIL replicas consist of discontinuous rows of IMPs and pits on both P- and E-faces, with most IMPs on E-faces and most pits on P-faces (Meier et al., 2004).
In samples double-labeled for Cx36 and Cx43 (Fig. 4), immunogold beads for Cx43 repeatedly drew attention to small gap junctions between tight junction strands linking SCs, but never to gap junctions involving ORNs. Cx43 was labeled using both 6 nm and 18 nm gold beads, whereas Cx36 was labeled by 12 nm gold beads (none present). Cx43-containing gap junctions ranged from four connexons (not shown, but see Fig. 5(C), discussed below) to approximately 80 connexons (Fig. 4(C)). In all cases, these polygonal gap junctions were between or adjacent to the interlocking tight junction strands that characterize the apical plasma membranes of SCs (Fig. 4(D)). Overall, 31 Cx43-labeled gap junctions were found in SCs, but none were labeled for Cx36.
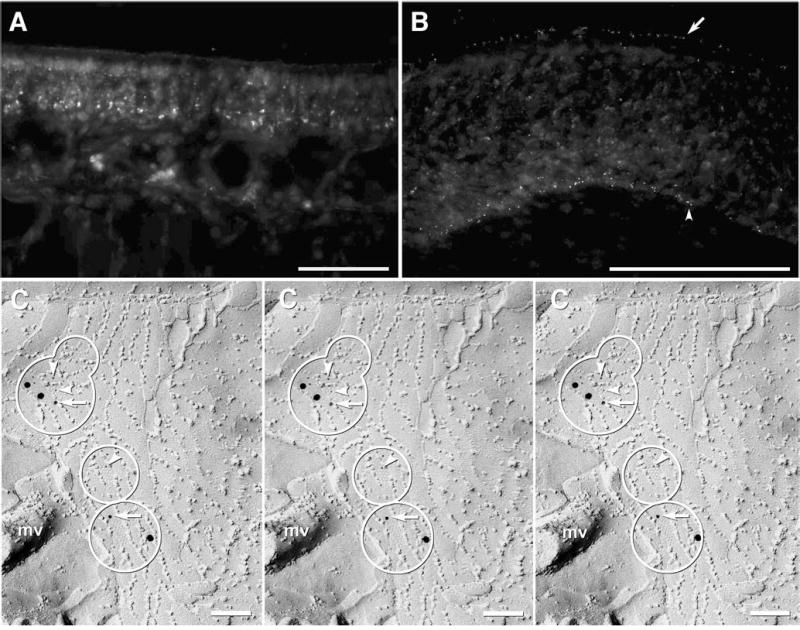
Fig. 5.
Immunofluorescence and immunogold labeling for Cx45 in adult mouse OE. (A) In most areas of the OE, a few Cx45 puncta are visible at low magnification. (B) In higher magnification images, tiny fluorescent puncta are present at the apical margin of the OE (arrow), and at the base of the OE (arrowhead). (C) Stereoscopic images (left pair) and reverse stereoscopic images (right pair) of gap junctions (circles) between tight junction strands in SC. This sample was double-labeled for Cx45 + AQP4 (6 and 18 nm gold beads; arrowheads = 6 nm gold beads) and Cx43 (12 nm gold beads; arrow). AQP4 labeling not detected in this image. mv = microvillus. Scale bars: A–B, 50 μm.
Cx45 IN OLFACTORY EPITHELIUM
Immunofluorescence staining for Cx45 was much less intense and much more localized in olfactory mucosa than was immunofluorescence for Cx43. In a view similar to Fig. 1(F) (above), Cx45 immunofluorescence was irregularly distributed, with some regions showing no distinct immunofluorescence (Fig. 5(A)). In other regions, Cx45 labeling was detected as very fine puncta localized to the apical region of the OE (Fig. 5(B), arrow), as well as to presumptive basal cells near the basal lamina (Fig. 5(B), arrowhead). However, the puncta for Cx45 were only about 1/10 as abundant as Cx43 puncta (Figs. 1(F), (G)), and most were near the detection limit for immunofluorescence labeling (Meier et al., 2004). In companion FRIL images (Fig. 5(C)) labeled for both Cx43 (12 nm gold beads, white arrows) and Cx45 (6 nm gold beads [white arrowheads] and 18 nm gold beads [self evident]), immunogold labeling for Cx45 was detected in 15 small gap junctions between tight junction strands in SC, often without accompanying labels for Cx43 (Fig. 5(C), middle circle), but occasionally, SC gap junctions were labeled for both Cx43 and Cx45 (Fig. 5(C), upper and lower circles) or for Cx43 only (not shown).
NUMERICAL ANALYSIS OF LABELED GAP JUNCTIONS IN OLFACTORY MUCOSA
Sustentacular cells
Overall, 66 SC gap junctions were observed in seven samples labeled for various combinations of Cx36, Cx43, and Cx45 (Table 2), of which 31 were labeled for Cx43, nine for Cx45, six for Cx43 + Cx45, and 20 were unlabeled, but nevertheless were detected because of their relatively close proximity to labeled SC gap junctions. Lack of labeling of ca. 30% of SC gap junctions in samples labeled for Cx43 and Cx36 led us to investigate the possibility of additional connexins in SC gap junctions, including Cx45. Of the 34 labeled gap junctions in Cx43, Cx45 and AQP4 triple-labeled samples, 56% (19/34) were single-labeled for Cx43, 18% were double-labeled for Cx43 and Cx45 (6/34; Fig. 5(C)), and 26% (9/34) were labeled for Cx45 but not for Cx43 (Fig. 5(C)). Based on the number of gold labels for each connexin, this may suggest that Cx43-containing gap junctions were 2–5 times as abundant as Cx45-containing gap junctions, that Cx43 protein was 2–5 times as abundant as Cx45 protein in those junctions, or that the antibodies for Cx43 were 2–5 times as effective as the Cx45 antibodies.
In a sample double-labeled for Cx36 and Cx43, no SC gap junctions (0/18) were labeled for Cx36 but 67% (12/18) were labeled for Cx43 (Fig. 4) and six (33%) were unlabeled. Based on similar labeling efficiency (ca. 1:30) seen in gap junctions of astrocytes and ependymocytes in other brain areas, as well as in gap junctions of ensheathing cells, fibroblasts and Bowman’s glands of the olfactory mucosa, we conclude that Cx43 is the primary connexin of gap junctions linking SCs, but that Cx45 is also present in a significant subset of SC/SC and/or SC/BG gap junctions. Based on an immunofluorescence detection threshold of about 10–20 connexons (Meier et al., 2004), we also conclude that most SC gap junctions are too small to be reliably detected by LM.
Basal cells
Basal cells were not separately examined for gap junctions by FRIL, in part because the presence or absence of gap junctions on basal cells was not at issue in this study. However, no gap junctions labeled for Cx43 were found in that region during relatively extended initial searches, suggesting that they are relatively rare compared with gap junctions in other cell types. Nevertheless, we note that a few Cx45 immunofluorescence puncta were present at the border between the basal cells and the lamina propria (Fig. 5(B), arrowhead), suggesting that Cx45 may be present in gap junctions between basal cells or Bowman’s gland cells, both of which may be present in that location.
Absence of Cx36 immunofluorescence in olfactory epithelium
The report of Zhang et al. 2003 suggested that over large expanses of the OE, ORN somata and axons contained abundant Cx36 immunoreactivity. Thus, attempts were made to detect Cx36 by immunofluorescence in any of the cells present in the OE of CD1 and C57BL/6 mice using each of five different monoclonal and polyclonal antibodies, as described in the Materials and Methods section. In all regions, sections of OE consistently had no detectable Cx36-immunofluorescent puncta, even when photographically processed to reveal substantial autofluorescence of cell nuclei (Fig. 6(A)). No specific labeling for Cx36 was evident on cell bodies or dendrites in OE or on their axons within OE or olfactory mucosa. This conclusion was based on comparisons with levels of background fluorescence observed in sections of OE obtained from Cx36 knockout mice. As positive and negative controls for testing the efficacy of these antibodies, sections of inferior olive (whose gap junctions contain Cx36; Rash et al., 2000; Nagy et al., 2004) showed robust labeling for Cx36 in tissue from wild-type mice (Fig. 6(B)) and an absence of Cx36 labeling in tissue from Cx36 knockout mice (Fig. 6(C)). (These two images were photographically intensified similar to Fig. 6(A).) Thus, using improved immunofluorescence methods and Cx36 KO controls, we found no support for previous descriptions of abundant Cx36 immunofluorescence localization anywhere in olfactory mucosa.
Fig. 6.
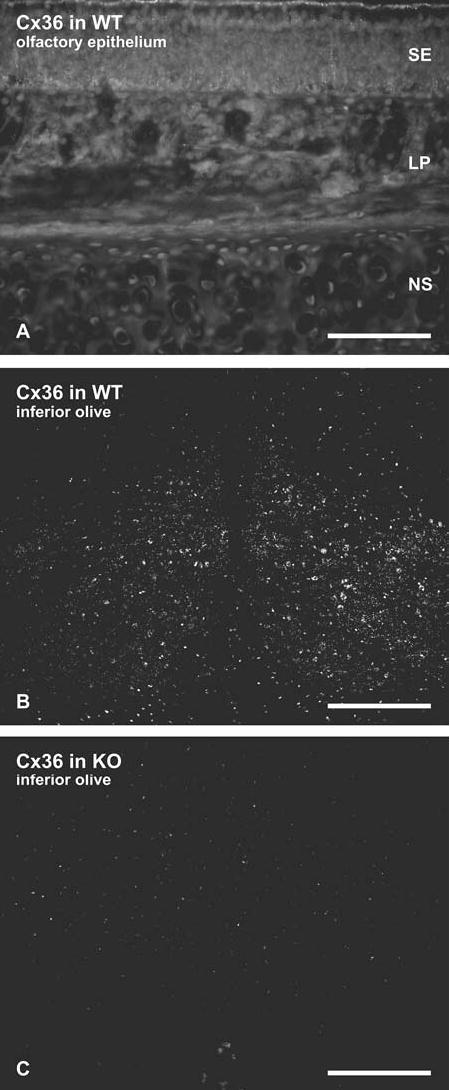
Immunofluorescence micrographs of Cx36 immunore-activity in OE and inferior olive of adult mouse. (A) Section of OE containing sensory epithelium (SE), lamina propria (LP) and nasal septum (NS) showing absence of labeling for Cx36 in all areas, enhanced to the level that autofluorescence is visible. (B, C) Sections through the inferior olive in wild-type mice (B) used as a positive control for detectability of Cx36, seen as densely distributed puncta with anti-Cx36 antibody on neurons for comparative level of labeling (also see Fig. 5 in Rash et al., 2001), and specificity control (C) for absence of labeling for Cx36 in Cx36 knockout mice, also enhanced to a level that reveals non-specific labeling. Scale bar: A, 100 μm; B, C, 200 μm.
FRIL analysis of olfactory receptor neurons
ORNs in four replicas double-labeled for Cx36 and Cx43, in one replica double-labeled for Cx43 and Cx45, and in one replica triple-labeled for Cx43, Cx45 and AQP4 (Table 1) were searched for immunogold labeling (Fig. 7). Each replica included about the same area as a low magnification light micrograph (ca. 2 × 2 mm to 3x3 mm), and each included replicated portions of several hundred ORNs, typically including 10–30 olfactory knobs. Each replica contained thousands of square micrometers of freeze-fractured ORN plasma membrane, virtually all of which were examined for gap junctions and for immunogold labels. Three replicas contained primarily ventrolateral areas of the epithelium that previous studies had indicated most likely to contain Cx43-LacZ-positive olfactory receptor neurons (Zhang et al., 2000), but several also contained dorsome-dial and dorsolateral areas. Although the fracture plane is essentially random through each tissue slice, it preferentially follows plasma membranes of the cells that are in or near the path of the fracture plane—neither selecting for nor against one type of epithelial cell or the other. Thus, each replica represents a relatively unbiased sampling of the cell membranes that are present over large areas of the OE.
Fig. 7.
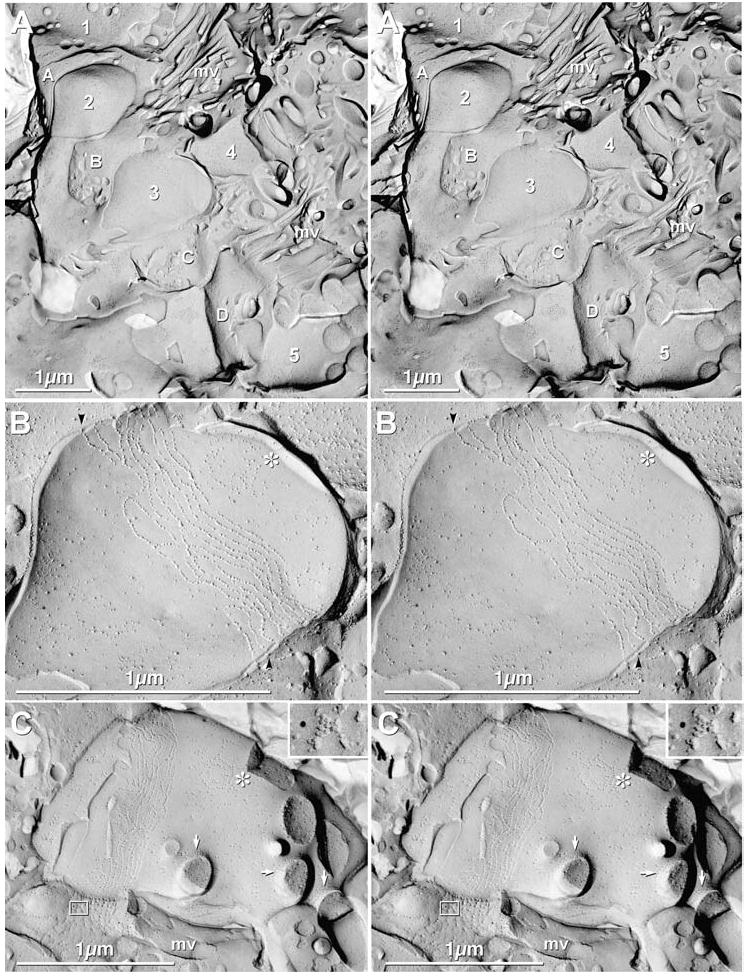
Stereoscopic images of ORNs in adult mice. (A) Low magnification overview of five ORNs (1–5) interspersed with four SCs (A–D). mv = microvilli. (B) Stereoscopic image of ORN #3 (from “A”), showing tight junctions as parallel strands of primarily E-face IMPs. A target array (*) also identifies this as an ORN. Extracellular space is narrowed at tight junction strands (black arrowheads). (C) Stereoscopic images of olfactory knob, showing cross-fractured cilia, with ciliary necklaces (arrows) at the apex of the knob. “*” = target array. The adjacent sustentacular cell has a Cx43-labeled gap junction (inscribed box and inset, upper right). (Inset) The SC gap junction is immunogold labeled for Cx43 by one 12 nm gold bead. mv = microvillus.
