Abstract
Appropriation of signalling pathways facilitates poxvirus replication. Poxviruses, as do most viruses, try to modify the host cell environment to achieve favourable replication conditions. In the present study, we show that the early growth response 1 gene (egr-1) is one of the host cell factors intensely modulated by the orthopoxviruses VV (vaccinia virus) and CPV (cowpox virus). These viruses stimulated the generation of both egr-1 mRNA and its gene product, throughout their entire replication cycles, via the requirement of MEK [mitogen-activated protein kinase/ERK (extracellular-signal-regulated kinase) kinase]/ERK pathway. We showed that, upon VV infection, EGR-1 translocates into the nucleus where it binds to the EBS (egr-1-binding site) positioned at the 5′ region of EGR-1-regulated genes. In spite of both viruses belonging to the same genus, several lines of evidence, however, revealed a remarkable contrast between them as far as the roles played by the MEK/ERK/EGR-1 pathway in their biological cycles are concerned. Hence (i) the knocking-down of egr-1 by siRNA (small interfering RNA) proved that this transcription factor is of critical relevance for VV biology, since a decrease of about one log cycle in virus yield was verified, along with a small virus plaque phenotype, whereas the gene silencing did not have a detrimental effect on either CPV multiplication or viral plaque size; (ii) while both pharmacological and genetic inhibition of MEK/ERK resulted in a significant decrease in VV yield, both approaches had no impact on CPV multiplication; and (iii) CPV DNA replication was unaffected by pharmacological inhibition of MEK/ERK, but phosphorylation of MEK/ERK was dependent on CPV DNA replication, contrasting with a significant VV DNA inhibition and VV DNA replication-independence to maintain ERK1/2 phosphorylation, observed under the same conditions.
Keywords: cowpox virus, early growth response 1 (EGR-1), mitogen-activated protein kinase/extracellular-signal-regulated kinase kinase (MEK), orthopoxvirus, vaccinia virus, virus–host cell interaction
Abbreviations: Ara C, cytosine arabinoside; BR, Brighton Red; cDNA, complementary DNA; CHX, cycloheximide; CPV, cowpox virus; egr-1, early growth response 1; EBS, egr-1-binding site; EEV, extracellular enveloped virus; EGF, epidermal growth factor; EMSA, electrophoretic mobility-shift assay; EMV, extracellular mature virus; ERK, extracellular-signal-regulated kinase; FBS, foetal bovine serum; GFP, green fluorescent protein; hpi, hours post-infection; HSP-90, heat-shock protein of 90 kDa; HSV, herpes simplex virus; IFN, interferon; IMV, intracellular mature virus; LAT, latency-associated transcript; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; MOI, multiplicity of infection; MV, myxoma virus; NF-κB, nuclear factor κB; PAK-1, p21-activated kinase 1; RSK, p90 ribosomal S6 kinase; siRNA, small interfering RNA; siEGR1, EGR-1 siRNA; siIRNA, irrelevant siRNA; TBP, TATA-box-binding protein; TK, thymidine kinase; VITF, vaccinia virus-intermediate transcription factor; VLTF, vaccinia virus-late transcription factor; VV, vaccinia virus; WT, wild-type
INTRODUCTION
The orthopoxvirus genus encompasses eight members of the Poxviridae family of viruses, from which VV (vaccinia virus) is the prototypic virus. VV shares with its closely related virus CPV (cowpox virus) its capacity to infect a wide range of hosts, among them humans, cows, rodents and zoo animals [1]. Edward Jenner pioneered, in 1796, human inoculation with a cow-derived poxvirus, which protected against smallpox, and, because of the global and large-scale utilization of VV, in 1980 the World Health Organization declared smallpox to be eradicated [2]. VV and CPV are complex double-stranded DNA viruses that have the potential capacity of encoding more than 200 gene products along their ∼200 kb linear genomes. Their replication cycles occur entirely within the cytoplasmic compartment of infected host cells [1]. Poxviruses present a genetic repertory, whose gene products enable them to efficiently evade the immune and inflammatory host defences [3–5]. Although these mechanisms operate mostly at the extracellular environment, they only facilitate viruses to approach the cells. Nonetheless, these viruses have also evolved intracellular mechanisms, the environment where replication will finally occur, to counteract the antiviral effects associated with IFNs (interferons) [6–8], and the innate responses elicited by Toll-like receptors [6–10].
