Abstract
Yeast Nhx1 [Na+(K+)/H+ exchanger 1] is an intracellular Na+(K+)/H+ exchanger, localizing to the late endosome where it is important for ion homoeostasis and vesicle trafficking. Phylogenetic analysis of NHE (Na+/H+ exchanger) sequences has ident-ified orthologous proteins, including HsNHE6 (human NHE6), HsNHE7 and HsNHE9 of unknown physiological role. These appear distinct from well-studied mammalian plasma membrane isoforms (NHE1–NHE5). To explore the differences between plasma membrane and intracellular NHEs and understand the link between ion homoeostasis and vesicle trafficking, we examined the consequence of replacing residues in the intramembranous H10 loop of Nhx1 between transmembrane segments 9 and 10. The critical role for the carboxy group of Glu355 in ion transport is consistent with the invariance of this residue in all NHEs. Surprisingly, residues specifically conserved in the intracellular isoforms (such as Phe357 and Tyr361) could not be replaced with closely similar residues (leucine and phenylalanine) found in the plasma membrane isoforms without loss of function, revealing unexpected side chain specificity. The trafficking phenotypes of all Nhx1 mutants, including hygromycin-sensitivity and missorting of carboxypeptidase Y, were found to directly correlate with pH homoeostasis defects and could be proportionately corrected by titration with weak base. The present study demonstrates the importance of the H10 loop of the NHE family, highlights the differences between plasma membrane and intracellular isoforms and shows that trafficking defects are tightly coupled with pH homoeostasis.
Keywords: endosomal pH, ion homoeostasis, Na+/H+ exchanger (NHE), salt tolerance, vesicle trafficking, yeast Na+(K+)/H+ exchanger 1 (Nhx1)
Abbreviations: APG, arginine phosphate glucose; BCECF, 2′,7′-bis-(2-carboxyethyl)-5(6)-carboxyfluorescein; CPY, carboxypeptidase Y; GFP, green fluorescent protein; HA, haemagglutinin; HSE, heat-shock element; NHE, Na+/H+ exchanger; HsNHE, human NHE; Nhx1, Na+(K+)/H+ exchanger 1; SC medium, synthetic complete medium; ScNHX, Saccharomyces cerevisiae NHX; TM10, transmembrane domain 10; TMA, tetramethylammonium
INTRODUCTION
NHEs (Na+/H+ exchangers) are involved in numerous pathophysiological processes including hypertension, post-ischaemic myocardial arrhythmia and regulation of aqueous humour secretion associated with glaucoma [1–3]; hence, study of their function and regulation is of prime interest. In mammals, there are nine members of the NHE gene family (NHE1–NHE9) with distinct tissue-specific distribution, subcellular localization and physiological function. Although derived from a common ancestral gene, recent phylogenetic analysis has revealed that the eukaryotic NHE fall into two major subgroups corresponding to plasma membrane (PM NHE) and intracellular transporters (IC NHE) [4]. Support for this classification comes from emerging experimental evidence that members of these two subgroups may be distinct from one another in cation selectivity, inhibitor-sensitivity and physiological function (reviewed in [4]). The PM NHE, represented by mammalian isoforms NHE1–NHE5, have been extensively characterized and implicated in the regulation of cytoplasmic pH, maintenance of cell volume, Na+ homoeostasis and transepithelial transport of electrolytes (reviewed in [5]). In contrast, much less is known about the properties of the IC NHE despite the discovery of numerous candidate genes from plants, model organisms and higher vertebrates, including mammalian isoforms NHE6–NHE9. For example, the endosomal exchanger NHE6 is highly expressed in human brain, skeletal muscle and heart, yet nothing is known about the physiological role of this isoform [4,6]. Molecular characterization of IC NHE family members is an important step towards understanding function; however, such studies have been hampered by difficulties in assessing ion transport activity and function within the intracellular compartments of mammalian cells. In the present study, we report on mutagenic analysis of Nhx1 [Na+(K+)/H+ exchanger 1], the closely related NHE6 homologue from Saccharomyces cerevisiae.
Yeast Nhx1 was the first member of the IC NHE subgroup to be identified and localizes to the membranes of the prevacuolar/late endosomal compartment [7,8]. Nhx1 has been shown to mediate vacuolar sequestration of Na+ and K+, coupling cation transport with the proton gradient established by the vacuolar H+-ATPase, to confer salt tolerance [7,9]. The use of specific probes of vacuolar and cytoplasmic pH demonstrated that Nhx1 is a major leak pathway for protons within the endosomal system, contributing to cellular pH regulation and weak acid tolerance [10,11]. The NHX1 gene was independently identified as VPS44, and shown to be required for vacuolar protein sorting [12]. Specifically, loss of Nhx1 leads to accumulation of cargo destined for the vacuole in an aberrant ‘Class E’ compartment, resulting in the missorting of proteins, such as CPY (carboxypeptidase Y), to the cell surface. Together, these observations have allowed us to establish a set of quantitative growth assays for Nhx1 activity as a means to screen mutations. We demonstrate here that the ease of phenotypic assessment in yeast makes this an excellent model system for structure–function analysis of NHEs.
Hydropathy analysis of Nhx1 reveals a domain organization similar to other NHE isoforms, suggesting that the structural features of intracellular exchangers are conserved across the family. In a prototypic member, there is a conserved hydrophobic domain consisting of 12 predicted transmembrane segments necessary for ion transport, and a divergent C-terminal hydrophilic domain that typically participates in numerous protein–protein interactions. An extensive topological characterization of NHE1 based on substituted cysteine accessibility analysis [13], followed by in vitro transcription and translation analysis [14], indicated that the extracellular loop region between transmembrane segments 9 and 10 is folded within the protein, analogous to the P loop of K+ channels that is critical for ion permeability. This region has been termed H10, due to its overall hydropathic nature. We therefore considered this stretch of polypeptide as a starting point for the mutagenic analysis of Nhx1, a model for the intracellular subgroup of the NHE family.