At low magnification, nuclei of ORNs and SCs were irregularly arrayed, as is characteristic of this pseudostratified columnar epithelium. Olfactory knobs extended above the apices of the SCs and were identified by the presence of cilia projecting into the mucous layer overlying the OE (Figs. 4(A), 7(A)). Cilia were identified by the presence of a “ciliary necklace” (Gilula & Satir, 1972; Menco, 1980c)—annular arrangements of membrane particles at the base of each cilium (arrows; Figs. 4(A), 7(C))—as well as by the two- to three-fold larger diameter of cilia (0.2 μm), as compared with SC microvilli (0.05–0.1 μm; Figs. 4(A), 7(A), (C)). Cilia were also distinguished from the larger microvilli of microvillar cells, which were approximately 0.1 μm in diameter (not shown). In addition, olfactory knobs had distinctive “target” arrays (Figs. 4(A), 7(B–C); “*”) of mixed P-face and E-face IMPs similar to those in ciliary necklaces (arrows; Figs. 4(A), 7(C)). Target arrays may represent the sites for formation of new cilia or retraction of damaged cilia (Menco, 1980c) and/or sites of centriole “docking”with the plasma membrane. The apical plasma membranes of ORNs were identified by their almost exclusively parallel and seldom-interlocking tight junction strands in the area immediately subjacent to their ciliated olfactory knobs (Figs. 7(B), (C)). As a final criterion, ORN cilia were relatively few and not parallel, whereas cilia in the respiratory epithelium were abundant and present in parallel bundles (not shown, but see images in Moran et al., 1982, for example).
For FRIL analysis of Cx36, Cx43, and Cx45, we utilized both the primary strategy of searching for immunogold labeling at low magnification (to maximize the probability of finding gap junctions), as well as a secondary search strategy of identifying cells at higher magnification based on ORN vs. SC markers (described above), then searching at high magnification for gap junctions. Utilizing the primary search strategy, we examined >10,000 μm2 of ORN and SC plasma membrane. Particular emphasis was placed on examining somatic and dendritic plasma membranes of ORNs, but >300 μm2 of olfactory knob and apical ends of >100 identified ORNs were also examined (Fig. 7(A)). More than 1000 μm2 of somatic plasma membranes of ORNs were examined for unlabeled as well as for immunogold-labeled gap junctions, but none were found. The area examined was equivalent to the entire surface of many dozens of ORNs, or 10–100 times the area of ORN plasma membrane in a typical TEM section (as in Fig. 2(A)).
In high-magnification searches of the tight junctional areas in the apical and dendritic plasma membranes of >100 identified ORNs labeled for Cx36, Cx43, and Cx45 (Figs. 7(B–C)), no ORN gap junctions were found; nor were ORN gap junctions found in replicas double-labeled for Cx43 and Cx45 or Cx43 and Cx36 (negative data not shown). In contrast, Cx43-labeled SC/SC gap junctions were often present immediately adjacent to ORNs (Figs. 4(A–C), 7(C), inset). Thus, FRIL data are consistent with data from conventional freeze fracture indicating that ORNs have no gap junctions on their dendritic knobs or somatic plasma membranes (Menco, 1980b). In contrast, >200 immunogold-labeled gap junctions were found in SCs, fibroblasts of the lamina propria, ensheathing cells surrounding axon bundles, and Bowman’s gland, and 157 of these junctions were photographed stereoscopically at low and high magnification, yielding >1150 TEM negatives. Notably, the levels of non-specific background immunolabeling for Cx36, Cx43 and Cx45 were not detectably higher in ORN plasma membranes, as compared to other membranes or to extracellular space. Therefore, with the demonstrated ability of FRIL to detect gap junctions containing as few as two connexons (Meier et al., 2004), it is unlikely that either Cx43, Cx45, or Cx36 was present above background, even as hemichannels. However, if present as single hemichannels, their connexins did not survive the SDS-washing step that is required in FRIL, and thus, were not detectable above the low non-specific background (i.e., typically, <0.1 gold bead per μm2).
OLFACTORY AXONS AND THEIR ENSHEATHING CELLS
Light microscopy
The report of Zhang and Restrepo (2003) suggested that most ORN axons in some olfactory nerve bundles contained abundant Cx36 immunoreactivity. Zhang et al. 2000 also showed labeling for Cx43 within the axon bundles located in the dorsal recess. In the current study, Cx36 immunoreactivity was not detected at the light level in axon bundles anywhere in the olfactory mucosa (Fig. 6(A)). Moreover, no large accumulations of Cx43 were detected in what might correspond to ORN axon bundles (Figs. 1(D), (E), (G)). Nevertheless, because the detection threshold is lower for FRIL than for LM (Meier et al., 2004), and because we could not identify cell types by TEM of unstained sections, we used FRIL to examine axon bundles in the OE, cribriform plate and OB.
FRIL analysis of axons
The unmyelinated axons of olfactory receptor neurons coalesce into axon bundles beneath the olfactory epithelium (Figs. 1(A), 2(A), 3(A), 8(A)), where they become invested with epithelioid ensheathing cells (Field et al., 2003). These axon bundles and their ensheathing cells traverse the cribriform plate to reach the olfactory bulb as the first cranial nerve (olfactory nerve), then penetrate and traverse the pia mater, forming the olfactory nerve layer (ONL) of the olfactory bulb. In FRIL replicas labeled for Cx36, Cx43, and Cx45, we examined several hundred unmyelinated olfactory axons within the olfactory mucosa (Fig. 8(A)), cribriform plate, and near the olfactory bulb, corresponding to >10,000 μm2 of axonal plasma membrane searched at high magnification. Even with high visibility and high detective efficiency of FRIL, no axonal gap junctions were detected, labeled or unlabeled; nor was immunogold labeling above background for any connexin. Similarly, other investigators using conventional freeze fracture did not detect gap junctions between axons or between axons and ensheathing cells (Blinder et al., 2003). Thus, we conclude that gap junctions do not occur (or are extremely rare) between adjacent olfactory axons within the olfactory mucosa, cribriform plate, or olfactory neuron layer of the OB. Consequently, we cannot account for the strong immunofluorescence labeling previously described (Zhang et al., 2000, 2003).
Ensheathing cells
In 12 samples of OB and OE labeled for Cx36, Cx43, and Cx45, >50 homologous gap junctions were found between ensheathing cells that surround axons in the olfactory mucosa, cribriform plate, approaching the OB, and in the OB nerve fiber layer, and 30 of these (27 labeled and three unlabeled) were photographed. Almost all were heavily labeled for Cx43 (Figs. 8(B), 10(C), 10(E)), but none between ensheathing cells were labeled for Cx45 or Cx36 (Table 2). Moreover, gap junctions were so rare between adjacent intrafascicular ensheathing cells that they could not contribute significantly to the Cx43 immunofluorescence previously reported.
Fig. 10.
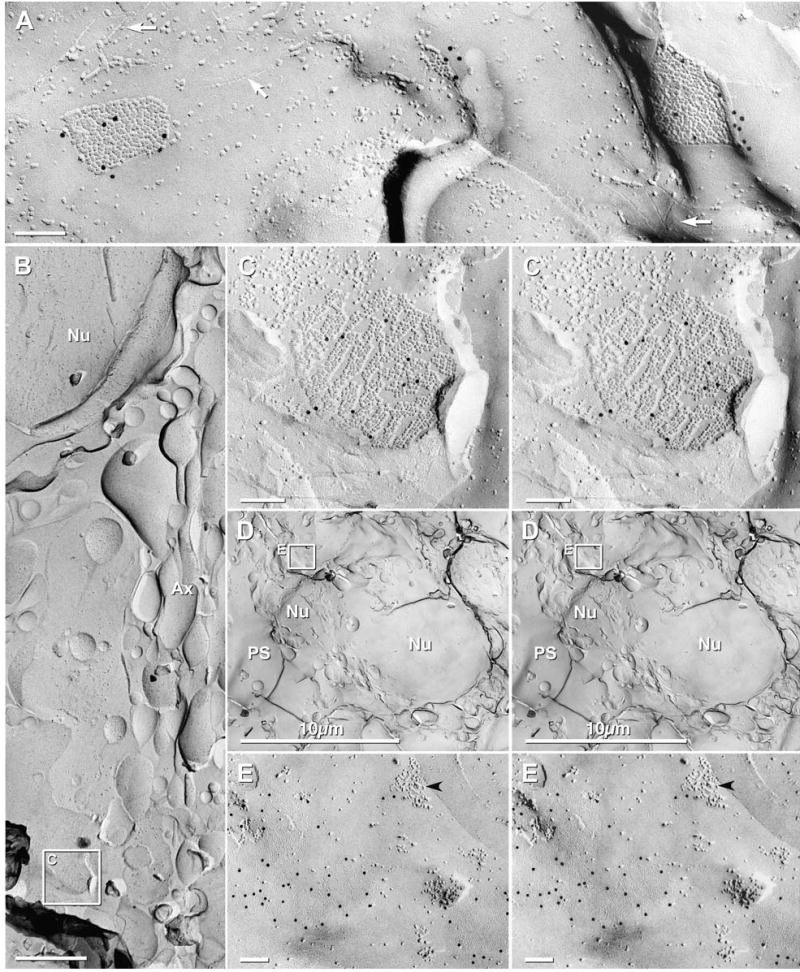
Immunogold labeling for Cx43 in gap junctions of the pia mater and periglomerular ensheathing cells of adult mouse. (A) Angular gap junctions between layers of the pia mater, labeled for Cx43 (12 nm gold beads) but not for Cx45 (6 nm and 18 nm gold beads, none present). White arrows point to furrows corresponding to P-face images of tight junctions. Tight junctions typically surround and delineate gap junctions. (B) Low magnification image of a periglomerular ensheathing cell. Axon bundles (Ax) on the right side; nucleus (Nu) is at top left, and a Cx43-labeled gap junction is within the inscribed box. (C) Higher magnification view of the labeled gap junction in B. Connexons are arranged in small domains. (D) Low magnification view of periglomerular ensheathing cells. Nu = nucleus, PS = periglomerular space. Inscribed area “E” shown at higher magnification below. (E) Cx43-labeled gap junctions (12 mm gold beads) and “reciprocal patches” (black arrowhead) in E-face of the plasma membrane of a periglomerular ensheathing cell.
OLFACTORY BULB
Overview
A confocal grid map of a FRIL sample of OB that was double-labeled for Cx36 and Cx45 (Figs. 9(A–C)) reveals the locations of all major anatomical components. The pia mater is faintly delineated at the margin of the OB (Fig. 9(A), arrow). Within the OB, the olfactory nerve layer, glomeruli, outer (or external) plexiform layer, mitral cell layer, inner (or internal) plexiform layer, and granule cell layer are easily discerned. However, during subsequent replica cleaning for FRIL, much of the outer plexiform, mitral cell, inner plexiform, and granule cell layers were destroyed (Figs. 9(C), (E); yellow overlay), leaving primarily portions of the glomeruli, olfactory nerve layer and pia mater. Within the remaining portions of 12 glomeruli, 25 Cx36- and/or Cx45-labeled gap junctions were found and mapped (see below). The locations of 21 of the labeled gap junctions are indicated (Fig. 9(D), arrows and arrowheads; color coded according to connexin composition). The merged stacks of confocal images reveal grid bar locations visible by LM (Fig. 9(B)), which were then used to map gap junctions by low-magnification TEM (Fig. 9(C)) and by LM (Fig. 9(D)). Arrows indicate locations of neuronal gap junctions shown in Figs. 12(B–D) (below). It should be noted, however, that the apparent localized concentrations in Cx36 labeling are due to an artifact of analyzing only the small intact portions of the replica; areas devoid of labeling were not examinable (yellow overlay).
Fig. 9.