Thus it is becoming apparent that poxvirus–host cell interaction results from a delicate balance between how viruses manipulate cellular functions associated with the generation of virus progeny while keeping the cells alive, and the avoidance of host responses. For instance, it has been demonstrated that activation of PAK-1 (p21-activated kinase 1) and Raf-1 upon MV (myxoma virus) (a rabbit-specific virus) infection renders mouse fibroblasts permissive for virus replication [11]. Furthermore, MV replication was made possible in non-permissive cells owing to disruption of the MEK [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase/ERK/IRF-3 (IFN regulatory factor 3)/IFN-β pathway [12]. VV also provides an attractive model, although divergent from MV [13]. By activating the MEK/ERK/RSK-2 (p90 ribosomal S6 kinase 2)/ELK-1 [ETS (E twenty-six)-like kinase 1] signalling pathway, VV facilitates its multiplication in mouse fibroblasts [14,15].
While appropriation of signalling pathways facilitates poxvirus replication, expression of intermediate and late VV genes results from the interplay between virus-encoded and cellular factors, whose association promote their transcription, as demonstrated for VV-intermediate or -late transcription factors, VITF [16] and VLTF [17–19] respectively. The requirement of other cellular proteins, such as the molecular chaperone HSP-90 (heat-shock protein of 90 kDa) [20], cyclophilin A [21], along with SP1, RNA polymerase II, or TBP (TATA-box-binding protein) [22], has also been described to benefit VV replication. Even though collectively those data suggest that some host factors could be beneficial for viral replication, definitive proof, nonetheless, awaits confirmation from in vivo experimentation.
The 82 kDa phosphoprotein EGR-1 (early growth response 1) belongs to a family of transcription factors that includes EGR-1–4 and NGFI-B (nerve growth factor inducible factor IB) [23,24]. It is a transcriptional regulator that presents a modular structure such as a DNA-binding domain, which binds to the consensus, GC-rich, DNA sequence 5′-GCG(G/T)GGGCG-3′ [11] and a transcription activation/repression domain, consistent with the diverse activities associated with the molecule [25,26]. Its activation moiety is equipped with three C2H2 zinc fingers, characteristic of a class of eukaryotic transcription factors [27]. EGR-1 couples extracellular stimulation elicited by growth factors, cytokines, hormones and environmental stress, to cellular responses associated with differentiation, proliferation, apoptosis and tissue injury [24,25]. Some viruses, such as HSV (herpes simplex virus), EBV (Epstein–Barr virus) and HIV, are also capable of activating EGR-1 [28–31].
We have shown previously, although to a limited time-frame of the viral replication cycle, that the abovementioned VV-stimulated pathway led to the expression of EGR-1 [14]. In the present study, we demonstrate that VV regulates EGR-1 expression from the very early until late stages of the virus infective cycle. Furthermore, our data also demonstrate that CPV shares with VV its ability in regulating this host factor via the MEK/ERK pathway. In a remarkable contrast, however, virus-regulation of MEK/ERK/EGR-1 appears to serve distinct viral biological needs. While the pathway seems to be of critical relevance for VV multiplication, loss-of-function experiments had no impact on CPV biology, emphasizing overlapping but yet distinct utilization of the signalling pathway by these closely related orthopoxviruses.
EXPERIMENTAL
Cell culture, antibodies and chemicals
A31 cells (a clone derived from mouse Balb/c 3T3 cells) or those stably expressing siRNA (small interfering RNA) [siIRNA (irrelevant siRNA) and siEGR1 (EGR-1 siRNA)] were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 7% (v/v) heat-inactivated FBS (foetal bovine serum; Cultilab), and antibiotics in 5% CO2 at 37 °C. Cells were starved after reaching 80–90% confluence by changing the medium to 1% FBS and incubating for 12 h. Egr-1 and viral TK (thymidine kinase) mRNAs were investigated by using specific probes as described [26,32] respectively. The anti-phospho-ERK1/2 and anti-(total ERK1/2) antibodies were purchased from Cell Signaling Technology, and rabbit polyclonal antibody against EGR-1 (SC110) was from Santa Cruz Biotechnology. Secondary antibody Texas Red-conjugated goat anti-(rabbit IgG) was from Jackson ImmunoResearch Laboratories. Chemicals and inhibitors used throughout the experiments were purchased either from Calbiochem or from Sigma. The pharmacological inhibitors used in the experiments were as follows: PD98059 and SB203580 are specific inhibitors of MEK and p38 MAPK respectively, and H89 acts by inhibiting PKA (protein kinase A) and RSK. The following inhibitors were used at the concentrations given: PD98059 (50 μM), SB203580 (10 μM), H89 (20 μM), CHX (cycloheximide) (100 μg/ml), actinomycin D (5 μg/ml) and Ara C (cytosine arabinoside) (40 μg/ml). The doses of drugs used throughout the experiments were established on the basis of experimental observations, without, nonetheless, causing any harm to the cells, given that no measurable effect on cell viability was verified by Trypan Blue dye exclusion.