A sequence alignment of the H10 region from representative members of the major phylogenetic clades of NHE, including yeast Nhx1, is shown in Figure 1. We hypothesized that invariant residues, exemplified by the phylogenetically conserved acidic residue Glu355 in yeast Nhx1, may be critical for function across all NHEs, whereas the non-conserved residues are likely to be more tolerant to substitution. Of particular interest are residues uniquely conserved only within the intracellular subgroup, such as Phe357 and Tyr361 in yeast Nhx1. We tested whether the IC NHE-specific residues Phe357 and Tyr361 were critical for Nhx1 function, and furthermore, whether replacement with the equivalent residues from the PM NHE could support function. Our findings reveal a surprisingly stringent requirement for these subgroup-specific residues and lend support to the new phylogenetic-based classification of NHE.
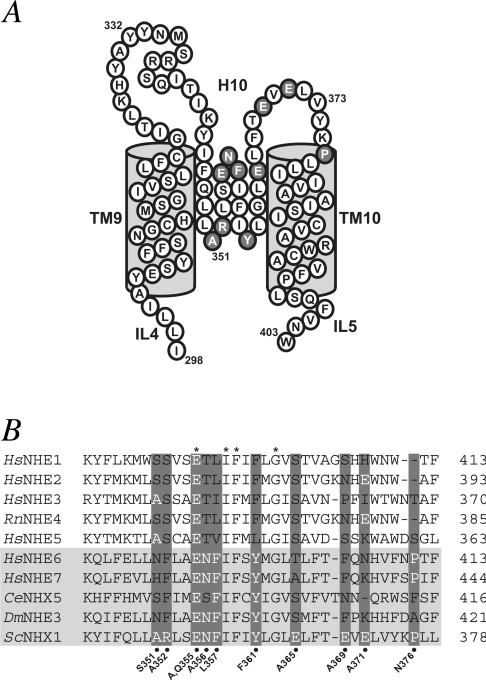
Figure 1. Sequence and predicted topology of the H10 region of NHE.
(A) Predicted topology of ScNhx1. The extracellular loop (H10) between transmembrane helices TM9 and TM10 is postulated to be inserted into the membrane based on the experimentally derived topology of HsNHE1 and Arabidopsis thaliana NHX1 [13,27]. Residues targeted for mutagenesis in the present study are shaded. IL4 (intracellular loop 4) and IL5 form the intracellular connections to the remainder of the polypeptide chain. (B) Alignment of NHE sequences in the H10 region. NHE1–NHE5 are plasma membrane isoforms from human (Hs) or rat (Rn). NHE6 and NHE7 from human, Caenorhabditis elegans NHX5 (CeNHX5) and Drosophila melanogaster NHE3 (DmNHE3) are more closely related to ScNHX1, and represent the intracellular isoforms. Residues conserved throughout the family are indicated (*) above the sequences. Residues chosen for mutagenesis (see text) are highlighted in grey, with the substitutions indicated below the ScNHX1 sequence.
EXPERIMENTAL
Yeast strains, media and growth conditions
All S. cerevisiae strains used were derivatives of BY4742 (ResGen; Invitrogen). Strains were grown at 30 °C in APG (arginine phosphate glucose), a synthetic minimal medium containing 10 mM arginine, 8 mM phosphoric acid, 2% (w/v) glucose, 2 mM MgSO4, 1 mM KCl and 0.2 mM CaCl2 and trace minerals and vitamins [7]. The pH was adjusted, by addition of phosphoric acid, to 4.0 or 2.7 as specified. Where indicated, NaCl, KCl or hygromycin was added. Seed cultures were grown in SC medium (synthetic complete medium) to saturation, washed three times in water and used to seed 200 μl of APG medium in 96-well plates to a starting attenuance of 0.05 D600 units/ml. Growth was monitored by measuring D600 after culturing for 17–24 h at 30 or 37 °C, where specified.
Plasmids and mutagenesis
A 1.2 kb BamHI fragment of NHX1 was subcloned into pBluescript SK+ (Stratagene) and used as a template for site-directed mutagenesis. All amino acid substitutions were generated by a one-step reverse cyclic PCR method [15] using the appropriate base changes in the synthetic oligonucleotides (results not shown). Mutagenesis was confirmed by sequencing and the fragment was cloned into the expression vector pRin71, using BamHI enzyme. pRin71 is a 2μ plasmid harbouring NHX1 tagged with a C-terminal triple HA (haemagglutinin) epitope (NHX1–HA) under control of tandem HSEs (heat-shock elements) in vector YCplac111 [8,16]. The mutations were also introduced into the 2μ plasmid pRin82, which contains NHX1–GFP as described earlier [8], by subcloning with appropriate restriction sites.
Measurement of vacuolar pH
Vacuolar pH measurements were performed using methods previously described [10,18]. Briefly, cells were grown in APG growth medium (pH 2.7) for 18 h at 30 °C, absorbance readings were taken at 600 nm to measure growth, and cultures were then incubated with 50 μM BCECF [2′,7′-bis-(2-carboxyethyl)-5(6)-carboxyfluorescein]-acetoxymethyl ester at 30 °C for 20–30 min, washed and suspended in APG medium at pH 2.7. Single fluorescence intensity and absorbance readings were taken at 485 and 600 nm respectively, and normalized background-subtracted fluorescence emission at 485 nm values were calculated [NI485 (normalized intensity at 485 nm)]. At least three independent calibration experiments were performed for each strain tested and vacuolar pH values were calculated as described previously [18]. Fluorescent intensity and absorbance values were acquired using a BMG FLUOstar Optima multimode plate reader with accompanying BMG FLUOstar Optima Version 1.20-0 software (BMG LabTechnologies, Durham, NC, U.S.A.). At the end of each experiment, a calibration curve of the ratio of fluorescence intensity values versus pH was obtained for each yeast strain as follows. Yeast cultures were incubated in 200 μl of experimental media containing 50 mM Mes buffer, 50 mM Hepes, 50 mM KCl, 50 mM NaCl, 0.2 M ammonium acetate, 10 mM NaN3, 10 mM 2-deoxyglucose, 75 μM monensin and 10 μM nigericin, titrated to eight different pH values within the range of 4.5–8.0 using 1 M NaOH (also see [10,18]). All experiments were performed at 30 °C.
Membrane preparation and biochemical methods
Total membrane preparations were prepared by disrupting yeast with glass beads, followed by collection of membranes by centrifugation as described in [19]. Protein concentrations were determined by method of Lowry [20]. Western blotting and SDS/PAGE were as described previously [19]. Antibodies were mouse anti-HA monoclonal 12CA5 (Roche Molecular Biochemicals) at 1:1000 dilution and horseradish peroxidase-coupled sheep anti-mouse antibody (Amersham Biosciences) at 1:10000 dilution.