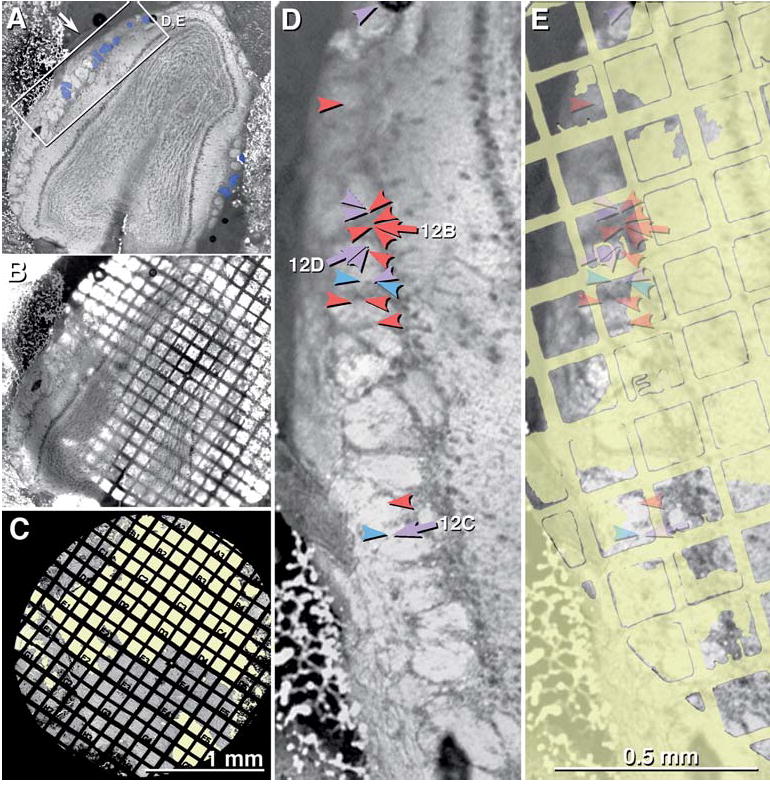
LM and TEM images of adult mouse olfactory bulb after freeze-fracture replication and while the tissue was still present on a TEM gold grid. (A) Brightfield confocal image of entire olfactory bulb. Arrow points to pia mater. Inscribed box is shown at higher magnification as 9D-E. Blue overlay indicates the glomeruli in which Cx36- and/or Cx45-labeled neuronal gap junctions were found (Fig. 12, below). (B) Confocal image stack showing same tissue slice, while bound to gold TEM “index”grid. (C) Very low magnification TEM image of the same tissue, now seen as a FRIL replica. Where the replica was destroyed, the unobstructed electron beam was seen (yellow overlay). The numbered grid openings within the “index” grid allow precise localization of labeled gap junctions mapped at high TEM magnification. (D) Higher magnification confocal image of the inscribed area in Fig. 9(A). Individual glomeruli are easily discerned (bright areas). Twenty-one overlay arrows and arrowheads are color-coded to indicate the connexins found in neuronal gap junctions in this photomapped FRIL replica. Red = Cx36; blue = Cx45; purple = Cx36 + Cx45. Numbered arrows (12B, 12C, and 12D) are shown in figures having the same number. (E) Yellow areas indicate regions of the replica that could not be examined because of fragmentation/destruction of the replica or presence of grid bars. The semitransparent overlay arrows indicates the locations of gap junctions (from “9D”) and their exact locations within individual grid openings.
Fig. 12.
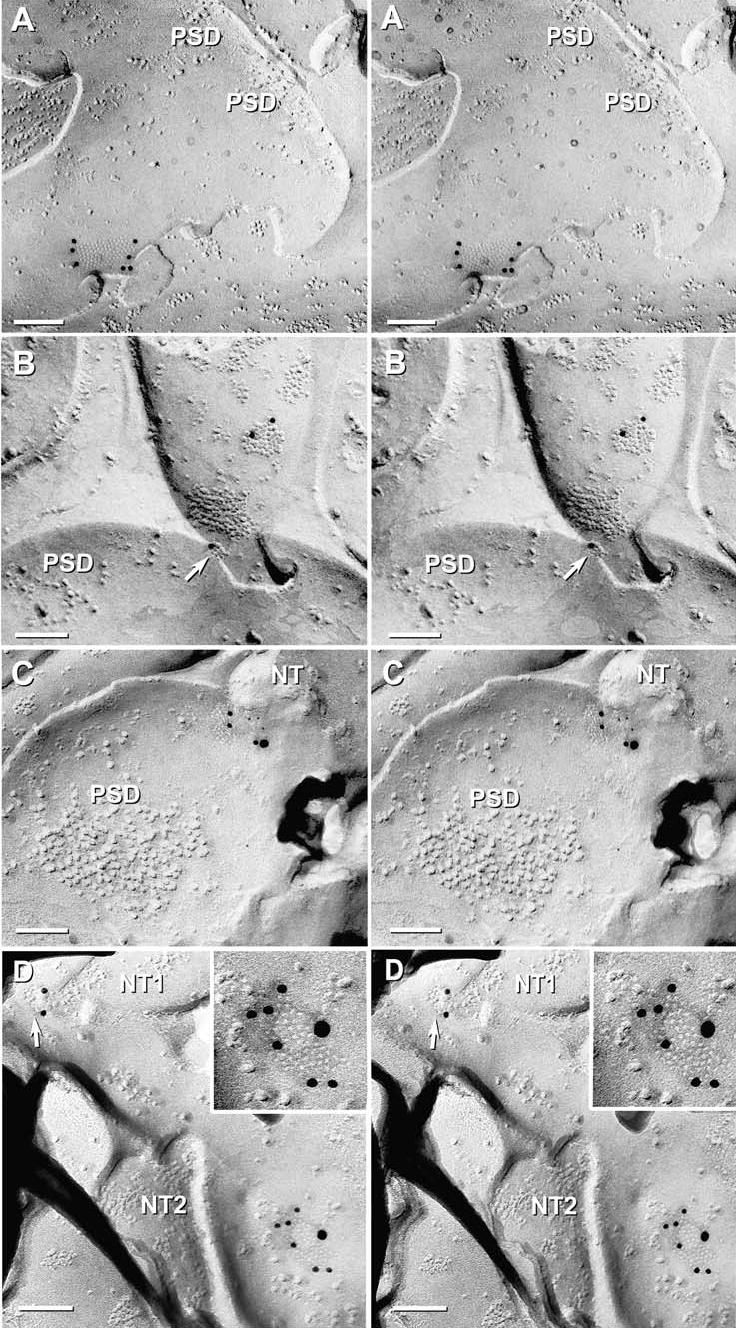
High-magnification stereoscopic images of Cx36 single-labeled and Cx36 and Cx45 double-labeled gap junctions at “mixed” synapses in adult mouse olfactory bulb. (A) E-face image of neuronal gap junction labeled for Cx36 (12 nm gold beads). PSD = postsynaptic density. (B) Two Cx36-labeled gap junctions on a presynaptic neurite. Note the narrowing of the extracellular space at the point of contact of the lower gap junction (arrow). PSD = postsynaptic density. (C) Gap junction double-labeled for Cx36 (12 nm gold beads) and Cx45 (6 nm and 18 nm gold beads). NT = nerve terminal. PSD = postsynaptic density. (D) Two gap junctions linking two neurites (NT1 and NT2) to a postsynaptic dendrite. Upper gap junction (arrow) is single-labeled for Cx36, whereas the lower gap junction (shown at higher magnification in the inset) is double labeled for Cx36 (six 12 nm gold beads) and Cx45 (one 18 nm gold bead).
Pia mater and olfactory bulb ensheathing cells
The olfactory bulb is surrounded by a modified pia/glia limitans (Doucette, 1991). As in other areas of the CNS, the outer layer of the OB is composed of flattened cells of the pia mater and arachnoid mater; however, a conventional glia limitans composed of flattened astrocyte processes is absent in most areas (Doucette, 1991). The pia mater is separated from the olfactory nerve layer by numerous arterioles, venules, and capillaries; multiple layers of axon bundle ensheathing cells, and occasional flattened astrocyte end-feet that are richly endowed with AQP4 square arrays (Rash et al., 1998b, 2004a). As the axon bundles cross the pia mater, their ensheathing cells are almost indistinguishable from the epithelioid cells of the pia mater, identifiable primarily by the more abundant tight junctions linking multiple cell layers of the pia mater (Fig. 10(A)) vs. the lower density of tight junctions in ensheathing cells (Fig. 10(B)) Both ensheathing cells and cells of the pia mater have abundant gap junctions, but in pia mater, they are often closely encircled by tight junction strands (Nagy et al., 2001). Gap junctions of both pia mater (Fig. 10(A)) and ensheathing cells (Figs. 10(B–E)) were well labeled for Cx43 but were not labeled for Cx45 or Cx36 (Fig. 10; triple-labeled for Cx43, Cx45, and AQP4; also see Table 2).
Astrocytes
Astrocytes within and near glomeruli were identified by the presence of distinctive glial fibrillary acidic protein (GFAP) filaments in their cytoplasms (Fig. 11(A)) and by square arrays in their plasma membranes (Figs. 11(B–C)). Immunogold labeling for Cx43 was abundant in astrocyte gap junctions (Fig. 11(A)), whereas labeling for Cx36 and Cx45 was absent from astrocytes, both within and outside the glomeruli. Likewise, in a sample that was quadruple-labeled for Cx36 + NMDAR1 and AQP4 + Cx45, AQP4 labeling was robust beneath square array P-faces (Fig. 11(B); 6 nm and 18 nm gold), whereas labeling for Cx36 and Cx45 was not present in astrocyte gap junctions (Fig. 11(B)).
Fig. 11.
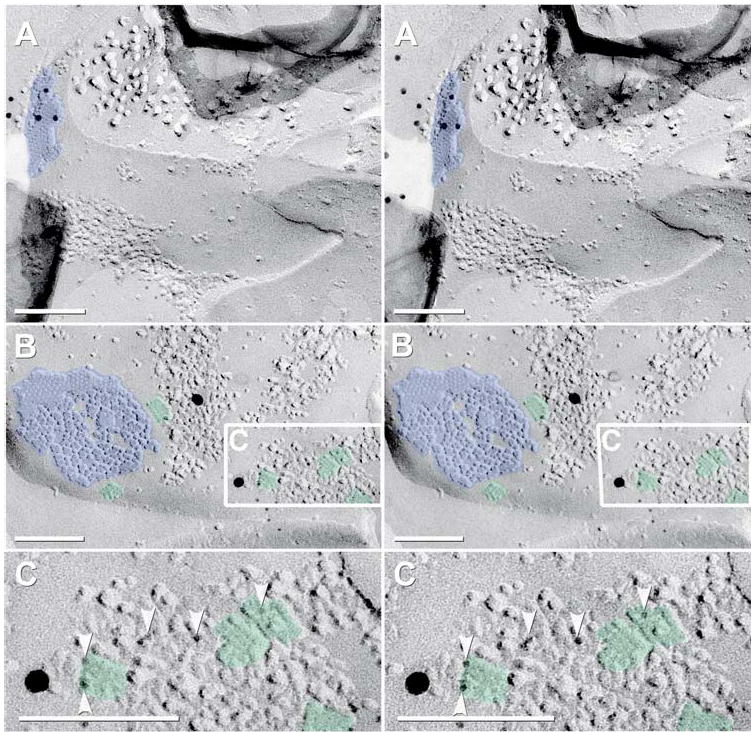
Stereoscopic FRIL images of immunogold-labeled gap junctions and AQP4 arrays in intraglomerular astrocytes of adult mouse. (A) Cross-fractured astrocyte (identified by GFAP filaments in the cytoplasm) and Cx43-labeled gap junction (12 mm gold beads). (B) “Square arrays” immunogold labeled for AQP4 (6 nm and 18 nm gold beads). (C) High-magnification reverse stereoscopic image of the inscribed area in “B”. Arrowheads point to 6 nm gold beads, which now appear on “top”of the replica. One 18 nm gold bead also is present. Blue overlay = astrocyte gap junctions; green overlay = AQP4 array.
Juxtaglomerular neurons
Within the olfactory bulb, glomeruli are delineated by a shell of neuronal somata—the “periglomerular neurons” and external tufted cells, collectively called “juxtaglomerular neurons”—plus ensheathing cells (Figs. 10(D–E)). Neither gap junctions nor immunogold labeling was found on the somata or neurites of the juxtaglomerular neurons. However, periglomerular neurons were recently shown to share gap junctions with mitral/tufted cells within the glomerular neuropil (Kosaka & Kosaka, 2005) (see next). Without cell-specific markers for FRIL, we were not able to separately identify juxtaglomerular neurons or their gap junctions. Nevertheless, several Cx36-labeled gap junctions were found at the margins of glomeruli (Fig. 9(D)), consistent with observations by Kosaka and Kosaka (2005).
Intraglomerular neuronal gap junctions contain Cx36 and lesser amounts of Cx45 but no Cx43
ORN axons form chemical synapses within spheroidal complexes of neuropil called olfactory glomeruli, which consist primarily of axon terminals of ORNs and the dendrites of mitral/tufted cells and periglomerular neurons (Shepherd & Greer, 1998). Previous experiments suggested the presence of Cx36 mRNA and Cx45 mRNA in ORNs, in mitral/tufted cells, and in juxtaglomerular neurons (Condorelli et al., 1998, 2000; Zhang & Restrepo, 2002, 2003; Degen et al., 2004; Christie et al., 2005; Zufall, 2005), and in addition, Cx43 mRNA was reported in ORN somata (Zhang et al., 2000) and in mitral cells (Miragall et al., 1996a). Thus, we tested whether Cx36, Cx43, and/or Cx45 (all reported to be synthesized in ORNs in the OE) might be exported to ORN nerve terminals, where any or all of those connexins might form ORN gap junctions with mitral/tufted cells or with juxtaglomerular cells. In single-, double-, and triple-labeled samples, no neuronal gap junctions were labeled for Cx43 (Table 2), even though many dozens of gap junctions in several other cell types were well labeled for Cx43. Thus, as in other areas of the CNS (Rash et al., 2001), we found no evidence for Cx43 as a neuronal connexin.