Viruses and virus infection
WT (wild-type) VV strain WR, recombinant VV vF13L–GFP (green fluorescent protein) chimaera [33] and CPV strain BR (Brighton Red) were propagated into Vero cells and were highly purified by sucrose gradient sedimentation as described in [34]. There are two infective forms of VV/CPV, the IMV (intracellular mature virus) and the EEV (extracellular enveloped virus), which use, at least for VV, distinct mechanisms to enter the target cells. Whereas the IMV form requires a signalling-dependent mechanism for entry, the EEV form seems to be independent [35]. It is believed that the IMV remains inside the cells, being released upon cell lysis. The experiments of the present study were carried out with the IMV form of VV or CPV. VV and CPV were UV-inactivated after exposure of the viruses stocks for 5 min to a UV lamp producing irradiation predominantly at 365 nm. After that, the UV-irradiated viruses were tested for virus infectivity. Virus that no longer was able to form plaques as compared with the non-irradiated virus was assumed to be UV-inactivated. VV or CPV infection of A31 cells was carried out when the cultures reached 80–90% confluence. Cells were infected in the absence of FBS, at the indicated MOI (multiplicity of infection), and for the times shown. Cells were incubated with the indicated pharmacological inhibitor for 30 min before VV infection which was maintained throughout the infection.
Virus infectivity assays
A31 cells stably expressing siEGR1, WT MEK1 or MEK1 dominant-negative mutation, were cultured as described above at a density of 4.5×105 cells per well, in a six-well culture dish and then VV- or CPV-infected. Infections were carried out at an MOI ranging from 1.0 to 10.0, as indicated, for the times shown. Cultures were then washed with cold PBS, followed by three freezing/thawing cycles. Virus was collected from the supernatant of centrifuged cells and then assayed for infectivity as described in [32]. Each experiment was run in duplicate, and the results are the means. Data were confirmed by at least three independent experiments with similar results.
RNA isolation and Northern blotting
Cells (3×106) were cultured and starved as described above. Then the cells were incubated with the inhibitors actinomycin D, CHX or PD98059 at the indicated concentrations, before virus infection at the indicated MOI for the times shown. Total RNA was isolated as described in [36], and 15 μg of RNA per sample was loaded, electrophoresed on a 1.5% denaturing agarose–formaldehyde gel, transferred on to a nylon membrane (Amersham Biosciences) and UV cross-linked for 2 min. Membranes were then probed with egr-1 cDNA (complementary DNA) or viral TK labelled with [α-32P]dCTP (Amersham Biosciences), to a specific activity of (1–5)×108 c.p.m./μg of DNA, by using a multiprime DNA labelling system. Hybridization and washing procedures were carried out as described in [36]. The membranes were then stripped of the probe and re-probed with 18 S rRNA, labelled with [γ-32P]ATP by using phage T4 polynucleotide kinase (Promega), which was used as an internal control for RNA loading.
EMSA (electrophoretic mobility-shift assay)
A31 cells were cultured and starved as above and then VV-infected at MOI of 3.0 for the indicated times. When appropriate, cells were pre-incubated for 30 min with PD98059 and then infected. EMSAs were carried out essentially as described in [37]. Whole-cell extracts were prepared by a modification of the method described in [38]. Briefly, frozen cell pellets were thawed on ice and lysed with an equal volume of lysis buffer (100 mM Tris/HCl, pH 8.0, 0.2 mM EDTA, 1% Triton X-100, 10% glycerol, 5 mM sodium pyrophosphate, 4 μg/ml leupeptin and 1 mM sodium orthovanadate). Lysates were scraped and collected into Eppendorf tubes and then centrifuged at 13000 g for 20 min at 4 °C. Protein concentration was determined by using a Bio-Rad assay. Protein (10 μg) was pre-incubated with 1.2 μl of poly(dI–dC)·(dI-dC) (5.4 mg/ml) (Amersham Biosciences) at room temperature (25 °C) for 10 min, followed by addition of 1.25 μg of BSA, 0.125 μg of Escherichia coli DNA, 0.25 μg of yeast tRNA, 2% Ficoll 400 and 0.32 ng of labelled probe. The reaction mixtures were incubated at room temperature for 15 min and then analysed by 6% PAGE. The 5′ 32P-end-labelled double-stranded probes (only one strand is shown) corresponding to the consensus cis-acting elements, EBS (EGR-1-binding site): 5′-GGATCCAGCGGGGGCGAGCGGGGGCGA-3′ or the unrelated probe NF-κB (nuclear factor κB) 5′-GTTGAGGGGACTTTCCCAGGC-3′, were used in the assays. The supershift experiments were carried out by incubation of the cell extracts with anti-EGR-1 antibody for 1 h before mixing it with the labelled probe for 15 min. Competition assays were carried out by incubating a 50-fold molar excess of unlabelled homologous or unrelated probe with the proteins for 10 min, before adding the labelled probe.