CPY secretion
CPY secretion from yeast cultures was detected as follows. Freshly grown seed cultures were washed in water and resuspended to a starting D600 of 0.05 ml−1 in SC medium in the presence or absence of TMA (tetramethylammonium; 600 mM final concentration) and grown at 30 °C for 21 h. Cultures from 1.5 D600 units of cells were centrifuged for 2 min at 16.1 relative centrifugal units, and the supernatants (25–800 μl) were applied to Immobilon (Millipore) membranes using a dot-blot apparatus. The membrane was dried overnight and CPY was detected by immunoblotting using monoclonal anti-CPY antibody (Molecular Probes; 1:1000 dilution) and horseradish peroxidase-coupled sheep anti-mouse antibody (Amersham Biosciences) at 1:10000.
Confocal microscopy
Yeast cells were grown in selective medium at 30 °C to exponential phase. The culture (1 ml) was briefly centrifuged to pellet cells. The cells were then washed twice with water and then resuspended in 50 μl of the same solution. Cell suspensions of 4 μl were dropped on poly(L-lysine)-treated coverslips and placed on slides. Samples were viewed on a Zeiss Axiovert 200 microscope equipped with an ultraview confocal scanner from PerkinElmer, using Zeiss ×100 oil-immersion lens. Digitized images (16-bit) were acquired with a Hamamatsu ORCA-ER camera and Ultraview imaging software (PerkinElmer).
Statistical analysis
Data are reported as means±S.E.M. and statistical comparisons were performed with Student's two-tailed t tests (paired or unpaired, as appropriate); significance was assumed at the 5% level.
RESULTS
Strategy for assessment of Nhx1 mutants
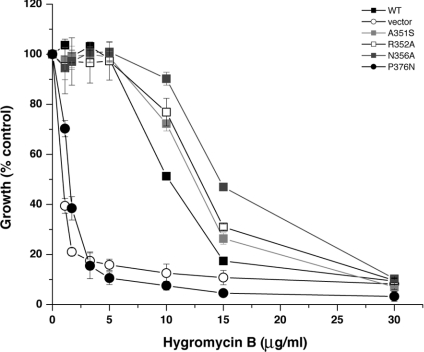
Previous studies have demonstrated an important role for Nhx1 in cellular Na+ and K+ homoeostasis, pH regulation and vesicle trafficking [7–12]. Each of these physiological functions can be separately monitored using quantitative liquid growth assays, allowing for a simple phenotypic assessment of mutants. Thus the nhx1Δ null mutant shows growth-sensitivity to salt stress (high NaCl or KCl), acid stress (pH 2.7 or weak acids) and hygromycin B toxicity, a common phenotype of vps (vacuolar protein sorting) mutants. Salt-sensitivity of the nhx1Δ strain is additionally exacerbated by deletion of NHA1, the phylogenetically distinct plasma membrane antiporter [11]; therefore, use of the nhx1Δnha1Δ double deletion in the host strain allowed for more sensitive evaluation of mutant phenotypes. NHX1 mutations were introduced into plasmids behind tandem HSEs and expressed in cultures maintained at 30 °C, or behind the endogenous NHX1 promoter, as specified. These constructs direct moderate levels of mutant protein expression suitable for screening of growth phenotypes. Control strains carried plasmid-encoded wild-type NHX1 or empty vector. Figure 1 identifies the ten amino acids within the H10 region of yeast Nhx1 targeted for substitution in the present study. Some of the targeted residues, including Ala351, Arg352 and Glu371, were widely divergent among the NHE sequences and were therefore predicted to be insensitive to substitution. Others were partially conserved (Asn356) or invariant (Glu355), and two hydrophobic residues (Phe357 and Tyr361) were uniquely invariant among the IC NHEs. We also targeted a proline residue (Pro376) at the beginning of TM10 (transmembrane domain 10), likely to be important for structural stability. In most cases, residues were changed to alanine, while Ala351, Phe357 and Tyr361 were changed to the corresponding residue in the human plasma membrane isoform NHE1 (Figure 1).
Role of acidic residues in the H10 region of Nhx1
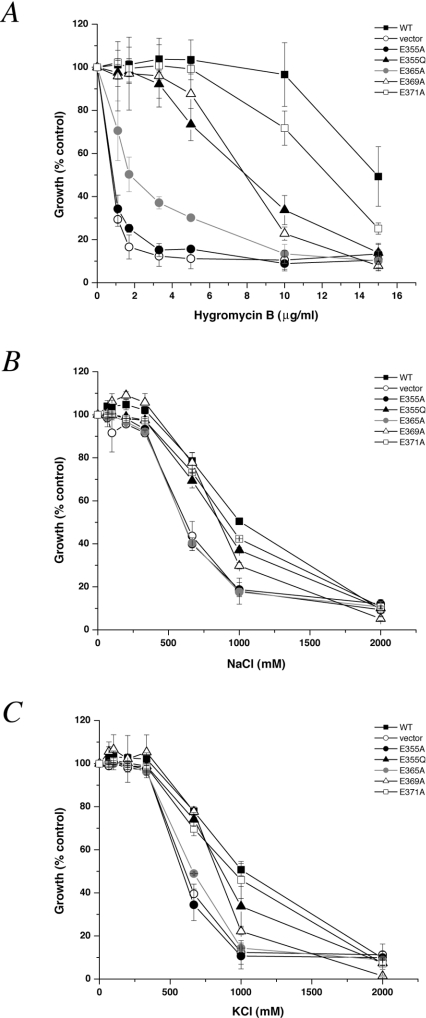
The importance of acidic residues in cation binding and translocation has been well established in studies of ion pumps and transporters; therefore each of the four glutamic residues within the H10 region was replaced with alanine, expressed in the nhx1Δnha1Δ host strain and subjected to phenotype screening. Mutant E371A was largely similar to wild-type in growth-sensitivities to hygromycin B, NaCl and KCl (Figures 2A–2C). In contrast, replacement of the invariant Glu355 residue with the neutral alanine residue resulted in severe impairment of growth under all three conditions of cell stress, such that it was indistinguishable from the antiporter null host strain. To test for the importance of the carbonyl oxygen at this position, we made the more conservative substitution to glutamine. Figure 2 shows that the E355Q substitution results in partial recovery of phenotype-specific growth, relative to E355A. We found that mutant E369A also retained partial tolerance to the stress conditions tested, whereas mutant E365A was relatively more severely impaired. Overall, three of four acidic residues in this stretch of polypeptide appear to contribute, at least in part, to Nhx1 function.