FRIL analysis of samples quadruple-labeled for Cx36 + NMDAR1 and AQP4 + Cx45, as well as double-labeled for Cx36 and Cx45 revealed extensive Cx36-immunolabeling of gap junctions in the olfactory bulb. Overall, a total of 34 immunogold-labeled neuronal gap junctions were found, including 23 that were single-labeled for Cx36, eight that were double-labeled for Cx36 and Cx45, and three that were single-labeled for Cx45 (Fig. 12). In a quadruple-labeled replica, four neuronal gap junctions were found – all labeled for Cx36 but not for Cx45 (Fig. 12(A)). In another replica double-labeled for Cx36 and Cx45, 25 immunogold-labeled gap junctions were found, 14 of which were labeled for Cx36 (Fig. 12(B)), eight were double-labeled for Cx36 and Cx45 (Figs. 12(C–D)), and three were single-labeled for Cx45 (not shown). Of these, 50% (17–21 out of 34) were at identifiable “mixed” (chemical plus electrical) synapses within photomapped glomeruli (range based on application of different criteria). Mixed synapses were identified based on the presence of clusters of distinctive 10 nm E-face IMPs closely resembling postsynaptic clusters of IMPs at presumptive glutamatergic synapses (Harris & Landis, 1986), some of which were within 0.2 μm of gap junctions (Fig. 12(C); also see below).
Mitral cell layer, plexiform layers and granule cell layer
Due to technical problems (replica fragmentation and undigested debris in the central area of the olfactory bulb; Figs. 9(C), (E); yellow overlay), we were unable to examine central areas of five replicas of OB. Consequently, we were unable to identify gap junctions or to determine their composition in the outer plexiform, mitral cell, inner plexiform, or granule cell layers. Additional FRIL experiments are in progress to obtain replicas from those areas.
IMMUNOGOLD IDENTIFICATION OF NMDA AND AMPA GLUTAMATE RECEPTORS
Both NMDA and AMPA glutamate receptors are abundant in OB (Sassoe-Pognetto & Ottersen, 2000; Sassoe-Pognetto et al., 2003). In other brain regions, distinctive clusters of E-face IMPs have been shown by FRIL to contain NMDA glutamate receptors (Pereda et al., 2003, 2004; Rash et al., 2004b), usually commingled with AMPA receptors. In ongoing studies of hippocampus, for example, we found that >90% of all clusters of 10 nm E-face IMPs were immunogold labeled for glutamate receptors, with most clusters containing both NMDA and AMPA subunits (Figs. 13(A–D)), albeit in highly variable ratios. NMDAR1 labeling of similar postsynaptic densities (PSDs) in thin sections of olfactory bulb (Sassoe-Pognetto & Ottersen, 2000; Sassoe-Pognetto et al., 2003) and in dense clusters of E-face IMPs that correspond to transmembrane components of the PSDs seen in thin sections (Fig. 13) are consistent with early suggestions that most or all such PSD-related E-face IMP clusters correspond to glutamate receptors (Harris & Landis, 1986). Because apparently identical clusters of 10 nm E-face IMPs were seen closely associated with neuronal gap junctions in OB (e.g., Fig. 12(C)), we used FRIL to analyze the composition of those distinctive PSDs. In one sample of OB that was double-double-labeled for Cx36+NMDAR1 and Cx45+AQP4, one large dendrite exhibited three Cx36-labeled gap junctions and five NMDAR1-labeled PSDs (Figs. 14(A–E)), one within 2 μm of the gap junction. (The rationale for using the same size of gold bead to label distinctly different structures, particularly those on different fracture faces, is described in Materials and methods.) Because of its large size and presence of multiple asymmetric synapses, this process was tentatively identified as a mitral/tufted cell dendrite, consistent with evidence from thin-section TEM that Cx36 occurs primarily or exclusively at MC/MC dendritic gap junctions (Christie et al., 2005; see also Chen & Shepherd, this issue p. 353). Moreover, that same report used physiological monitoring strategies to reveal that electrical coupling occurred at AMPA glutamatergic mixed synapses (Christie et al., 2005), perhaps co-localized with NMDA receptors (Christie et al., 2001, 2005; Schoppa & Westbrook, 2002; Zufall, 2005). However, because mitral-cell-specific markers are not yet available for FRIL, we were not able to positively identify either pre- or postsynaptic cells at these synapses. Nevertheless, we note that because there are four potential fracture faces at each conventional synapse (i.e. presynaptic E-face, presynaptic P-face, postsynaptic P-face and postsynaptic E-face), each fracture has a 25% probability of revealing the postsynaptic E-faces. This is significant because postsynaptic E-faces are recognized by their content of 10 nm IMPs corresponding to glutamate receptors. Thus, the observation of >50% of fracture faces having glutamate receptor-like IMP clusters (17–21 out of 34, or more than twice the expected value calculated above) suggested that most Cx36- and Cx36+Cx45-containing gap junctions were at glutamate mixed synapses. This high ratio is also consistent with proposals that many of these mixed synapses correspond to “reciprocal” glutamatergic synapses between large dendrites (Christie et al., 2005; Kosaka & Kosaka, 2005).
Fig. 13.
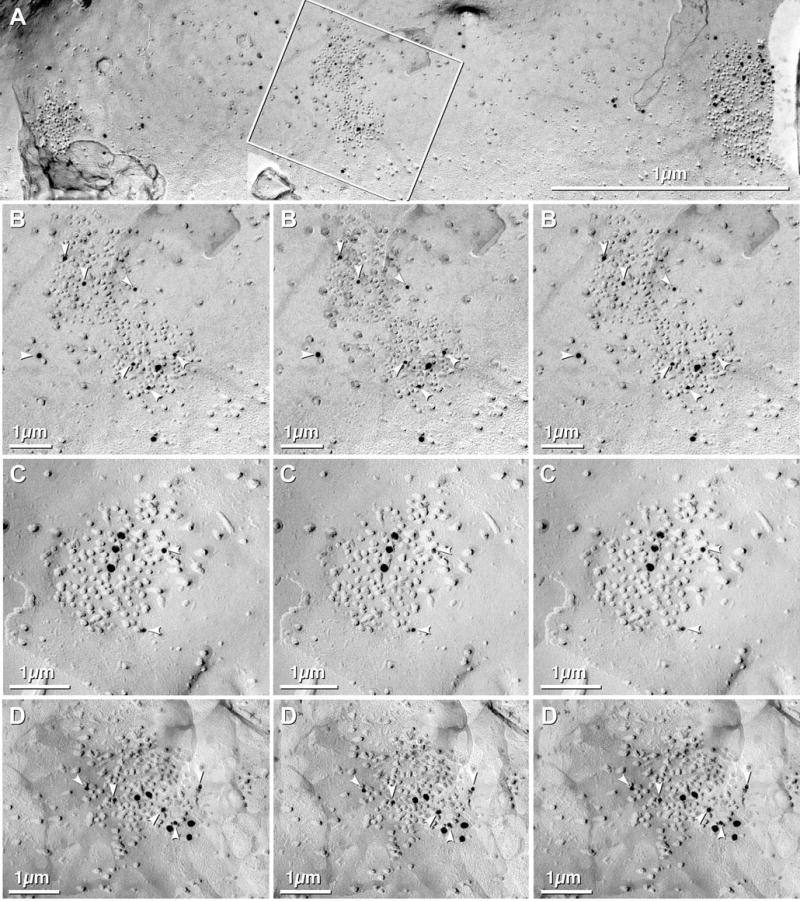
E-face image of large dendrite in adult rat hippocampus after double-labeling for NMDA and AMPA glutamate receptors. (A) Three large PSDs consisting primarily of 10 nm IMPs, plus some widely distributed 10 nm IMPs. PSDs are well labeled for panAMPA (10 nm gold beads) and NMDAR1 (20 nm gold beads). Inscribed area (middle PSD) is shown at higher magnification in B. (B) High-magnification stereoscopic and reverse stereoscopic image of one of three PSDs double-labeled for NMDA and AMPA receptors. White arrowheads point to 10 nm gold beads, labeling AMPA glutamate receptors. (C–D) High-magnification stereoscopic and reverse stereoscopic image of PSD labeled for NMDA and AMPA receptors, but with reversed sizes of gold beads (10 nm = NMDAR1; 20 nm = panAMPA). White arrowheads = 10 nm gold beads.
Fig. 14.
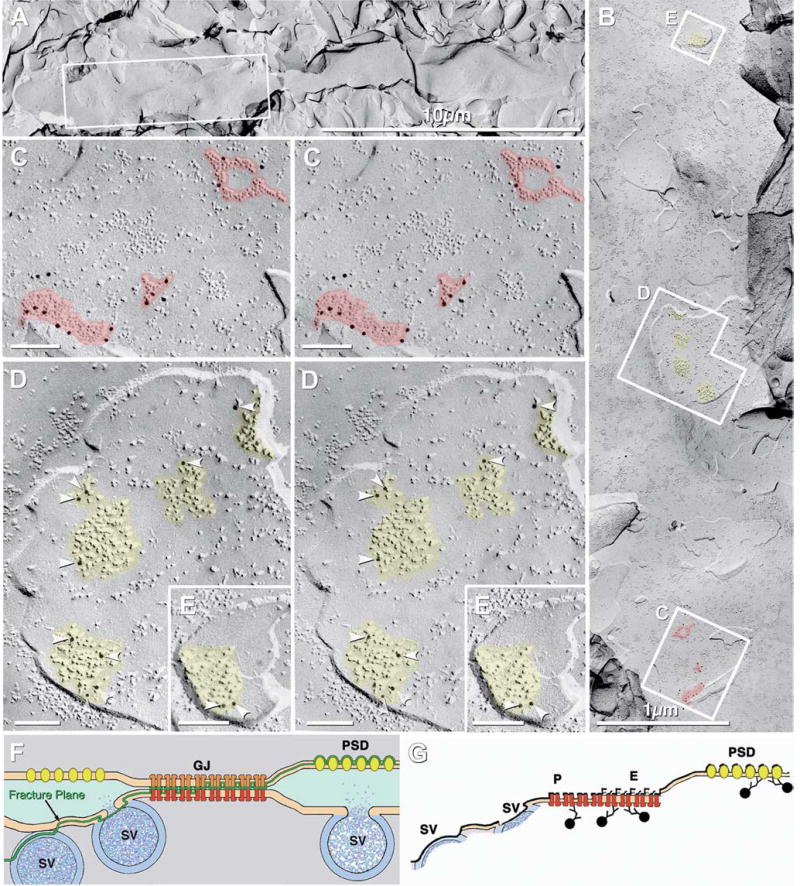
Large dendrite with multiple postsynaptic terminals in a sample of adult mouse that was quadruple-labeled (“double-double” labeled) for Cx36+NMDAR1 (both labeled with 12 nm gold beads), and Cx45+AQP4 (both labeled with 6 nm and 18 nm gold beads; none present). (A) Low-magnification image of dendrite. Inscribed area shown at higher magnification in “B”. (B) Intermediate-magnification view of the same dendrite, rotated counter clockwise about 90′. Inscribed areas are shown at higher magnification in images C–E. Yellow overlay marks PSDs labeled for NMDAR1. Red overlay = gap junctions labeled for Cx36. (C) Higher-magnification view of three gap junctions (red overlay) immunolabeled for Cx36 (12 nm gold beads). (D–E) PSDs immunogold labeled for NMDAR1 (12 nm gold beads, arrowheads). (F) Drawing of fracture plane (green line) through mixed synapse. Glutamate receptor PSDs are revealed as E-face particles (yellow), with immunogold-labeled extracellular epitopes remaining beneath E-face IMPs. (G) Drawing of immunogold- labeled gap junction and glutamate receptor PSD. SV = synaptic vesicle; GJ = gap junction; P = P-face; E = E-face; PSD = postsynaptic density.
GENOMIC ANALYSIS: IDENTIFICATION OF miRNA “SEED MATCHING” SEQUENCES IN mRNAS FOR Cx36, Cx43, AND Cx45
In situ hybridization suggested the presence of mRNAs for Cx36, Cx43, and Cx45 in ORNs, and histochemical staining revealed the expression of β-galactosidase in ORNs of transgenic mice that contain the Cx43- or Cx36-promoter driven LacZ reporter gene (Zhang et al., 2000; Zhang & Restrepo, 2002, 2003). In contrast, current LM and FRIL data cast doubt on the presence of Cx36, Cx43, and Cx45 proteins in ORN somata, dendrites, axons or axon terminals. One possible explanation that was considered was that the connexin genes were transcribed into mRNA but that the mRNA was not translated into protein. Consequently, we examined genomic data for evidence consistent with post-transcriptional repression of protein synthesis.
Background
MicroRNAs (miRNAs) are small non-coding, endogenous RNAs, typically 21–22 nucleotides (22nt) long, that play important roles in gene regulation, especially in development and in tissue-type and cell subtype specification (reviewed by Bartel, 2004; Wienholds et al., 2005). miRNAs are thought to bind mainly to complementary sites on 3′ untranslated regions (3′ UTR) of target mRNAs and repress protein synthesis at the stage of ribosomal translation (Bartel, 2004). In many cases, the level of mRNAs is not substantially affected by miRNAs, whereas in other cases (Lim et al., 2005) reduction of transcript levels is observed. The complementarity of miRNAs to target sites is usually imperfect except in plant cells (Rhoades et al., 2002). The pairing of the miRNA 5′ region to its target mRNA is believed to be most important and essential for target recognition; therefore the nucleotide positions 2–8 at the 5′ regions of miRNAs are called the miRNA “seed sequences” (Lewis et al., 2003). miRNA binding sites that give rise to comparable pairing energies at 5′ seed regions are known to show considerable differences in targeting efficiency depending on the pairing position. There is no clear correlation between pairing energy at 5′ seed regions vs. targeting efficiency, and any mismatch or G:U wobble base-pairing in this region is highly detrimental beyond the prediction based on pairing energies (Doench & Sharp, 2004; Brennecke et al., 2005). The latter report classified miRNA binding sites into two broad categories; 5′ dominant sites and 3′ compensatory sites. 5′ dominant sites depend primarily on seed matching, with varying degrees of 3′ pairing thought not to be essential for targeting. 3′ compensatory sites have imperfect complementarity with most seed sequences; hence they require extensive pairing at 3′ regions of miRNAs to be effective. 3′ compensatory sites are suggested to be present in few miRNAs because the more extensive base-pairing to mRNAs would be more difficult to maintain over the course of evolution (Brennecke et al., 2005; Lewis et al., 2005).