MEK1 dominant-negative cell lines
A31 cells were transfected with 10 μg of plasmid DNA carrying either dominant-negative mutant MEK1 or WT MEK1 cDNA [39] using standard calcium phosphate protocols [40]. Transfectants were ring-cloned after selection with 800 μg/ml Geneticin (G418; Invitrogen) for at least 21 days and then tested for MEK1 unresponsiveness, as evaluated by ERK1/2 phosphorylation, after stimulation with 50 μg/ml EGF (epidermal growth factor; Sigma–Aldrich) or virus infection at an MOI of 3.0. Unresponsive clones to both stimuli above were then used to carry out the experiments.
Western blotting: whole-cell lysate preparation
Cells were left untreated or incubated with the specific inhibitor for 30 min before VV or CPV infection at the indicated MOI. Cells were then washed twice with cold PBS and lysed on ice with lysis buffer (100 mM Tris/HCl, pH 8.0, 0.2 mM EDTA, 1% Triton X-100, 10% glycerol, 5 mM sodium pyrophosphate, 4 μg/ml leupeptin and 1 mM sodium orthovanadate). Lysates were scraped and collected into Eppendorf tubes and then centrifuged at 13000 g for 15 min at 4 °C. Protein concentration was determined by using the Bio-Rad assay.
Western blotting: electrophoresis and immunoblotting
Whole-cell lysates (25–30 μg) were separated by electrophoresis on an SDS/10% polyacrylamide gel and then transferred on to nitrocellulose membranes as described in [14]. Membranes were blocked overnight at 4 °C with PBS containing 5% (w/v) non-fat dried milk and 0.1% (v/v) Tween 20. The membranes were washed three times with PBS containing 0.1% (v/v) Tween 20 and then incubated with the specific primary polyclonal antibody (1:1500) in PBS containing 5% (w/v) BSA and 0.1% (v/v) Tween 20. After washing, the membranes were incubated with horseradish-peroxidase-conjugated secondary anti-rabbit antibody (1:3000 dilution). Immunoreactive bands were visualized by using ECL® (enhanced chemiluminescence) detection system as described in the manufacturer's instructions (Amersham Biosciences).
siRNA targeted to egr-1
A DNA construct from which egr-1 siRNA could be generated in vivo was designed to specifically target the egr-1 mRNA, therefore excluding any other egr family member, according to Ambion's siRNA target finder (http://www.ambion.com/techlib/misc/siRNA_finder.html). Only the 19 nucleotides of sense-strand sequence is described: 5′-GGTGGTTTCCAGGTTCCCA-3′, which corresponds to positions +2404 to +2422 of the egr-1 mRNA (Gene ID 13653). BamHI and HindIII restriction endonuclease recognition sequences were added to the ends of the siRNA. Sense and antisense strands were annealed and then cloned into the plasmid pSilencer™ 3.1-Hi neo (Ambion). Egr-1 recombinants were confirmed by DNA sequencing and were purified, and then 10 μg of DNA was used to transfect A31 cells using standard calcium phosphate protocols. Cell clones were allowed to grow under selection with 800 μg/ml G418 for 2–3 weeks, and individual clones were ring-cloned and then checked for functionality. As a control, 12 clones generated by stable transfection of the pSilencer™ 3.1-Hi neo vector expressing a hairpin siRNA with limited homology (irrelevant) to any known sequences of human and mouse genomes were ring-cloned and designated siIRNA.
Immunofluorescence microscopy
Cells were grown on coverslips and infected with VV vF13L–GFP at an MOI of 10.0 for 12 h. Detection of EGR-1 was performed in 4% (w/v) paraformaldehyde-fixed and 0.2% (v/v) Triton X-100-permeabilized cells, after incubation with specific rabbit polyclonal serum coupled with a secondary Texas Red-conjugated goat anti-rabbit antibody. Fluorescently labelled cells were visualized using a Zeiss (LSM 510 META) confocal laser-scanning microscope.