Figure 2. Growth phenotypes of mutations at acidic residues in the H10 region of Nhx1.
(A) Growth-sensitivity to hygromycin B. The host strain (nhx1Δnha1Δ) was transformed with empty vector or plasmid pRin71 expressing wild-type Nhx1 or the indicated mutants. Cultures were inoculated with equal numbers of cells in APG medium (pH 4.0), and hygromycin B was added at the concentrations indicated, as described in the Experimental section. Growth (D600) was measured after 17 h at 30 °C and is expressed as percentage of growth in the absence of hygromycin, which was nearly identical for all strains (see Figure 7). Results shown are averages of quadruplicate determinations and are representative of at least three independent experiments. (B) Growth-sensitivity to NaCl. Cultures, as in (A), were grown in a medium supplemented with NaCl as indicated, and incubated for 24 h at 30 °C. Cell growth (D600) was expressed as percentage of growth in unsupplemented medium. (C) Growth-sensitivity to KCl. Cultures, as in (A), were grown in a medium supplemented with KCl, as indicated. Results are expressed as percentage of growth in unsupplemented medium.
Importance of IC NHE-specific residues in Nhx1 function
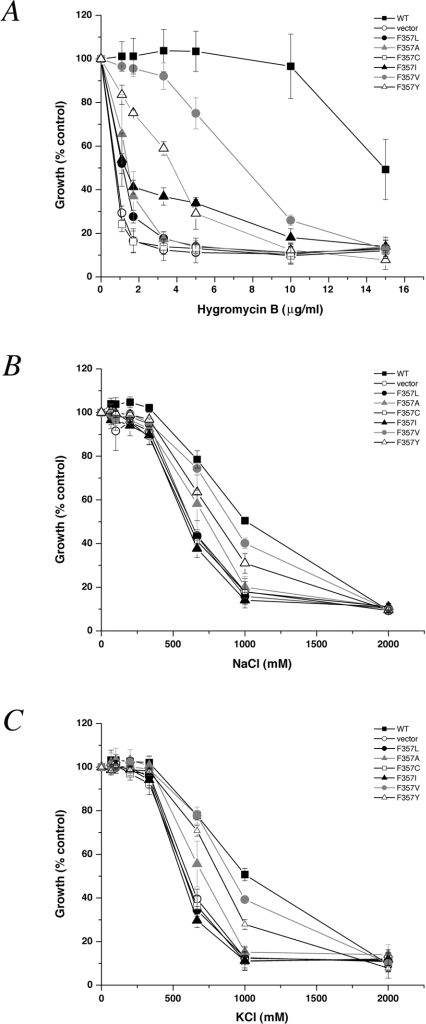
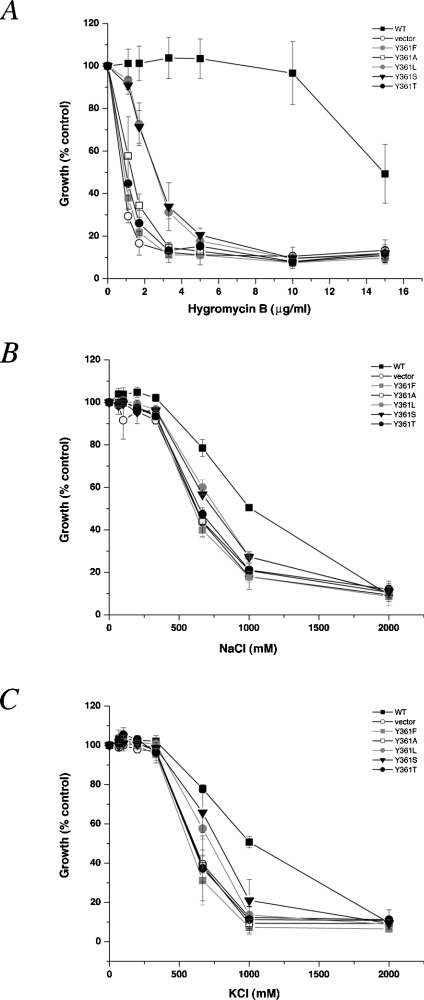
Sequence alignment of the H10 region reveals some residues that are conserved only within the PM NHE or the IC NHE, but not both. To probe the functional importance of such residues, we first replaced Phe357 and Tyr361 with the small, neutral side chain alanine. Since mutants F357A and Y361A displayed nhx1Δ null phenotypes in assays of hygromycin B, Na+ or K+ tolerance (Figures 3A–3C and 4A–4C), we next substituted a variety of amino acids at each position, including the equivalent residues found in the PM NHE. Surprisingly, such conservative substitutions as F357L, F357I and Y361F did not support phenotype-specific growth, indicating severe loss of Nhx1 function, despite their occurrence at the equivalent position in the PM NHE. Of the remaining substitutions tested, only F357V could partially support Nhx1 growth phenotypes, whereas others gave little (F357Y, Y361L and Y361S) or no (F357C and Y361T) evidence of function. Taken together, these findings indicate an unexpected stringency in the requirement for IC NHE-specific residues for Nhx1 function.
Figure 3. Growth phenotypes of substitutions at Phe357 of Nhx1.
Mutants were assessed for growth in medium supplemented with (A) hygromycin B, (B) NaCl and (C) KCl exactly as described in the legend of Figure 2. Only mutants F357V and F357Y showed significant phenotype-specific growth, whereas the remaining substitutions resulted in apparent loss of Nhx1 function.
Figure 4. Growth phenotypes of substitutions at Tyr361 of Nhx1.
Mutants were assessed for growth in a medium supplemented with (A) hygromycin B, (B) NaCl and (C) KCl exactly as described in the legend of Figure 2. All substitutions resulted in major loss of phenotype-specific growth similar to the null strain.
Lack of functional effects of mutations in non-conserved residues
To test the efficacy of the phenotype screening assays, we also generated several mutations in non-conserved or partially conserved residues. We found that mutations A351S, R352A and N356A did not significantly diminish phenotype-specific growth in hygromycin B (Figure 5) or salt (high NaCl and KCl; results not shown). Only the substitution of proline (P376N), predicted to be at the start of transmembrane helix 10, led to complete loss of phenotype similar to the antiporter null host strain, as seen in Figure 5.