Most miRNAs are brain-specific or brain-enriched, and many have been identified as neuron-specific (Sempere et al., 2004), with some considered to regulate neuronal development (Smirnova et al., 2005) and others to maintain neuronal differentiation (Wienholds et al., 2005). Based on extensive bioinformatics studies, it was suggested that each mRNA can be regulated by multiple miRNAs simultaneously (Lewis et al., 2003). Moreover, it has been suggested that at least three miRNA binding sites are required for complete gene silencing (Doench et al., 2003). However, a single 8nt seed match was shown to be sufficient to confer strong regulation (Brennecke et al., 2005).
Genomic searches
Based on the above, and to identify 5′ dominant sites, we performed genomic searches for possible miRNA “seed matching” in the 3′ UTR for all mouse connexins reported in ORNs. The annotated 3′ UTRs for mCx36, mCx43, and mCx45 were retrieved from Ensembl genome database using MartView (see Materials and methods for access information) and extended to 2000 nucleotide bases by including downstream flanking sequences. Mouse miRNA sequences were retrieved from miRBase. TargetScan currently predicts 9 miRNAs for Cx43 but none for Cx36 or Cx45, whereas Miranda predicted 8 miRNAs for Cx36, 15 miRNAs for Cx43 and 7 miRNAs for Cx45, consistent with the possibility that the synthesis of these three connexins may be down-regulated by miRNA mechanisms. Thus, we searched the 3′ UTRs for seed matching against a total of 233 mouse mature miRNAs. We found 25 miRNAs that have at least one 7nt seed match in the 3′ UTR for mCx36, 49 miRNAs for mCx43, and 38 miRNAs for mCx45 (Tables 3–5), indicating the existence of additional, previously unpredicted miRNA binding sites in each of these mRNAs. Many of these sequences correspond to the most effective 8nt seed sequences (including 11 for Cx36 (Table 4), 18 for Cx43 (Table 3), and 10 for Cx45 (Table 5)), any one of which may be able to strongly suppress synthesis of the target protein (Brennecke et al., 2005). In addition, two 7nt seed sequences may be as effective as one 8nt seed sequence in repressing protein expression (Brennecke et al., 2005). Some of the additional sequences identified by us were not detected in previous predictions because they used algorithms that filtered out some target sites as potential false positives that were not identically shared by humans, mice and rats (TargetScan) or shared by humans and rats or humans and mice (Miranda), or because they used different strategies for searching target sequences. For example, Miranda requires additional pairing at 3′ region of miRNA rather than complete seed matching. In our searches, about half of the seed matches for Cx36, Cx43, Cx45 were found to be conserved between mouse and human, and half were not present in humans, and thus would have been rejected by TargetScan.
Table 3.
Potential miRNA binding sites for mouse Cx43a).
| miRNA ID | Seedb) | Seed positionc) | Humand) | ||
|---|---|---|---|---|---|
| mmu-miR-1 | UGGAAUGU | 322 | 8nt | 478 | 8nt |
| ” | 1446 | 8nt | 1609 | 8nt | |
| mmu-miR-7 | UGGAAGAC | 524 | 8nt | – | |
| mmu-miR-7b | UGGAAGAC | 524 | 8nt | – | |
| mmu-miR-15a | UAGCAGCA | 1741 | – | ||
| mmu-miR-15b | UAGCAGCA | 1741 | – | ||
| mmu-miR-16 | UAGCAGCA | 1741 | – | ||
| mmu-miR-19a | UGUGCAAA | 1225 | 495, 1377 | ||
| mmu-miR-19b | UGUGCAAA | 1225 | 495, 1377 | ||
| mmu-miR-23a | AUCACAUU | 1057 | 1211 | ||
| mmu-miR-23b | AUCACAUU | 1057 | 1211 | ||
| mmu-miR-25 | CAUUGCAC | 1727 | 8nt | – | |
| mmu-miR-30a-3p | CUUUCAGU | 969 | – | ||
| mmu-miR-30a-5p | UGUAAACA | 822 | 8nt | 971 | 8nt |
| mmu-miR-30b | UGUAAACA | 822 | 8nt | 971 | 8nt |
| mmu-miR-30c | UGUAAACA | 822 | 8nt | 971 | 8nt |
| mmu-miR-30d | UGUAAACA | 822 | 8nt | 971 | 8nt |
| mmu-miR-30e | UGUAAACA | 822 | 8nt | 971 | 8nt |
| mmu-miR-30e* | CUUUCAGU | 969 | – | ||
| mmu-miR-32 | UAUUGCAC | 1727 | – | ||
| mmu-miR-92 | UAUUGCAC | 1727 | – | ||
| mmu-miR-101a | UACAGUAC | 1032 | 8nt | 1186 | 8nt |
| mmu-miR-101b | UACAGUAC | 1032 | 8nt | 1186 | 8nt |
| mmu-miR-130a | CAGUGCAA | 1226 | 496, 1378 | ||
| mmu-miR-130b | CAGUGCAA | 1226 | 496, 1378 | ||
| mmu-miR-135a | UAUGGCUU | 641 | 793 | ||
| mmu-miR-135b | UAUGGCUU | 641 | 793 | ||
| mmu-miR-141 | UAACACUG | 328 | – | ||
| ” | 768 | – | |||
| ” | 1848 | – | |||
| mmu-miR-150 | UCUCCCAA | 1953 | 8nt | 2306,2338 | |
| mmu-miR-154 | UAGGUUAU | 1570 | – | ||
| mmu-miR-195 | UAGCAGCA | 1741 | – | ||
| mmu-miR-199a* | UACAGUAG | 1461 | – | ||
| mmu-miR-200a | UAACACUG | 328 | – | ||
| ” | 768 | – | |||
| ” | 1848 | – | |||
| mmu-miR-206 | UGGAAUGU | 322 | 8nt | 478 | 8nt |
| ” | 1446 | 8nt | 1609 | 8nt | |
| mmu-miR-216 | UAAUCUCA | 227 | 2075 | 8nt | |
| mmu-miR-218 | UUGUGCUU | 859 | 1005 | ||
| mmu-miR-221 | AGCUACAU | 949 | – | ||
| mmu-miR-222 | AGCUACAU | 949 | – | ||
| mmu-miR-224 | UAAGUCAC | 1337 | 8nt | – | |
| mmu-miR-290 | CUCAAACU | 1860 | – | ||
| mmu-miR-301 | CAGUGCAA | 1226 | 496, 1378 | ||
| mmu-miR-322 | AAACAUGA | 1381 | 8nt | – | |
| ” | 1467 | – | |||
| mmu-miR-323 | GCACAUUA | 1056 | 1210 | ||
| mmu-miR-363 | AUUGCACG | 845 | e) | ||
| mmu-miR-377 | AUCACACA | 1585 | 8nt | 2255 | 8nt |
| mmu-miR-381 | UAUACAAG | 1176 | 1323 | ||
| mmu-miR-412 | ACUUCACC | 1259 | – | ||
| mmu-miR-424 | CAGCAGCA | 1741 | – | ||
| mmu-miR-434-3p | UUUGAACC | 453 | 8nt | e) | |
| mmu-miR-466 | AUACAUAC | 55 | e) | ||
Ensemble Gene ID : ENSMUSG00000050953
UCUUUGGU Incomplete underlining = 7nt seed match (bases 2–8 of the miRNA)
UCUUUGGU Complete underlining = 8nt seed match (bases 1–8 of the miRNA)
All are 7nt seed match unless designated as 8nt.
The first nucleotide of the seed match position in the 3′ UTR.
Number is relative to the start (+1) of the 3′ UTR.
Seed conservation with human Cx43 (Ensemble Gene ID : ENSG00000152661)
Ortholog of the miRNA has not been found in human.
”Additional matches for same miRNA
– Not present in the first 2500nt
= asterisk is part of the name of the miRNA
Table 5.
Potential miRNA binding sites for mouse Cx45.a)
| miRNA ID | Seeda) | Seed positionc) | Humand) | ||
|---|---|---|---|---|---|
| mmu-miR-7 | UGGAAGAC | 235 | 8nt | 236 | 8nt |
| mmu-miR-7b | UGGAAGAC | 235 | 8nt | 236 | 8nt |
| mmu-miR-15a | UAGCAGCA | 697 | – | ||
| ” | 1835 | – | |||
| mmu-miR-15b | UAGCAGCA | 697 | – | ||
| ” | 1835 | – | |||
| mmu-miR-16 | UAGCAGCA | 697 | – | ||
| ” | 1835 | – | |||
| mmu-miR-30a-3p | CUUUCAGU | 1024 | – | ||
| mmu-miR-30e* | CUUUCAGU | 1024 | – | ||
| mmu-miR-125a | UCCCUGAG | 129 | 129 | ||
| mmu-miR-125b | UCCCUGAG | 129 | 129 | ||
| mmu-miR-126-5p | CAUUAUUA | 1210 | – | ||
| ” | 1731 | – | |||
| mmu-miR-137 | UUAUUGCU | 202 | 8nt | e) | |
| mmu-miR-141 | UAACACUG | 178 | 179 | ||
| ” | 1220 | 1556 | |||
| mmu-miR-193 | AACUGGCC | 1128 | – | ||
| mmu-miR-194 | UGUAACAG | 638 | – | ||
| mmu-miR-195 | UAGCAGCA | 697 | – | ||
| ” | 1835 | – | |||
| mmu-miR-199a | CCCAGUGU | 1251 | – | ||
| mmu-miR-199b | CCCAGUGU | 1251 | – | ||
| mmu-miR-200a | UAACACUG | 178 | 179 | ||
| ” | 1220 | 1556 | |||
| mmu-miR-200b | UAAUACUG | 300 | 8nt | 301 | 8nt |
| ” | UAAUACUG | 1149 | 1191 | ||
| mmu-miR-200c | UAAUACUG | 300 | 8nt | 301 | 8nt |
| ” | UAAUACUG | 1149 | 1191 | ||
| mmu-miR-204 | UUCCCUUU | 1971 | 8nt | 168 | 8nt |
| mmu-miR-207 | GCUUCUCC | 851 | e) | ||
| mmu-miR-211 | UUCCCUUU | 1971 | 8nt | 168 | 8nt |
| mmu-miR-217 | UACUGCAU | 209 | – | ||
| mmu-miR-290 | CUCAAACU | 1517 | 8nt | e) | |
| mmu-miR-291-5p | CAUCAAAG | 709 | 8nt | e) | |
| mmu-miR-292-5p | ACUCAAAC | 1518 | e) | ||
| mmu-miR-300 | UAUGCAAG | 1550 | e) | ||
| mmu-miR-322 | AAACAUGA | 1281 | 8nt | e) | |
| mmu-miR-324-5p | CGCAUCCC | 103 | 103 | ||
| ” | 748 | – | |||
| mmu-miR-330 | GCAAAGCA | 707 | 735 | ||
| ” | 1054 | – | |||
| mmu-miR-351 | UCCCUGAG | 129 | 129 | ||
| mmu-miR-376b | AUCAUAGA | 722 | – | ||
| mmu-miR-424 | CAGCAGCA | 697 | 8nt | – | |
| ” | 1835 | 8nt | – | ||
| mmu-miR-431 | UGUCUUGC | 1390 | 8nt | – | |
| mmu-miR-465 | UAUUUAGA | 1121 | e) | ||
| mmu-miR-466 | AUACAUAC | 293 | e) | ||
| mmu-miR-468 | UAUGACUG | 1131 | e) | ||
Ensemble Gene ID : ENSMUSG00000034520
UCUUUGGU: 7nt seed match (bases 2–8 of the miRNA)
UCUUUGGU: 8nt seed match (bases 1–8 of the miRNA)
All are 7nt seed match unless designated as 8nt.
The first nucleotide of the seed match position in the 3′ UTR.
Number is relative to the start (+ 1) of the 3′ UTR.
Seed conservation with human Cx45 (Ensemble Gene ID : ENSG00000182963)
Ortholog of the miRNA has not been found in human.
” Additional matches for same miRNA
– Not present in the first 2500nt
Table 4.
Potential miRNA binding sites for mouse Cx36.1)
| miRNA ID | Seedb) | Seed positionc) | Humand) | ||
|---|---|---|---|---|---|
| mmu-miR-9 | UCUUUGGU | 1158 | 1181 | ||
| mmu-miR-29a | UAGCACCA | 684 | 709 | ||
| mmu-miR-29b | UAGCACCA | 684 | 709 | ||
| mmu-miR-29c | UAGCACCA | 684 | 709 | ||
| mmu-miR-31 | AGGCAAGA | 1406 | 8nt | – | |
| mmu-miR-128a | UCACAGUG | 1249 | 1275 | ||
| mmu-miR-128b | UCACAGUG | 1249 | 1275 | ||
| mmu-miR-130a | CAGUGCAA | 678 | 8nt | – | |
| mmu-miR-130b | CAGUGCAA | 678 | 8nt | – | |
| mmu-miR-136 | ACUCCAUU | 877 | – | ||
| mmu-miR-148a | UCAGUGCA | 679 | 704 | ||
| mmu-miR-148b | UCAGUGCA | 679 | 704 | ||
| mmu-miR-152 | UCAGUGCA | 679 | 704 | ||
| mmu-miR-181a | AACAUUCA | 1476 | 8nt | 1644 | 8nt |
| mmu-miR-181b | AACAUUCA | 1476 | 8nt | 1644 | 8nt |
| mmu-miR-181c | AACAUUCA | 1476 | 8nt | 1644 | 8nt |
| mmu-miR-183 | UAUGGCAC | 1225 | 8nt | 1251 | 8nt |
| mmu-miR-190 | UGAUAUGU | 1536 | 176 | 8nt | |
| mmu-miR-301 | CAGUGCAA | 678 | 8nt | – | |
| mmu-miR-320 | AAAAGCUG | 476 | 8nt | 494 | 8nt |
| mmu-miR-350 | UUCACAAA | 366 | – | ||
| mmu-miR-365 | UAAUGCCC | 1834 | – | ||
| mmu-miR-383 | AGAUCAGA | 1979 | 8nt | – | |
| mmu-miR-410 | AAUAUAAC | 1377 | 8nt | – | |
| mmu-miR-433-5p | UACGGUGA | 755 | e) | ||
Ensemble Gene ID : ENSMUSG00000058516
UCUUUGGU: 7nt seed match (bases 2–8 of the miRNA)
UCUUUGGU: 8nt seed match (bases 1–8 of the miRNA)
All are 7nt seed match unless designated as 8nt.