Dot-blot assays
A31 cells (3×106) were cultured and starved as described above and then were infected with CPV at an MOI of 10.0 for 3, 5, 7 or 9 h, in either the presence or the absence of 50 μM PD98059. After the infections, the cells were scraped from the dishes and collected by centrifugation at 800 g as described in [14]. In brief, after centrifugation, the cells were washed with cold PBS and were resuspended in 0.3 ml of loading buffer [10× SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate) and 1 M ammonium acetate). The cells were then frozen and thawed three times, followed by the addition of 0.45 ml of loading buffer. A 25 μl volume of each sample was applied under vacuum to Hybond-N membranes (Amersham Bioscience) using a HYBRI·DOT Manifold apparatus (BRL Life Technology). The DNA was denatured for 30 min with 500 mM NaOH and 1.5 M NaCl, and the membrane was washed twice with 10× SSC in situ for 10 min. DNA was then cross-linked by exposure of the membrane to UV light for 2 min. Hybridization conditions were as described for Northern blotting.
Densitometric analysis
Dot-blot assays were quantified by using a densitometer (Typhoon 9210 phosphoimager; GE Healthcare). Results are the means for duplicate samples (arbitrary units) representing each time post-infection.
RESULTS
Analysis of egr-1 expression upon VV and CPV infection
Our previous studies had shown that the VV-stimulated MEK/ERK signalling pathway led to the expression of EGR-1 [14]. Since EGR-1 expression paralleled the sustained activation of MEK/ERK upon viral infection, our present analysis attempted to extend our studies to the entire virus replication cycle and tried to get some insights into its biological significance. We first observed that VV infection stimulated a prolonged and yet sustained accumulation of the egr-1 transcript. The steady-state levels of egr-1 mRNA were apparent 1 hpi [hour(s) post-infection] and remained elevated for up to 9 hpi (Figure 1A, upper panel). As shown in Figure 1(B), we also observed that VV-induced egr-1 mRNA is MOI-dependent, and reached a plateau at an MOI of 1.0. In addition, gene stimulation seems to rely on both ongoing protein synthesis (Figure 1C, lanes 3 and 5) and active transcription (Figure 1D, lanes 3–8), since incubation with CHX or actinomycin D respectively resulted in a remarkable reduction in VV-induced egr-1 transcript. Our data also demonstrated that virus multiplication was required for VV-induced egr-1 transcription, (Figure 1C, lanes 6–8), since UV-inactivated viruses were no longer able to stimulate egr-1 transcription. Moreover, our findings provided evidence that ERK1/2 activation was required for egr-1 expression (Figure 1C, lane 4), because incubation with PD98059 before VV infection caused a significant reduction in egr-1 mRNA accumulation. Finally, VV stimulated EGR-1 protein accumulation up to 36 hpi, i.e. the late stages of the viral life cycle, and the pathway associated with this stimulation appears to recruit MEK, since its pharmacological inhibition resulted in an accentuated decrease of EGR-1 accumulation (Figures 1E and 1F).
Figure 1. VV-stimulated EGR-1 expression.
Northern blot assays (A–D) carried out as described in the Experimental section. Cells were serum-starved and then were either mock- (MO) or VV-infected at an MOI of 3.0 or as stated otherwise. (A) Time course of egr-1 expression (upper panel). Lane 1, MO; lane 2, stimulated with 10% FBS for 30 min; lanes 3–11, VV-infected for 1–9 h respectively. (B) VV-induced egr-1 expression is MOI-dependent (upper panel). Lane 1, MO; lane 2, incubated with 10% FBS for 30 min; lanes 3–6, VV-infected with MOIs of 0.01, 0.1, 1.0 and 10.0 respectively. (C) VV-induced egr-1 expression relies on ongoing protein synthesis, ERK1/2 activation and virus multiplication (upper panel). Lanes 1 and 6, MO; lane 2, stimulated with 10% FBS for 30 min; lanes 3–5 and 7, VV-infected at an MOI of 3.0 for 4 h; lanes 4 and 5, pre-incubated with PD98059 (50 μM) or CHX (100 μg/ml) respectively for 30 min; lane 8, incubated with UV-inactivated VV for 4 h. (D) VV-induced egr-1 expression is dependent on active gene transcription (upper panel). Lanes 1 and 5, MO-infected for 3 or 5 h respectively; lanes 2 and 6, VV-infected for 3 or 5 h; lanes 3, 4, 7 and 8, VV-infected for 4, 5, 6 and 7 h respectively. Actinomycin D (5 μg/ml) was added to the culture at 3 or 5 hpi and culture continued for an additional 1 or 2 h respectively. Lane 9, VV-infected for 7 h. Lower panels of (A–D) show membranes re-probed with 18 S rRNA as an internal control for RNA loading. (E and F) Upper panels: VV-stimulated EGR-1 accumulation. Western blot analysis of VV-infected cells at MOIs and times indicated. Cells were either MO-infected, or infected with VV, blotted and then probed with anti-EGR-1 antibody. Where indicated, cells were incubated with PD98059 (50 μM) before virus infection. Lower panels: blots were re-probed with anti-total ERK1/2 antibody as a control for protein loading. Experiments were carried out independently three times with similar results. Molecular masses are given in kDa.