Figure 5. Hygromycin B-sensitivity of mutations in non-conserved residues in the H10 region of Nhx1.
Mutants were assessed for growth in the presence of hygromycin B, as described in the legend of Figure 2(A). Only mutant P376N showed a loss-of-function phenotype that was confirmed in growth assays in NaCl and KCl (results not shown).
Expression and localization of H10 mutants
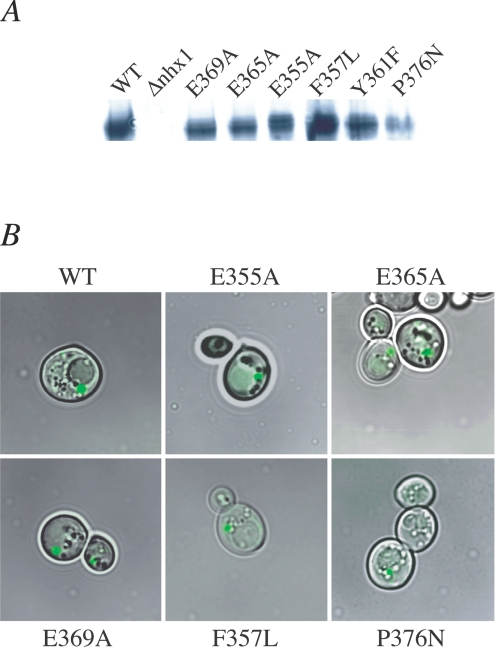
We considered whether the null phenotypes of non-functional Nhx1 mutants could be due to failure to express and correctly localize the mutant proteins. Previous studies have established that introduction of a C-terminal GFP (green fluorescent protein) tag does not alter growth phenotypes of Nhx1 or interfere with subcellular targeting of the protein to the late endosome [8] and, indeed, we were able to confirm that both untagged and C-terminally GFP-tagged H10 mutants had identical growth phenotypes (results not shown). Expression levels of mutant Nhx1 protein varied in membrane preparations, but were similar to wild-type with the exception of P376N, which was significantly lower (Figure 6A). Densitometric analysis of two independent membrane preparations showed average levels ranging from 60 to 75% for all mutants, whereas P376N had an average expression level of 18% of wild-type. Both mutant and wild-type Nhx1 show the same multibanded appearance on the Western blot, indicating normal post-translational processing of the proteins [19]. Similar to wild-type Nhx1, the GFP-tagged mutants examined were found to localize to one to two discrete fluorescent dots per cell, directly abutting on the vacuolar membrane (Figure 6B). This unique localization is characteristic of the late endosome and has been shown to co-localize with markers of the prevacuolar compartment [8]. Since this is distinct from the appearance of endoplasmic reticulum where misfolded proteins are typically retained, our observations suggest that non-functional mutants, including the P376N mutant with decreased expression levels, appeared to correctly traffic to the late endosomal compartment, although further studies and more quantitative analysis would be required to rule out some misfolding or mislocalization of mutant proteins.
Figure 6. Expression and subcellular localization of Nhx1 mutants.
(A) Western blot, using anti-GFP antibody, of total membranes (100 μg) from yeast strains (Δnhx1Δnha1) expressing wild-type Nhx1–HA (WT), empty vector (Δnhx1) or individual H10 mutants that showed significant loss-of-function phenotypes. Only mutant P376N showed consistently lower levels of Nhx1 expression. (B) Confocal fluorescence images of Nhx1–GFP overlaid on contrast image of living yeast cells. Wild-type and a set of H10 mutants are shown. In each case, Nhx1–GFP localizes to one to two discrete dots directly abutting on the vacuolar membrane, characteristic of the late endosome in yeast [8].
Growth phenotypes of Nhx1 mutants relate directly to pH homoeostasis
Correlation of low pH-sensitive growth with vacuolar pH
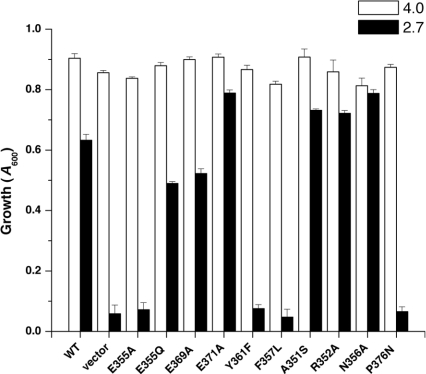
H+ transport activity of Nhx1 mutants was evaluated in situ by measuring vacuolar pH in response to acid stress using the pH-sensitive fluorescent dye BCECF, which preferentially accumulates inside the vacuoles of yeast cells [10,18]. We have previously shown that the nhx1Δ strain is hypersensitive to growth in low pH medium (pH 2.7) or in weak acids (acetic or propionic acids) and that this is accompanied by hyperacidification of both vacuolar and cytoplasmic compartments [11]. We confirm these findings here by showing that plasmid-encoded expression of wild-type Nhx1 confers growth tolerance to acid stress (Figure 7) and protection against hyperacidification of vacuolar pH (pHv 5.11; Table 1) relative to the nhx1Δnha1Δ null strain (pHv 3.87), again demonstrating the major contribution of Nhx1 to vacuolar pH homoeostasis. Next, we assessed vacuolar pH in Nhx1 mutants to determine how in situ H+ transport activity relates to their acid-sensitive growth phenotypes. The results show that growth-sensitivity to acid stress in Nhx1 mutants correlates with hyperacidification of the vacuole (Figure 7 and Table 1). Thus the growth phenotypes appear to be excellent indicators of the in situ H+ transport activity of the exchanger.
Figure 7. Growth-sensitivity of H10 mutants to low pH.
Cultures expressing wild-type Nhx1 (WT), empty vector or individual mutants were grown in APG medium buffered to pH 4.0 or 2.7 for 21 h as described in the legend of Figure 2. The average of quadruplicate determinations from two independent experiments is shown.
Table 1. Yeast vacuolar pH.