The first nucleotide of the seed match position in the 3′ UTR.
Number is relative to the start (+ 1) of the 3′ UTR.
Seed conservation with human Cx36 (Ensemble Gene ID : ENSG00000159248)
Ortholog of this miRNA has not been found in humans.
”Additional matches for same miRNA
– Not present in the first 2500nt
It is not yet clear if non-conserved sequences are relevant to differences in CNS cell-type specification in mouse vs. humans. If so, the other methods will likely have improved signal-to-noise ratios in target prediction for “universal” genes but may miss important regulatory mechanisms that may differ in human vs. rodent CNS. A recent study showed that non- conserved sites also potentially mediate repression by miRNAs as well as conserved sites (Farh et al., 2005). Regardless, we have identified miRNAs that are capable of down-regulating the synthesis of connexins in ORNs and for providing differential regulation of connexin synthesis in closely-related neurons. However, there is as yet no experimental evidence that any of the identified miRNAs perform those functions in OB or OE.
Discussion
OVERVIEW
This report addressed major discrepancies regarding gap junctions, their connexin proteins and their mRNAs in ORNs and SCs of the OE. It also further clarifies the composition of mixed synapses in OB. By both confocal immunofluorescence microscopy and by FRIL, we show that Cx36, Cx43, and Cx45 are not present in detectable amounts in ORN dendrites or somata, and that to the limit of FRIL, ORNs in OE do not possess gap junctions, consistent with the proposed absence of electrical coupling between ORNs (Delay & Dionne, 2003) and between ORNs and SCs (Vogalis et al., 2005a). By FRIL, Cx43 plus Cx45 (but not Cx36) were present in gap junctions of SCs, thereby providing additional immunocytochemical evidence consistent with SC/SC coupling (Vogalis et al., 2005a, b) via Cx43 and Cx45-containing gap junctions. Cx43 (without Cx45 or Cx36) was present in ensheathing cells surrounding axon bundles and in fibroblasts of the lamina propria. However, none of these connexins were detected in unmyelinated ORN axons in the olfactory mucosa, cribriform plate or olfactory nerve layer. No differences were detected in cell-specific labeling for Cx43 labeling in OE of adult mouse and 4 day postnatal rat. In the OB, Cx43 was found in gap junctions of pia mater, glomerular ensheathing cells, and astrocytes but not in any neurons. In contrast, Cx36 and, less abundantly, Cx45 were found in gap junctions linking large dendrites at mixed synapses in OB.
In attempting to reconcile previous data for detection of other mRNAs without detection of the corresponding protein (Willecke et al., 1991; Hennemann et al., 1992; Miragall et al., 1996a; Simberger et al., 1997), we assembled genomic data suggesting regulated repression of connexin synthesis by miRNA-dependent post-transcriptional control mechanisms. These data point to an additional mechanism to consider when accounting for mRNA/protein discrepancies. Thus, our genomic analysis is consistent with an emerging concept for generalized post-transcriptional repressor mechanisms that coordinately limit biological expression of specific groups of genes in diverse cells of the same lineage (C. Anderson and R. Werner, unpublished observations, International Gap Junction Conference, 2005, Whistler, Canada; Abstract, p. 82).
Cx43 PROTEIN NOT DETECTED IN ORNs IN OLFACTORY EPITHELIUM OR OLFACTORY BULB
A previous report suggesting the presence of Cx43 protein in ORNs, presumably within gap junctions in ORN dendrites, somata and axons (Zhang et al., 2000), was not supported by current FRIL labeling with any of a battery of antibodies. Likewise, neither Cx43 nor gap junctions were found in ORN somata or axons within the olfactory mucosa. Although Cx43 was widespread and abundant in epithelioid cells of the pia mater, periglomerular ensheathing cells and astrocytes, Cx43 either was not present or was present in such low amounts in neurons that we were unable to detect any Cx43-labeled neuronal gap junctions in OE or OB. If Cx43 protein occurs in ORNs, those hypothetical Cx43-containing gap junctions must be at least two orders of magnitude fewer than those in SCs and ensheathing cells, and several orders of magnitude less abundant than in fibroblasts of the lamina propria.
Cx36 PROTEIN NOT DETECTED IN ORNs
By both FRIL and LM immunocytochemistry, Cx36 was not detected in ORNs, their axons or axon terminals. In previous demonstrations of LacZ staining in ORNs in mice having a LacZ marker protein under the control of a Cx36 promoter (Zhang & Restrepo, 2003), this LacZ-expression system lacked the regulatory sequences in the 3′ UTR that are likely to be essential in regulating post-translational Cx36 gene expression (see below).
ANALYSIS OF Cx36, Cx43 AND Cx45 IN FIBROBLASTS, ENSHEATHING CELLS, AND BOWMAN’S GLANDS
By immunofluorescence and FRIL, Cx43 is the primary connexin in gap junctions between SCs, confirming previous reports (Miragall et al., 1992 Miragall et al., 1993, 1996b). In single-, double-, and triple-labeling experiments, Cx43 also was abundant in gap junctions between fibroblasts of the lamina propria, and between ensheathing cells surrounding axon bundles. However, even in the largest of these gap junctions, neither Cx36 nor Cx45 were detected. In addition, immunogold labeling for Cx45 was also demonstrated in ca. 30% of SC gap junctions. Because false-positive labeling for Cx45 was nonexistent in the largest gap junctions in lamina propria, pia mater, and ensheathing cells, and because non-specific “background” was low in all other areas, “false-positive” labeling for Cx45 is an unlikely explanation for the labeling seen in the much smaller gap junctions in SCs.
In SCs, gap junctions have been proposed to conduct calcium waves as one aspect of functional coupling of the cells forming the primary trans-epithelial barrier of the OE (Vogalis et al., 2005b). This form of intercellular signaling has been proposed to regulate cell division for rapid cell replacement when the trans-epithelial osmotic and electrical barrier is damaged (Larsen, 1983) Our data are consistent with the hypothesis that Cx43 is the primary connexin responsible for transmission of these calcium waves between SCs, but that Cx45 may also participate.
Cx43 and Cx45 apparently were co-localized in two E-face images of gap junctions in Bowman’s gland cells, suggesting that the BG cell coupling partner (either an SC or another BG cell) express Cx45 (see above). However, the identities of the cellular coupling partners at these heterotypic gap junctions were not determined (i.e., BG/SC or BG/BG). Because of possible proximity of similar sizes of gold beads for AQP4 in BG cells, reservations remain regarding apparent FRIL labeling of Cx45 in BG gap junction coupling partners.
NEITHER CONNEXINS NOR GAP JUNCTIONS DETECTED BY FRIL IN ORN SYNAPSES
In the OB, Cx36- and Cx36 + Cx45-labeled gap junctions were relatively abundant and easily detected, with an average of 3.5 gap junctions detected in each grid opening that contained replicated glomeruli (see Fig. 9(D)). However, none were detected at identifiable ORN terminals, consistent with all previous ultra-structural studies that have been unable to detect gap junctions in freeze-fractured ORN terminals (Miragall et al., 1996a) or in ORN terminals identified by serial section reconstruction (Kosaka & Kosaka, 2003, 2004, 2005; Christie et al., 2005). Those studies and the current data substantially diminish the possibility of ORN gap junctions as a common component in information processing in the OB, but support Cx36 (and Cx45) in mitral/tufted cell gap junctions.
Cx36 IN MITRAL CELL/MITRAL CELL GLUTAMATERGIC MIXED SYNAPSES
This report confirms and extends prior elegant thin-section immunogold labeling data indicating that Cx36-containing gap junctions occur at mixed synapses between mitral/tufted cells and between mitral/tufted cells and periglomerular cells (Christie et al., 2005). In those thin-section serial reconstruction studies, gap junctions were not detected between ORN terminals, or between ORNs and any other neuronal cell type in the OB (Kosaka & Kosaka, 2003, 2004, 2005; Christie et al., 2005). Our FRIL evidence for glutamate receptors at mixed synapses in OB are also consistent with electrophysiological data for coordinate activation and direct feedback between gap junctions and excitatory glutamate receptors during high-frequency spike synchronization of mitral/tufted cell pairs during olfactory information encoding in the CNS (Christie et al., 2005; also Schoppa et al., 1998; Schoppa & Westbrook, 2001, 2002). Most of the Cx36- and Cx36 + Cx45-containing gap junctions found by FRIL were at dendrodendritic mixed synapses, where large dendrites (which therefore, presumably represent mitral/tufted cells) were presynaptic to membranes containing clusters of E-face PSDs that closely resembled glutamate receptor clusters (Harris & Landis, 1986). Identification of the E-face IMP clusters as glutamate receptors was obtained by FRIL labeling of NMDA receptor subtypes in synaptic contacts closely adjacent to Cx36-labeled gap junctions. In companion FRIL studies, we showed generalized co-localization of AMPA and NMDA glutamate receptors in hippocampus, similar to that previously described in abstract form (Utvik et al., 2000). Similar co-localization of NMDA and AMPA receptor subtypes may also occur in the OB at PSDs of mitral/tufted cell pairs, where NMDAR1 was demonstrated. Precedent for such co-localization of NMDA and AMPA receptor subtypes in glutamatergic synapses in other CNS regions has been provided based on post-embedding immunogold labeling (Kharazia et al., 1996; He et al., 1998; Takumi et al., 1999; Racca et al., 2000). Nevertheless, in most cases, ORNs (always presynaptic) were excluded as either synaptic component based on identification of the large presynaptic dendrite with multiple active zones as a mitral/tufted cell dendrite. Consequently, the smaller postsynaptic components with glutamate receptor PSDs could not be ORN presynaptic terminals.
Cx45 IN MIXED SYNAPSES OF OB GLOMERULI
By FRIL, we demonstrated Cx36, and in less abundance, Cx45 in presumptive MC/MC gap junctions in the olfactory bulb of adult mice. However, in the absence of definitive markers for identifying individual neuronal cell types in OB, it is not yet possible by FRIL to identify either the pre- or postsynaptic neurons forming these mixed synapses.
In the retina, Cx45 mRNA (i.e., Cx45 promoter activity) has been detected in retinal ganglion cells and some amacrine cells (Güldenagel et al., 2000), and Cx45 protein has been found in retinal gap junctions by FRIL (Kamasawa et al., 2004; Li et al., unpublished observations). Cx45 was not found in any type of glial cell in the OB but somewhat surprisingly, Cx45 was found in SCs in the OE. SCs are thought to play a supporting role akin to that of glia in the central nervous system (Vogalis et al., 2005a). If Cx45 is confirmed to be expressed both in neurons in the CNS and in glia-like supporting cells in OE, it would be an unusual connexin, as other CNS connexins that have been studied by FRIL have been found to be exclusively either neuronal or glial (Rash et al., 2001).
DISCREPANCY BETWEEN DETECTIONS OF CONNEXIN mRNA vs. PROTEIN
Evidence for mRNAs of Cx36, Cx43, and Cx45 were reported in many/most ORNs (Zhang et al., 2000; Zhang & Restrepo, 2002, 2003), but these connexin proteins were not detected in ORNs by FRIL. The discrepancy between detection of mRNA vs. the lack of detection of the corresponding protein is not particular to ORNs. In brain and lung, too, there is a lack of correspondence between the amount of mRNA and the protein level for several different connexins, including Cx43 (Willecke et al., 1991; Hennemann et al., 1992; Miragall et al., 1996a; Simberger et al., 1997). Those other authors speculated on causes for these discrepancies. As an additional potential explanation, a novel computer algorithm recently revealed that Cx43 mRNA is a potential target for multiple mammalian miRNAs, suggesting the possibility in ORNs of inhibition or complete blockade of Cx43 protein translation by miRNA (Lewis et al., 2003).
In this study, we found multiple, previously unrecognized potential “seed matching sequences” in the 3′ UTR on mouse Cx43 mRNA (Table 3). Thus, in ORNs, as in other cells, detection of a particular mRNA by in situ hybridization or RT-PCR may not constitute direct evidence for synthesis of the corresponding protein. Our miRNA data also suggest the need for caution in interpreting previous reporter gene experiments, either because some plasmid constructs did not contain the complete 3′ UTR of the original gene, or because the 3′ UTR was separated from the expressed portion of the gene by insertion of inappropriate termination sequences. It is now clear that more attention must be paid to the importance of 3′ UTRs, as well as 5′ UTRs, because both may be essential for cell-specific regulation of gene expression based on their influences on mRNA stability and post-transcriptional regulation of protein synthesis (Mignone et al., 2002; Pfeifer et al., 2004). Finally, it is increasingly apparent that expression of many genes may be hindered by endogenous miRNA mechanisms that are just beginning to be understood. Inhibition of Cx43 translation by ORN miRNA would not only account for the apparent discrepancy in expression levels between mRNA and protein, but it would also account for the high level of expression of LacZ marker protein in some ORNs in a transgenic mouse in which the proximal 6.5 kb Cx43 promoter drove expression of a LacZ marker (Zhang et al., 2000). Apparently, that LacZ construct did not include the putative 3′ site for binding endogenous miRNAs. However, to our knowledge, the possibility that Cx43 protein translation in neurons is regulated by miRNA has not been studied experimentally.