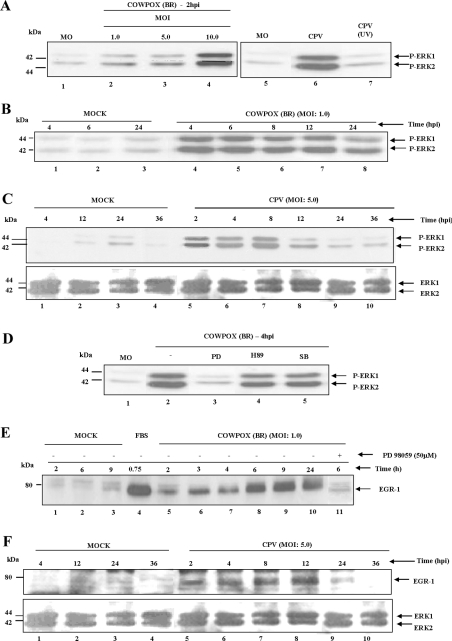
CPV activates ERK1/2 leading to EGR-1 expression
Since VV regulates the expression of EGR-1 via MEK/ERK, we hypothesized that the same regulation might be a common theme utilized by another orthopoxvirus, CPV. Indeed, our findings showed that CPV stimulated ERK1/2 phosphorylation in an MOI-dependent manner (Figure 2A, lanes 1–4). Furthermore, replication-competent CPV was absolutely required to phosphorylate the kinases, because UV-inactivated virus was not allowed to do so (Figure 2A, lanes 5–7). We next examined the time course of phospho-ERK1/2 stimulation upon viral infection. Our findings (Figures 2B and 2C) showed that the kinases were stimulated from early (∼2 hpi) until late times (∼24–36 hpi) during the virus infection cycle, in accordance with the dependence on the MOI observed in Figure 2(A) and therefore only kinetic differences were verified. Our data also showed that virusstimulation of ERK1/2 was specifically regulated by MEK, since pre-incubation with its pharmacological inhibitor PD98059 completely blocked ERK1/2 phosphorylation (Figure 2D, lane 3), whereas PKA (H89) and p38 MAPK (SB203580) inhibitors were not effective (Figure 2D, lanes 4 and 5). We next investigated whether the signal generated through the MEK/ERK pathway upon CPV infection was delivered to egr-1. Figure 2(E) shows that CPV regulated EGR-1 expression with kinetics that paralleled that of virus-activation of MEK/ERK and was equally inhibited by PD98059, characterizing MEK/ERK as the virus-activated kinases recruited to regulate EGR-1 expression. Once again, the kinetics of EGR-1 expression upon CPV infection at MOI of 5.0 (Figure 2F) were paralleled to that verified with viral activation of MEK/ERK at the same MOI (Figure 2C). Thus Figures 1 and 2 showed that both VV and CPV activated the MEK/ERK pathway, which in turn targeted the expression of EGR-1, during their entire life cycle.
Figure 2. CPV activates ERK1/2 leading to EGR-1 expression.
Cells were either mock-infected (MO/MOCK) or infected with CPV at an MOI of 1.0 or 5.0 as indicated, blotted and then probed with anti-phospho-ERK1/2 antibody (A–D) or with anti-EGR-1 antibody (E and F). (A) Effects of MOI and UV-inactivated CPV on virus-stimulated ERK1/2 activation. Lanes 2–4, infected with CPV for 2 h; lanes 6–7, incubated with CPV at an MOI of 5.0 or the same amount of UV-inactivated virus for 4 h respectively. (B and C) Time courses of CPV-stimulated ERK1/2 activation at an MOI of 1.0 or 5.0 respectively. (D) Viral-activation of ERK1/2 is specifically affected by PD98059. Cells were incubated with PD98059 (50 μM), H89 (20 μM) or SB203580 (10 μM) before virus infection (MOI of 5.0) as indicated. (E and F) Time courses of CPV-stimulated EGR-1 expression at an MOI of 1.0 or 5.0 respectively. (E) Lane 4, incubated with 10% FBS. Data were consistently confirmed in three independent experiments. Molecular masses are given in kDa.