The antiporter null host strain nhx1Δnha1Δ was transformed with plasmid expressing wild-type Nhx1 (WT), empty vector (Vector) or the indicated mutants. Vacuolar pH (pHv) was estimated using BCECF fluorescence (Experimental section) in cells subjected to acid stress, as shown in Figure 7. Values for mutant E365A were more variable and are therefore not reported.
| Yeast strain | pHv±S.E.M. |
|---|---|
| WT | 5.11±0.13 |
| Vector | 3.87±0.20 |
| E355A | 4.29±0.19 |
| E355Q | 4.75±0.24 |
| E369A | 4.62±0.04 |
| E371A | 4.95±0.19 |
| Y361F | 4.17±0.04 |
| F357L | 3.87±0.06 |
| A351S | 4.86±0.13 |
| R352A | 4.81±0.40 |
| N356A | 4.99±0.22 |
| P376N | 4.20±0.05 |
Correlation of hygromycin B toxicity with dose-dependent recovery by weak base
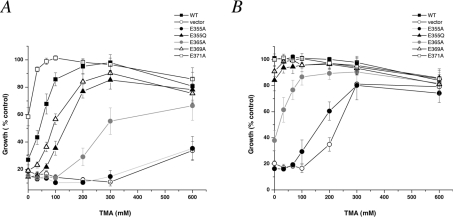
A prominent phenotype of nhx1Δ strains is their striking hypersensitivity to hygromycin B, a well-known inhibitor of protein synthesis. Many vps mutants also share this phenotype [21], suggesting that hygromycin B-sensitivity of the nhx1Δ is chiefly the result of defective biogenesis of the vacuole, potentially a site for detoxification of this drug. We have previously suggested that the role of Nhx1 in vesicle trafficking must be related to its role in pH regulation since the weak base TMA was able to compensate for cellular acidification in nhx1Δ null strain and relieve hygromycin B-sensitivity [11]. We were interested in determining whether hygromycin B-sensitivity in the Nhx1 mutants would correlate directly with defects in cellular pH homoeostasis. To test this, we titrated the ability of TMA added to the growth medium to relieve growth-sensitivity to hygromycin B. At both high (15 μg/ml; Figure 8A) and low (5 μg/ml; Figure 8B) concentrations of hygromycin B, there was a dose-dependence rescue of growth by TMA that was directly proportional to the severity of the mutant growth phenotype. This suggests that trafficking defects of Nhx1 mutants are a consequence of defects in pH homoeostasis.
Figure 8. Dose-dependent correction of hygromycin B-sensitivity of Nhx1 mutants by TMA.
Yeast strains expressing the indicated Nhx1 mutants were grown in APG (pH 4.0) medium supplemented with (A) 15 μg/ml or (B) 5 μg/ml hygromycin B as described in the legend of Figure 2(A). TMA was added at the concentrations indicated, and growth was assessed after 21 h at 30 °C. All mutants showed dose-dependent growth recovery of hygromycin B-sensitive growth that was proportional to their growth phenotypes in hygromycin B (Figure 2A) and acid pH (Figure 7).
Correlation of growth phenotypes with CPY secretion in Nhx1 mutants
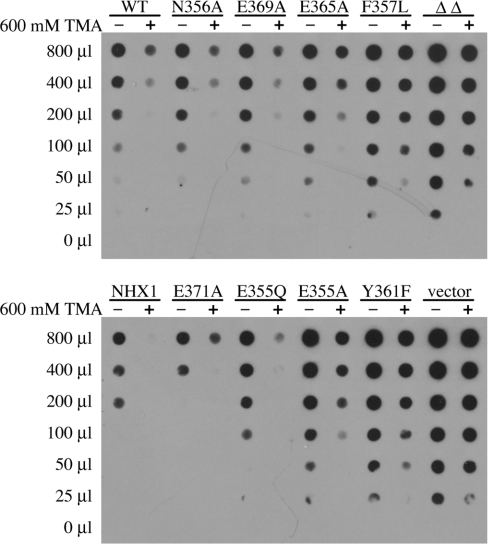
We monitored missorting of CPY to the cell surface, a well-characterized vacuolar targeting defect previously reported in nhx1 and other vacuolar biogenesis mutants [10,12]. As shown in Figure 9, wild-type yeast (upper panel) or nhx1nha1 mutants transformed with NHX1 plasmid (lower panel) secrete low levels of CPY that can be abolished by including TMA in the medium. In contrast, the antiporter null strains secrete high levels of CPY, which can also be attenuated, albeit less effectively, with TMA. These findings indicate for the first time that missorting of CPY, like hygromycin-sensitivity, also relates to pH homoeostasis. We show that levels of CPY secretion and efficacy of TMA in attenuating CPY secretion, correlate with the severity of growth phenotypes observed in the Nhx1 mutants. Mutants F367L, Y361F and E355A most closely resembled the null strain, consistent with results from multiple growth assays. Moderate CPY secretion and more effective correction by TMA were observed in mutants with partial growth phenotypes (E365A, E369A and E355Q) and mutants with normal growth phenotypes closely resembled wild-type in CPY secretion (N356A and E371A). Overall, there was a good correlation between the range of growth phenotypes observed in the mutants and the severity of CPY missorting, again consistent with the trafficking role of Nhx1 being directly related to H+ transport activity.
Figure 9. Missorting of CPY.
Extracellular CPY in culture supernatants (0–800 μl) was assessed by Western blotting of dot-blots as described in the Experimental section. Upper panel shows wild-type yeast (BY4742; WT) and antiporter null strain nhx1Δnha1Δ (ΔΔ) transformed with plasmids expressing the indicated mutants. Lower panel shows the antiporter null strain transformed with plasmid expressing WT NHX1 (NHX1), empty vector or the indicated mutants. TMA was added to the cultures at a final concentration of 600 mM, where indicated (+).