Acknowledgments
This work was supported by NIH grants NS31027, NS44010, and NS 44395 (JER), DC00566, DC04657(DR) and DC04952 (CZ) and by CIHR (JIN). We thank B. McLean for excellent technical assistance, Dr. D. Paul (Harvard University) for providing us with breeding pairs of wild-type and Cx36 knockout mice, Brian Gebhardt and Jennifer A. Sampson for assistance in preparing stereoscopic images, Elek Molnar (University of Bristol) for providing antibodies to AMPA receptors, and Elliot Hertzberg (Albert Einstein School of Medicine) for providing anti-connexin Cx43 antibody Ab18A. We also thank Dr. Torgeir Holen (University of Oslo) for useful comments regarding miRNA.
References
- BARTEL DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- BLINDER KJ, PUMPLIN DW, PAUL D, KELLER A. Intercellular interactions in the mammalian olfactory nerve. The Journal of Comparative Neurology. 2003;466:230–239. doi: 10.1002/cne.10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRANTON D, BULLIVANT S, GILULA NB, KARNOVSKY MJ, MOOR H, NORTHCOTE DH, PACKER L, SATIR B, SATIR P, SPETH V, STAEHELIN LA, STEERE RL, WEINSTEIN RS. Freeze-etching nomenclature. Science. 1975;190:54–56. doi: 10.1126/science.1166299. [DOI] [PubMed] [Google Scholar]
- BRENNECKE J, STARK A, RUSSELL RB, COHEN SM. Principles of microRNA-target recognition. PLoS Biology. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCK LB. The molecular architecture of odor and pheromone sensing in mammals. Cell. 2000;100:611–618. doi: 10.1016/s0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- BUCK LB, AXEL R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- CHRISTIE JM, BARK C, HORMUZDI SG, HELBIG I, MONYER H, WESTBROOK GL. Connexin36 mediates spike synchrony in olfactory bulb glomeruli. Neuron. 2005;46:761–772. doi: 10.1016/j.neuron.2005.04.030. [DOI] [PubMed] [Google Scholar]
- CHRISTIE JM, SCHOPPA NE, WESTBROOK GL. Tufted cell dendrodendritic inhibition in the olfactory bulb is dependent on NMDA receptor activity. Journal of Neurophysiology. 2001;85:169–173. doi: 10.1152/jn.2001.85.1.169. [DOI] [PubMed] [Google Scholar]
- CONDORELLI DF, BELLUARDO N, TROVATO-SALINARO A, MUDO G. Expression of Cx36 in mammalian neurons. Brain Research Reviews. 2000;32:72–85. doi: 10.1016/s0165-0173(99)00068-5. [DOI] [PubMed] [Google Scholar]
- CONDORELLI DF, PARENTI R, SPINELLA F, SALINARO AT, BELLUARDO N, CARDILE V, CICIRATA F. Cloning of a new gap junction gene (Cx36) highly expressed in mammalian brain neurons. European Journal of Neuroscience. 1998;10:1202–1208. doi: 10.1046/j.1460-9568.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- COPELLI M, OLIVEIRA RF, ROQUE AC, KINOUCHI O. Signal compression in the sensory periphery. Neurocomputing. 2005;65–66:691–696. [Google Scholar]
- DEGEN J, MEIER C, VAN DER GIESEN RS, SOHL G, PETRASCH-PARWEZ E, URSCHEL S, DERMIETZEL R, SCHILLING K, DE ZEEUW CI, WILLECKE K. Expression pattern of lacZ reporter gene representing connexin36 in transgenic mice. The Journal of Comparative Neurology. 2004;473:511–525. doi: 10.1002/cne.20085. [DOI] [PubMed] [Google Scholar]
- DELAY RJ, DIONNE VE. Coupling between sensory neurons in the olfactory epithelium. Chemical Senses. 2003;28:807–815. doi: 10.1093/chemse/bjg074. [DOI] [PubMed] [Google Scholar]
- DHALLAN RS, MACKE JP, EDDY RL, SHOWS TB, REED RR, YAU KW, NATHANS J. Human rod photoreceptor cGMP-gated channel: Amino acid sequence, gene structure, and functional expression. Journal of Neuroscience. 1992;12:3248–3256. doi: 10.1523/JNEUROSCI.12-08-03248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DINCHUK JE, JOHNSON TJA, RASH JE. Postreplication labeling of E-leaflet molecules: Membrane immunoglobulins localized in sectioned labeled replicas examined by TEM and HVEM. Journal Of Electron Microscopy Technique. 1987;7:1–16. doi: 10.1002/jemt.1060070102. [DOI] [PubMed] [Google Scholar]
- DOENCH JG, PETERSEN CP, SHARP PA. siRNAs can function as miRNAs. Genes & Development. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOENCH JG, SHARP PA. Specificity of microRNA target selection in translational repression. Genes & Development. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUCETTE R. PNS-CNS transitional zone of the first cranial nerve. The Journal of Comparative Neurology. 1991;312:451–466. doi: 10.1002/cne.903120311. [DOI] [PubMed] [Google Scholar]
- FARH KK, GRIMSON A, JAN C, LEWIS BP, JOHNSTON WK, LIM LP, BURGE CB, BARTEL DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- FIELD PM, LI Y, RAISMAN G. Ensheathment of the olfactory nerves in the adult rat. Journal of Neurocytology. 2003;32:317–324. doi: 10.1023/B:NEUR.0000010089.37032.48. [DOI] [PubMed] [Google Scholar]
- FIRESTEIN S. A Nobel nose: The 2004 Nobel Prize in Physiology and Medicine. Neuron. 2005;45:333–338. doi: 10.1016/j.neuron.2005.01.021. [DOI] [PubMed] [Google Scholar]
- FRISCH D. Ultrastructure of mouse olfactory mucosa. American Journal of Anatomy. 1967;121:87–120. doi: 10.1002/aja.1001210107. [DOI] [PubMed] [Google Scholar]
- FUJIMOTO K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. Journal of Cell Science. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- FUJIMOTO K. SDS-digested freeze-fracture replica labeling electron microscopy to study the two-dimensional distribution of integral membrane proteins and phospholipids in biomembranes: Practical procedure, interpretation and application. Histochemistry and Cell Biology. 1997;107:87–96. doi: 10.1007/s004180050092. [DOI] [PubMed] [Google Scholar]
- GILULA NB, SATIR P. The ciliary necklace. A ciliary membrane specialization. The Journal of Cell Biology. 1972;53:494–509. doi: 10.1083/jcb.53.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GÜLDENAGEL M, SÖHL G, PLUM A, TRAUB O, TEUBNER B, WEILER R, WILLECKE K. Expression patterns of connexin genes in mouse retina. The Journal of Comparative Neurology. 2000;425:193–201. [PubMed] [Google Scholar]
- HARRIS KM, LANDIS DMD. Membrane structure at synaptic junctions in area CA1 of the rat hippocampus. Neuroscience. 1986;19:857–872. doi: 10.1016/0306-4522(86)90304-0. [DOI] [PubMed] [Google Scholar]
- HE Y, JANNSEN WG, MORRISON JJ. Synaptic coexistence of AMPA and NMDA receptors in the rat hippocampus: A postembedding immunogold study. Journal of Neuroscience Research. 1998;55:444–449. doi: 10.1002/(SICI)1097-4547(19981115)54:4<444::AID-JNR2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- HENNEMANN H, SUCHYNA T, LICHTENBERGFRATE H, JUNGBLUTH S, DAHL E, SCHWARZ J, NICHOLSON BJ, WILLECKE K. Molecular cloning and functional expression of mouse connexin40, a second gap junction gene preferentially expressed in lung. The Journal of Cell Biology. 1992;117:1299–1310. doi: 10.1083/jcb.117.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HSU YC, BASKAR J, STEVENS LC, RASH JE. Development in vitro of mouse embryos from the two-cell egg stage to the early somite stage. Journal of Embryology and Experimental Morphology. 1974;31:235–245. [PubMed] [Google Scholar]
- HULL BE, STAEHELIN LA. The terminal web. A reevaluation of its structure and function. The Journal of Cell Biology. 1979;81:67–82. doi: 10.1083/jcb.81.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAMASAWA N, SIK A, MORITA M, YASUMURA T, DAVIDSON KGV, NAGY JI, RASH JE. Connexin47 and connexin32 in gap junctions of oligodendrocyte somata, myelin sheaths, paranodal loops, Schmidt-Lanterman incisures: Implications for ionic homeostasis and potassium siphoning. Neuroscience. 2005;136:65–86. doi: 10.1016/j.neuroscience.2005.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAMASAWA N, YASUMURA T, DAVIDSON KGV, FURMAN CS, NAGY JI, RASH JE. Differential distribution of connexin45 (Cx45) and Cx36 in neuronal gap junctions of adult rat retina, olfactory bulb and cerebral cortex. Abstracts, ASCB Annual Meeting. 2004;44:110. [Google Scholar]
- KERJASCHKI D, HÖRANDNER H. The development of mouse olfactory vesicles and their cell contacts: A freeze-etching study. Journal of Ultrastructure Research. 1976;54:420–444. doi: 10.1016/s0022-5320(76)80027-5. [DOI] [PubMed] [Google Scholar]
- KHARAZIA VN, PHEND KD, RUSTIONI A, WEINBERG RJ. EM colocalization of AMPA and NMDA receptor subunits at synapses in rat cerebral cortex. Neuroscience Letters. 1996;210:37–40. doi: 10.1016/0304-3940(96)12658-6. [DOI] [PubMed] [Google Scholar]
- KOSAKA T, KOSAKA K. Neuronal gap junctions in the rat main olfactory bulb, with special reference to intraglomerular gap junctions. Neuroscience Research. 2003;45:189–209. doi: 10.1016/s0168-0102(02)00222-5. [DOI] [PubMed] [Google Scholar]
- KOSAKA T, KOSAKA K. Neuronal gap junctions between intraglomerular mitral/tufted cell dendrites in the mouse main olfactory bulb. Neuroscience Research. 2004;49:373–378. doi: 10.1016/j.neures.2004.04.003. [DOI] [PubMed] [Google Scholar]
- KOSAKA T, KOSAKA K. Intraglomerular dendritic link connected by gap junctions and chemical synapses in the mouse main olfactory bulb: Electron microscopic serial section analyses. Neuroscience. 2005;131:611–625. doi: 10.1016/j.neuroscience.2004.11.050. [DOI] [PubMed] [Google Scholar]
- LARSEN WJ. Biological implications of gap junction structure, distribution and composition: A review. Tissue & Cell. 1983;15:645–671. doi: 10.1016/0040-8166(83)90041-1. [DOI] [PubMed] [Google Scholar]
- LEWIS BP, BURGE CB, BARTEL DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- LEWIS BP, SHIH IH, JONES-RHOADES MW, BARTEL DP, BURGE CB. Prediction of Mammalian MicroRNA Targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- LIM LP, LAU NC, GARRETT-ENGELE P, GRIMSON A, SCHELTER JM, CASTLE J, BARTEL DP, LINSLEY PS, JOHNSON JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- MACK A, WOLBURG H. Heterogeneity of glial membranes in the rat olfactory system as revealed by freeze-fracturing. Neuroscience Letters. 1986;65:17–22. doi: 10.1016/0304-3940(86)90113-8. [DOI] [PubMed] [Google Scholar]
- MAUL, G. G. (2005) Temperature-dependent changes in intramembrane particle distribution. In Freeze Fracture: Methods, Artifacts and Interpretations (edited by RASH, J. E. & Hudson, C. S.), pp. 37–42. New York: Raven Press.
- MAXEINER S, DEDEK K, JANSSEN-BIENHOLD U, AMMERMULLER J, BRUNE H, KIRSCH T, PIEPER M, DEGEN J, KRUGER O, WILLECKE K, WEILER R. Deletion of connexin45 in mouse retinal neurons disrupts the rod/cone signaling pathway between AII amacrine and ON cone bipolar cells and leads to impaired visual transmission. Journal of Neuroscience. 2005;25:566–576. doi: 10.1523/JNEUROSCI.3232-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAXEINER S, KRUGER O, SCHILLING K, TRAUB O, URSCHEL S, WILLECKE K. Spatiotemporal transcription of connexin45 during brain development results in neuronal expression in adult mice. Neuroscience. 2003;119:689–700. doi: 10.1016/s0306-4522(03)00077-0. [DOI] [PubMed] [Google Scholar]
- MEIER C, DERMIETZEL R, DAVIDSON KGV, YASUMURA T, RASH JE. Connexin32-containing gap junctions in Schwann cells at the internodal zone of partial myelin compaction and in Schmidt-Lanterman incisures. Journal of Neuroscience. 2004;24:3186–3198. doi: 10.1523/JNEUROSCI.5146-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENCO BPHM. Qualitative and quantitative freeze-fracture studies on olfactory and nasal respiratory structures of frog, ox, rat, and dog. I. A general survey. Cell & Tissue Research. 1980a;207:183–209. doi: 10.1007/BF00237805. [DOI] [PubMed] [Google Scholar]
- MENCO BPHM. Qualitative and quantitative freeze-fracture studies on olfactory and nasal respiratory epithelial surfaces of frog, ox, rat, and dog. III. Tight-junctions. Cell & Tissue Research. 1980b;211:361–373. doi: 10.1007/BF00234393. [DOI] [PubMed] [Google Scholar]
- MENCO BPHM. Qualitative and quantitative freeze-fracture studies on olfactory and respiratory epithelial surfaces of frog, ox, rat, and dog. IV. Ciliogenesis and ciliary necklaces (including high-voltage observations) Cell & Tissue Research. 1980c;212:1–16. doi: 10.1007/BF00234028. [DOI] [PubMed] [Google Scholar]
- MIGNONE F, GISSI C, LIUNI S, PESOLE G. Untranslated regions of mRNAs. Genome Biology. 2002;3:reviews0004.1–reviews0004.10. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIRAGALL F, BREIPOHL W, NAGURO T, VOSSWERMBTER G. Freeze-fracture study of the plasma membranes of the septal olfactory organ of Masera. Journal of Neurocytology. 1984;13:111–125. doi: 10.1007/BF01148321. [DOI] [PubMed] [Google Scholar]
- MIRAGALL F, HWANG TK, TRAIB P, HERTZBERG EL, DERMIETZEL R. Expressions of connexins in developing olfactory system of the mouse. The Journal of Comparative Neurology. 1992;325:359–378. doi: 10.1002/cne.903250304. [DOI] [PubMed] [Google Scholar]
- MIRAGALL, F., KREMER, M. & DERMIETZEL, R. (1994) Intercellular communications via gap junctions in the olfactory system. In Olfaction and TasteXI (edited by KURIHARA, K., SUZUKI, N. & OGAWA, H.), pp. 32–35. Tokyo: Springer-Verlag.