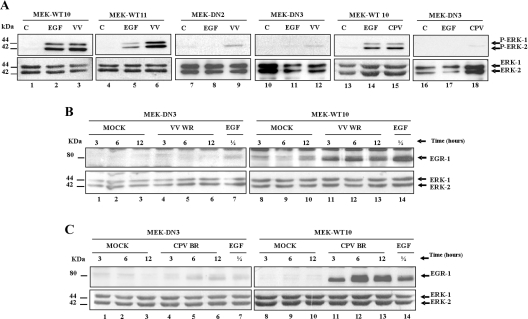
Cell lines expressing MEK1 dominant-negative mutation interfere with both VV- and CPV-stimulated ERK1/2 activation and egr-1 expression
On the basis of pharmacological inhibition of MEK (Figure 1F) and on our previous results [14], we demonstrated that VV-stimulated EGR-1 expression relies on MEK/ERK pathway. However, to rule out the possibility of non-specific inhibition of MEK, we generated cell lines stably expressing either WT MEK1 or MEK1 dominant-negative mutation. Figure 3(A) illustrates this analysis. As shown, while cells expressing WT MEK1 associated EGF stimulation and VV infection with ERK1/2 phosphorylation (lanes 1–6), cells expressing dominant-negative MEK1 mutation responded neither to EGF nor to the infection (lanes 7–12). This analysis was also performed with CPV infection with very similar results (lanes 13–18). Expression of EGR-1 was then investigated in these lines. Figures 3(B) and 3(C) demonstrate that MEK/ERK was specifically required to mediate EGR-1 expression either upon VV or CPV infection respectively; the same requirement was also observed with the control EGF (lanes 1–14). Taken together, results from Figures 1–3 demonstrated that both viruses regulated the MEK/ERK/EGR-1 pathway throughout their entire replication cycles. The data were confirmed not only by pharmacological but also by genetic inhibition of the pathway.
Figure 3. MEK dominant-negative mutation affects both VV- and CPV-stimulated ERK1/2 phosphorylation and EGR-1 expression.
Upper panels: cell lines expressing WT MEK or MEK dominant-negative mutation (DN) were either stimulated with EGF (50 μg/ml) for 30 min or infected with VV or CPV at an MOI of 10.0 for 4 h (A) or at an MOI of 10 for the indicated times (B and C). Whole-cell lysates were prepared and immunoblotted with anti-phospho-ERK1/2 (A) or anti-EGR-1 (B and C) antibodies. Lower panels: blots were re-probed with anti-(total ERK1/2) antibody as an internal control for protein loading. Blots are representative of at least three independent experiments with similar results.
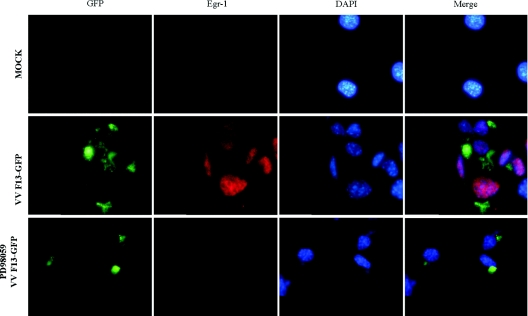
EGR-1 translocates into the nucleus upon VV infection
EGR-1 is a transcriptional regulator associated with a number of biological processes, such as proliferation/differentiation, apoptosis and pathogenesis of vascular disease [24]. To investigate whether viral infection would be able to stimulate EGR-1 to translocate into the nucleus, immunofluorescence microscopy was carried out. Figure 4 demonstrates that VV stimulated the nuclear translocation of EGR-1 (middle row). It also shows that the translocation relied upon the MEK/ERK signalling pathway, since its pharmacological inhibition resulted in a complete block of EGR-1 expression (bottom row). CPV infection was also able to stimulate EGR-1 nuclear translocation (results not shown).
Figure 4. VV stimulates EGR-1 nuclear translocation.
Immunofluorescence microscopy of EGR-1 cellular localization in virus-infected cells. Cells growing on coverslips were mock-infected (top row) or infected with VVF13-GFP (green) at an MOI of 10.0 for 12 h and then stained with anti-EGR-1 antibody (red), either in the absence (middle row) or in the presence of PD98059 (50 μM) (bottom row). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Merged pictures show the DAPI-stained image superimposed on the EGR-1-stained image. Results were confirmed by three independent assays with similar results.