DISCUSSION
The intracellular subgroup of NHE includes numerous examples from plants, fungi and animals that localize to various compartments of the endomembrane system, including the trans-Golgi network {HsNHE7 (human NHE7) [22]}, endosomes (HsNHE6 and yeast Nhx1, [4,8,17]) and the vacuole (Arabidopsis NHX1, [23]). In these compartments, the transmembrane H+ gradient generated by the V-type H+ pumping ATPase serves as principal driving force for the antiport of cations. As shown here, and in previous studies [11,24], overexpression of the IC NHE leads to alkalinization of the compartmental lumen, and conversely, loss of Nhx1 function results in acidification of the vacuolar pH by nearly 1 pH unit. Thus the intracellular NHE must provide a leak pathway for protons out of the endosome and vacuole, and play a major role in regulating luminal pH. This function of the IC NHE differs from the established role of the PM NHE: at the plasma membrane of animal cells, the PM NHE couple H+ efflux with the Na+ gradient established by the sodium pump to alkalinize the cytosol. Mammalian NHE isoforms that reside at or continuously recycle to the plasma membrane (NHE1–NHE5) are highly selective for Na+ over K+ ions so as not to shunt the activity of the Na+, K+-ATPase. In contrast, there is growing evidence that intracellular NHE transports K+ as well as or even in preference to Na+ and may play a significant role in cellular K+ homoeostasis [22,25,26].
Herein, we have begun to investigate these differences at the amino acid level by mutagenic replacement. Such studies are greatly facilitated by the use of quantitative growth assays based on phenotype differences between wild-type and nhx1 null strains. Mutations may be readily screened in medium supplemented with salt (K+ or Na+), H+ (acidic pH) or hygromycin B to provide information on cation transport and selectivity, and vesicle trafficking, respectively. In the present study, we validate the use of growth phenotypes to evaluate Nhx1 function by demonstrating the excellent correlation between low pH-sensitive growth and vacuolar pH, measured in situ using a pH-sensitive fluorescent probe (BCECF) that localizes to the endosomal/vacuolar lumen [10,11,18]. Furthermore, we show that ability of the weak base TMA to compensate for hygromycin B-sensitive growth and missorting of CPY in a range of Nhx1 mutants is directly proportional to the severity of their growth phenotypes. Taken together, our findings suggest that vesicle trafficking and protein targeting defects in Nhx1 mutants relate directly to pH homoeostasis.
Alignment of the predicted membrane spans that constitute the N-terminal 500 amino acids of all NHEs reveals highly conserved residues throughout the family, potentially important for transport function. A recent, thorough analysis of Arabidopsis NHX1 supports the view that all NHEs share the same membrane topology, including the orientation of the TM9–H10–TM10 region [27]. Equally intriguing are the conserved differences between the intracellular and plasma membrane subgroups that may contribute to the newly recognized differences in ion selectivity, subcellular localization and regulation. In the present study, we examined the effect of replacing both conserved and divergent residues within the H10 region. A striking finding was that replacement of Phe357 and Tyr361 of Nhx1, conserved in all other members of this subgroup (Figure 1), with the closely similar residue in the equivalent position of plasma membrane NHE1 from human (leucine and phenylalanine respectively) led to loss of function. This would suggest that there is structural specificity within the H10 region that must be maintained for function. Equivalent replacements have not been tested in the plasma membrane isoforms and it will be of interest to determine if there is similar evidence for such specificity.
Not surprisingly, replacement of non-conserved residues in the H10 had little or no effect on function, in both ScNHX1 (S. cerevisiae NHX1) as well as HsNHE1. This includes the following replacements in our study, with the equivalent mutations analysed in HsNHE1 (see Figure 1) given in parentheses: R352A (S388A, C), N356A (T392V), E369A (S406A) and E371A (H408C). Additional substitutions studied in the H10 region that were not tested in our study but were without effect in HsNHE1 include Ser354 (S390A), Lys366 (T402V), Val370 (H407C) and Leu372 (W409C) [28]. Similarly, residues equivalent to Leu350 (W390L) and Leu353 (V393G) in RnNHE1 failed to alter Na+/H+ exchange activity when 22Na+ influx was measured in stably transfected cultured AP-1 cells [29]. A notable exception is E365A, which showed a significant loss of function despite the lack of conservation at this position among other members of the IC NHE. Replacement of the equivalent residue in NHE1 (S401A) was without effect [28].
The only acidic residue in H10 that is conserved throughout the NHE superfamily, E355A, is essential for Nhx1 function in yeast. We show that replacement of glutamic residue with alanine residue at this position leads to complete loss of function, whereas a more conservative replacement with glutamine largely restores function. An earlier analysis of CPY secretion and FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide] trafficking to the vacuole also indicated that the E355Q mutant resembled wild-type; however, mutant E355A was not assessed [12]. Similar to the findings in yeast Nhx1, Murtazina et al. [28] showed that the equivalent mutation in HsNHE1, E391Q, partially compromised pH recovery from acid loads in mammalian AP-1 cultured cells. Interestingly, replacement with asparagine at this position completely restored function, confirming that a carbonyl group is critical at this position.
Taken together, mutagenesis studies in yeast and mammalian NHEs are remarkably consistent and indicate the importance of the conserved glutamic residue in the H10, as well as the potential importance of conserved differences between intracellular and plasma membrane groups. These findings provide independent confirmation of the newly established phylogenetic classification of the NHE superfamily and pave the way for systematic structure–function analysis of the IC NHE family in the yeast model.
Acknowledgments
This work was supported by grant DK54214 from the National Institutes of Health (Bethesda, MD, U.S.A.) (to R.R.) and a predoctoral award from the American Heart Association Mid-Atlantic Affiliate (to C.L.B.).