- MIRAGALL F, SIMBURGER E, DERMIETZEL R. Mitral and tufted cells of the mouse olfactory bulb possess gap junctions and express connexin43 mRNA. Neuroscience Letters. 1996a;216:199–202. doi: 10.1016/0304-3940(96)13042-1. [DOI] [PubMed] [Google Scholar]
- MIRAGALL, F., TRAUB, O. & DERMIETZEL, R. (1996b) Gap junction expression in the olfactory system. In Gap Junctions in the Nervous System (edited by SPRAY, D. C. & DERMIETZEL, R.), pp. 243–260, New York, R. G. Landes, Co.
- MORAN DT, ROWLEY JC, JAFEK BW, LOVELL MA. The fine structure of the olfactory mucosa in man. Journal of Neurocytology. 1982;11:721–746. doi: 10.1007/BF01153516. [DOI] [PubMed] [Google Scholar]
- NAGY JI, DUDEK FE, RASH JE. Update on connexins and gap junctions in neurons and glia in the mammalian central nervous system. Brain Research Reviews. 2004;47:191–215. doi: 10.1016/j.brainresrev.2004.05.005. [DOI] [PubMed] [Google Scholar]
- NAGY JI, LI X, REMPEL J, STELMACK GL, PATEL D, STAINES WA, YASUMURA T, RASH JE. Connexin26 in adult rodent CNS: Demonstration at astrocytic gap junctions and co-localization with connexin30 and connexin43. The Journal of Comparative Neurology. 2001;441:302–323. doi: 10.1002/cne.1414. [DOI] [PubMed] [Google Scholar]
- NIELSEN S, NAGELHUS EA, AMIRYMOGHADDAM M, BOURQUE C, AGRE P, OTTERSEN OP. Specialized membrane domains for water transport in glial cells: High-resolution immunogold cytochemistry of aquaporin-4 in rat brain. Journal of Neuroscience. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREDA A, O’BRIEN J, NAGY JI, SMITH M, BUKAUSKAS F, DAVIDSON KGV, KAMASAWA N, YASUMURA T, RASH JE. Short-range functional interaction between connexin35 and neighboring chemical synapses. Cell Communication and Adhesion. 2003;10:419–423. doi: 10.1080/15419060390263254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREDA A, RASH JE, NAGY JI, BENNETT MVL. Dynamics of electrical transmission at club endings on the Mauthner cells. Brain Research Reviews. 2004;47:227–244. doi: 10.1016/j.brainresrev.2004.06.010. [DOI] [PubMed] [Google Scholar]
- PFEIFER I, ANDERSON C, WERNER R, OLTRA E. Redefining the structure of the mouse connexin43 gene: Selective promoter usage and alternative splicing mechanisms yield transcripts with different translational efficiencies. Nucleic Acids Research. 2004;32:4550–4562. doi: 10.1093/nar/gkh792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILLIPS TE, BOYNE AF. Liquid nitrogen-based quick freezing: Experiments with bounce-free delivery of cholinergic nerve terminals to a metal surface. Journal of Electron Microscopy Technique. 1984;1:9–29. [Google Scholar]
- RACCA C, STEPHENSON FA, STREIT P, ROBERTS JD, SOMOGYI P. NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. The Journal of Neuroscience. 2000;20:2512–2522. doi: 10.1523/JNEUROSCI.20-07-02512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, DAVIDSON KGV, YASUMURA T, FURMAN CS. Freeze-fracture and immunogold analysis of aquaporin-4 (AQP4) square arrays, with models of AQP4 lattice assembly. Neuroscience. 2004a;129:915–934. doi: 10.1016/j.neuroscience.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, PEREDA A, KAMASAWA N, FURMAN CS, YASUMURA T, DAVIDSON KGV, DUDEK FE, OLSON C, NAGY JI. High-resolution proteomic mapping in the vertebrate central nervous system: Close proximity of connexin35 to NMDA glutamate receptor clusters and co-localization of connexin36 with immunoreactivity for zonula occludens protein-1 (ZO-1) Journal of Neurocytology. 2004b;33:131–152. doi: 10.1023/B:NEUR.0000029653.34094.0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, SHAY JW, BIESELE JJ. Cilia in cardiac differentiation. Journal of Ultrastructure Research. 1969;29:470–484. doi: 10.1016/s0022-5320(69)90067-7. [DOI] [PubMed] [Google Scholar]
- RASH JE, STAINES WA, YASUMURA T, PATEL D, HUDSON CS, STELMACK GL, NAGY JI. Immunogold evidence that neuronal gap junctions in adult rat brain and spinal cord contain connexin36 (Cx36) but not Cx32 or Cx43. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7573–7578. doi: 10.1073/pnas.97.13.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, WHALEN LR, GUTHRIE PB, MORITA M, DILLMAN RK, YASUMURA T, FAYGUTHRIE D. Confocal “grid-mapped” freeze fracture of preselected neurons in spinal cord and brain. Society of Neuroscience Abstracts. 1993;19:1112. [Google Scholar]
- RASH JE, YASUMURA T. Direct immunogold labeling of connexins and aquaporin4 in freeze-fracture replicas of liver, brain and spinal cord: Factors limiting quantitative analysis. Cell & Tissue Research. 1999;296:307–321. doi: 10.1007/s004410051291. [DOI] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T, DUDEK FE. Ultrastructure, histological distribution, and freeze-fracture immunocytochemistry of gap junctions in rat brain and spinal cord. Cell Biology International. 1998a;22:731–749. doi: 10.1006/cbir.1998.0392. [DOI] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T, DUDEK FE, NAGY JI. Cell-specific expression of connexins, and evidence for restricted gap junctional coupling between glial cells and between neurons. Journal of Neuroscience. 2001;21:1983–2001. doi: 10.1523/JNEUROSCI.21-06-01983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASH JE, YASUMURA T, HUDSON CS, AGRE P, NIELSEN S. Direct immunogold labeling of aquaporin-4 in “square arrays” of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proceedings of the National Academy of Science of the United States of America. 1998b;95:11981–11986. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RHOADES MW, REINHART BJ, LIM LP, BURGE CB, BARTEL B, BARTEL DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- SASSOE-POGNETTO M, OTTERSEN OP. Organization of ionotropic glutamate receptors at dendro-dendritic synapses in the rat olfactory bulb. Journal of Neuroscience. 2000;20:2192–2201. doi: 10.1523/JNEUROSCI.20-06-02192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SASSOE-POGNETTO M, UTVIK JK, CARMOLETTO P, WATANABE M, STEPHENSON FA, BREDT DS, OTTERSEN OP. Organization of postsynaptic density proteins and glutamate receptors in ax-odendritic and dendrodendritic synapses of the rat olfactory bulb. The Journal of Comparative Neurology. 2003;463:237–248. doi: 10.1002/cne.10745. [DOI] [PubMed] [Google Scholar]
- SATIR, B. H. & SATIR, P. (1979) Partitioning of intramembrane particles during the freeze-fracture procedure. In Freeze Fracture: Methods, Artifacts, and Interpretations (edited by RASH, J. E. & HUDSON, C. S.), pp. 43–49. New York, NY: Raven Press.
- SCHLOSSER G. Evolutionary origins of vertebrate placodes: Insights from developmental studies and from comparisons with other deuterosomes. Journal of Experimental Zoology. 2005;304B:347–399. doi: 10.1002/jez.b.21055. [DOI] [PubMed] [Google Scholar]
- SCHOPPA NE, KINZIE JM, SAHARA Y, SEGERSON TP, WESTBROOK GL. Dendrodendritic inhibition in the olfactory bulb is driven by NMDA receptors. Journal of Neuroscience. 1998;18:6790–6802. doi: 10.1523/JNEUROSCI.18-17-06790.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHOPPA NE, WESTBROOK GL. Glomerulus-specific synchronization of mitral cells in the olfactory bulb. Neuron. 2001;31:639–651. doi: 10.1016/s0896-6273(01)00389-0. [DOI] [PubMed] [Google Scholar]
- SCHOPPA NE, WESTBROOK GL. AMPA autoreceptors drive correlated spiking in olfactory bulb glomeruli. Nature Neuroscience. 2002;5:1194–1202. doi: 10.1038/nn953. [DOI] [PubMed] [Google Scholar]
- SEMPERE L, FREEMANTLE S, PITHA-ROWE I, MOSS E, DMITROVSKY E, AMBROS V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biology. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEPHERD, G. M. & GREER, C. R. (1998) Olfactory bulb. In The Synaptic Organization of the Brain (edited by SHEPHERD, G. M.), pp. 159–20. New York: Oxford University Press.
- SIMBERGER E, STANG A, KREMER M, DERMIETZEL R. Expression of connexin43 mRNA in adult rodent brain. Histochemistry and Cell Biology. 1997;107:127–137. doi: 10.1007/s004180050097. [DOI] [PubMed] [Google Scholar]
- SMIRNOVA L, GRÄFE A, SEILER A, SCHUMACHER S, NITSCH R, WULCZYN FG. Regulation of miRNA expression during neural cell specification. European Journal of Neuroscience. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- STAEHELIN LA. Structure and function of intercellular junctions. International Review of Cytology. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- TAKUMI Y, RAMIREZ-LEON V, LAAKE P, RINVIK E, OTTERSEN OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nature Neuroscience. 1999;2:618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- THEIS M, JAUCH R, ZHUO L, SPEIDEL D, WALLRAFF A, DORING B, FRISCH C, SOHL G, TEUBNER B, EUWENS C, HUSTON J, STEINHAUSER C, MESSING A, HEINEMANN U, WILLECKE K. Accelerated hippocampal spreading depression and enhanced locomotory activity in mice with astrocyte-directed inactivation of connexin43. Journal of Neuroscience. 2003;23:766–776. doi: 10.1523/JNEUROSCI.23-03-00766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UTVIK JK, RASH JE, YASUMURA T, CHANDLER J, MULLER E, SEEBURG PH, KOEHR G, OTTERSEN OP. Organization of ionotropic glutamate receptors at hippocampal commissural-Schaffer collateral (SCC) synapses: Effects of C-terminal truncation of NR2A subunits. Society for Neuroscience Abtracts. 2000;26:1390. [Google Scholar]
- VENABLE JH, COGGESHALL RA. A simplified lead citrate stain for use in electron microscopy. The Journal of Cell Biology. 1965;25:407–408. doi: 10.1083/jcb.25.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGALIS F, HEGG CC, LUCERO MT. Electrical coupling in sustentacular cells of the mouse olfactory epithelium. The Journal of Neurophysiology. 2005a;94:1001–012. doi: 10.1152/jn.01299.2004. [DOI] [PubMed] [Google Scholar]
- VOGALIS F, HEGG CC, LUCERO MT. Ionic conductances in sustentacular cells of the mouse olfactory epithelium. The Journal of Physiology. 2005b;562:785–799. doi: 10.1113/jphysiol.2004.079228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIENHOLDS E, KLOOSTERMAN WP, MISKA E, ALVAREZ-SAAVEDRA E, BEREZIKOV E, DE BRUIJN E, HORVITZ HR, KAUPPINEN S, PLASTERK RHA. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- WILLECKE K, HEYNKES R, DAHL E, STUTENKEMPER R, HENNEMANN H, JUNGBLUTH S, SUCHYNA T, NICHOLSON BJ. Mouse connexin37: Cloning and functional expression of a gap junction gene highly expressed in lung. The Journal of Cell Biology. 1991;114:1049–1057. doi: 10.1083/jcb.114.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINKLER H, WILDHABER H, GROSS H. Decoration on the surface of a regular protein layer. Ultramicroscopy. 2002;16:331–339. [Google Scholar]
- YASUMURA T, RASH JE. Freeze fracture at the turn of the century: Techniques for visualizing and labeling tissues, cells, and molecules. Proceedings: Microscopy and Microanalysis 1997. 1997;3:345–346. [Google Scholar]
- ZHANG C, FINGER TE, RESTREPO D. Mature olfactory receptor neurons express connexin 43. The Journal of Comparative Neurology. 2000;426:1–12. doi: 10.1002/1096-9861(20001009)426:1<1::aid-cne1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- ZHANG C, RESTREPO D. Expression of connexin 45 in the olfactory system. Brain Research. 2002;929:37–47. doi: 10.1016/s0006-8993(01)03372-8. [DOI] [PubMed] [Google Scholar]
- ZHANG C, RESTREPO D. Heterogeneous expression of connexin 36 in the olfactory epithelium and glomerular layer of the olfactory bulb. The Journal of Comparative Neurology. 2003;459:426–439. doi: 10.1002/cne.10617. [DOI] [PubMed] [Google Scholar]
- ZUFALL F. Connexins and olfactory synchronicity: Toward the olfactory code. Neuron. 2005;46:693–694. doi: 10.1016/j.neuron.2005.05.015. [DOI] [PubMed] [Google Scholar]