References
- 1.Hayashi M., Yoshida T., Monkawa T., Yamaji Y., Sato S., Saruta T. Na+/H+-exchanger 3 activity and its gene in the spontaneously hypertensive rat kidney. J. Hypertens. 1997;15:43–48. [PubMed] [Google Scholar]
- 2.Karmazyn M. Amiloride enhances postischemic ventricular recovery: possible role of Na+–H+ exchange. Am. J. Physiol. 1988;255:H608–H615. doi: 10.1152/ajpheart.1988.255.3.H608. [DOI] [PubMed] [Google Scholar]
- 3.Counillon L., Touret N., Bidet M., Peterson-Yantorno K., Coca-Prados M., Stuart-Tilley A., Wilhelm S., Alper S. L., Civan M. M. Na+/H+ and Cl−/HCO3− antiporters of bovine pigmented ciliary epithelial cells. Pflugers Arch. 2000;440:667–678. doi: 10.1007/s004240000302. [DOI] [PubMed] [Google Scholar]
- 4.Brett C. L., Wei Y., Donowitz D., Rao R. Human Na+/H+ exchanger isoform 6 is found in recycling endosomes of cells, not in mitochondria. Am. J. Physiol. Cell Physiol. 2002;282:C1031–C1041. doi: 10.1152/ajpcell.00420.2001. [DOI] [PubMed] [Google Scholar]
- 5.Bobulescu I. A., Di Sole F., Moe O. W. Na+/H+ exchangers: physiology and link to hypertension and organ ischemia. Curr. Opin. Nephrol. Hypertens. 2005;14:485–494. doi: 10.1097/01.mnh.0000174146.52915.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Numata M., Orlowski J. Identification of a mitochondrial Na+/H+ exchanger. J. Biol. Chem. 1998;273:6951–6959. doi: 10.1074/jbc.273.12.6951. [DOI] [PubMed] [Google Scholar]
- 7.Nass R., Cunningham K. W., Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. Insights into mechanisms of sodium tolerance. J. Biol. Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 8.Nass R., Rao R. Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast. Implications for vacuole biogenesis. J. Biol. Chem. 1998;273:21054–21060. doi: 10.1074/jbc.273.33.21054. [DOI] [PubMed] [Google Scholar]
- 9.Quintero F. J., Blatt M. R., Pardo J. M. Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 2000;471:224–228. doi: 10.1016/s0014-5793(00)01412-5. [DOI] [PubMed] [Google Scholar]
- 10.Ali R., Brett C. L., Mukherjee S., Rao R. Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J. Biol. Chem. 2004;279:4498–4506. doi: 10.1074/jbc.M307446200. [DOI] [PubMed] [Google Scholar]
- 11.Brett C. L., Tukaye D. N., Mukherjee S., Rao R. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol. Biol. Cell. 2005;16:1396–1405. doi: 10.1091/mbc.E04-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowers K., Levi B. P., Patel F. I., Stevens T. H. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:4277–4294. doi: 10.1091/mbc.11.12.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakabayashi S., Pang T., Su X., Shigekawa M. A novel topology model of the human Na+/H+ exchanger isoform 1. J. Biol. Chem. 2000;275:7942–7949. doi: 10.1074/jbc.275.11.7942. [DOI] [PubMed] [Google Scholar]
- 14.Sato Y., Ariyoshi N., Mihara K., Sakaguchi M. Topogenesis of NHE1: direct insertion of the membrane loop and sequestration of cryptic glycosylation and processing sites just after TM9. Biochem. Biophys. Res. Commun. 2004;324:281–287. doi: 10.1016/j.bbrc.2004.09.054. [DOI] [PubMed] [Google Scholar]
- 15.Gama L., Breitwieser G. E. Generation of epitope-tagged proteins by inverse polymerase chain reaction mutagenesis. Methods Mol. Biol. 2002;182:77–83. doi: 10.1385/1-59259-194-9:077. [DOI] [PubMed] [Google Scholar]
- 16.Nakamoto R. K., Rao R., Slayman C. W. Expression of the yeast plasma membrane H+-ATPase in secretory vesicles. A new strategy for directed mutagenesis. J. Biol. Chem. 1991;266:7940–7949. [PubMed] [Google Scholar]
- 17.Miyazaki E., Sakaguchi M., Wakabayashi S., Shigekawa M., Mihara K. NHE6 protein possesses a signal peptide destined for endoplasmic reticulum membrane and localizes in secretory organelles of the cell. J. Biol. Chem. 2001;276:49221–49227. doi: 10.1074/jbc.M106267200. [DOI] [PubMed] [Google Scholar]
- 18.Plant P. J., Manolson M. F., Grinstein S., Demaurex N. Alternative mechanisms of vacuolar acidification in H+-ATPase-deficient yeast. J. Biol. Chem. 1999;274:37270–37279. doi: 10.1074/jbc.274.52.37270. [DOI] [PubMed] [Google Scholar]
- 19.Wells K. M., Rao R. The yeast Na+/H+ exchanger Nhx1 is an N-linked glycoprotein. Topological implications. J. Biol. Chem. 2001;276:3401–3407. doi: 10.1074/jbc.M001688200. [DOI] [PubMed] [Google Scholar]
- 20.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 21.Yoshida S., Anraku Y. Characterization of staurosporine-sensitive mutants of Saccharomyces cerevisiae: vacuolar functions affect staurosporine sensitivity. Mol. Gen. Genet. 2000;263:877–888. doi: 10.1007/s004380000255. [DOI] [PubMed] [Google Scholar]
- 22.Numata M., Orlowski J. Molecular cloning and characterization of a novel (Na+, K+)/H+ exchanger localized to the trans-Golgi network. J. Biol. Chem. 2001;276:17387–17394. doi: 10.1074/jbc.M101319200. [DOI] [PubMed] [Google Scholar]
- 23.Gaxiola R. A., Rao R., Sherman A., Grisafi P., Alper S. L., Fink G. R. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura N., Tanaka S., Teko Y., Mitsui K., Kanazawa H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J. Biol. Chem. 2005;280:1561–1572. doi: 10.1074/jbc.M410041200. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi T., Aharon G. S., Scottosanto J. B., Blumwald E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16107–16112. doi: 10.1073/pnas.0504437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venema K., Belver A., Marin-Manzano M. C., Rodriguez-Rozales M. P., Donaire J. P. A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J. Biol. Chem. 2003;278:22453–22459. doi: 10.1074/jbc.M210794200. [DOI] [PubMed] [Google Scholar]
- 27.Sato Y., Sakaguchi M. Topogenic properties of transmembrane segments of Arabidopsis thaliana NHX1 reveal a common topology model of the Na+/H+ exchanger family. J. Biochem. (Tokyo) 2005;138:425–431. doi: 10.1093/jb/mvi132. [DOI] [PubMed] [Google Scholar]
- 28.Murtazina R., Booth B. J., Bullis B. L., Singh D. N., Fliegel L. Functional analysis of polar amino-acid residues in membrane associated regions of the NHE1 isoform of the mammalian Na+/H+ exchanger. Eur. J. Biochem. 2001;268:4674–4685. doi: 10.1046/j.1432-1327.2001.02391.x. [DOI] [PubMed] [Google Scholar]
- 29.Khadilkar A., Iannuzzi P., Orlwoski J. Identification of sites in the second exomembrane loop and ninth transmembrane helix of the mammalian Na+/H+ exchanger important for drug recognition and cation translocation. J. Biol. Chem. 2001;276:43792–43800. doi: 10.1074/jbc.M106659200. [DOI] [PubMed] [Google Scholar]