Upon translocation to the nucleus, EGR-1 binds to the regulatory sequence EBS
We next examined whether EGR-1 was able to recognize and bind to the cis-acting element EBS found at the promoter region of putative EGR-1-regulated genes. Our findings showed that, upon nuclear translocation, EGR-1 binds to EBS as revealed by gel supershift assay (Figure 5, lane 18). The earliest DNA–protein interaction observed was at 3.0 hpi, reached its maximum at 12–18 hpi and declined thereafter. The EGR–DNA complex formed is specific and was dissociated after competition with a molar excess of unlabelled homologous probe, but was not dissociated after competition with the unrelated one, as shown in Figure 5 (lanes 16 and 17). Likewise, EGR–DNA interaction relied on the MEK/ERK transduction pathway, as demonstrated after its specific pharmacological blockade (Figure 5, lanes 11, 12 and 15).
Figure 5. Upon nuclear translocation, EGR-1 binds to the regulatory sequence EBS.
EMSA carried out with labelled EBS probe. Whole-cell extract (10 μg per sample) was mixed with the probe. Lanes 1–4 and 13, mock-infected; lane 5, stimulated with 10% FBS for 30 min; lanes 6–12 and 14–17, VV-infected for the times shown; lanes 11, 12 and 15, incubated with PD98059 (50 μM) before virus infection; lanes 16 and 17, competed with 50-fold molar excess of unlabelled EBS or NF-κB oligonucleotides respectively; lane 18, incubated with anti-EGR-1 antibody for 15 min before virus infection. Data were confirmed experimentally three times with similar results.
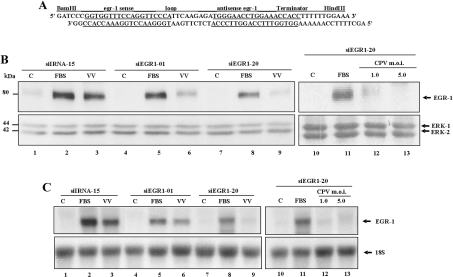
Efficient silencing of erg-1 by stable transfection with siRNA
Because (i) both VV and CPV stimulated EGR-1 expression from the very early until late stages of their replication cycle, and (ii) by knowing that EGR-1 is not the only downstream substrate of MEK/ERK, we decided to investigate the functional significance of this specific host factor in orthopoxvirus biology. To that end, we knocked-down the gene by using siRNA [41]. To examine the levels of egr-1 gene silencing, cell lines stably expressing either siEGR1 or the siIRNA were analysed. Figure 6(A) shows the DNA sequences designed to generate siEGR1 in vivo. A total of 28 G418-resistant clones expressing siEGR1 and 12 expressing siIRNA were analysed by immunoblot and Northern blot analysis. Figures 6(B) and 6(C) illustrate these analyses. Cells were grown to 80% confluence, serum-starved for 12 h and then stimulated with 10% FBS for 30 min, VV at an MOI of 3.0 for 4 h or CPV at an MOI of 1.0 or 5.0 for 6 or 4 h respectively, and then probed either with anti-EGR-1 antibody or with α-32P-labelled egr-1 probe. Figure 6(B) shows that egr-1 was knocked-down to a variable extent, compare the differences between clones siEGR1-01 and siEGR1-20 (lanes 4–6 and 7–9), with the latter presenting a higher level of gene silencing. In contrast, egr-1 expression in response to either FBS or VV was unaltered in the clone expressing siIRNA (siIRNA-15) (lanes 1–3). Of note, reduction in EGR-1 protein accumulation observed in Figure 6(B) was paralleled by a decrease in the steady-state levels of egr-1 mRNA as shown in Figure 6(C). Very similar results were obtained when the cell lines siIRNA-15 (not shown) and siEGR1-20 were infected with CPV (Figures 6B and 6C, lanes 10–13). It is important to point out that the levels of expression of both egr-1 mRNA and its gene product in the clone siIRNA-15, upon viral infection, were comparable with those ones observed in the untransfected cell line (A31) (results not shown).
Figure 6. Efficient siRNA-mediated silencing of EGR-1 expression.
(A) The DNA sequence designed to knock-down the gene is shown. (B and C) Cell lines stably expressing siEGR1 (lanes 4–13) or expressing siIRNA (lanes 1–3) were left unstimulated (C) or stimulated with 20% FBS, or infected either with VV at an MOI of 3.0 for 4 h, as indicated, or with CPV at an MOI of 1.0 (lane 12) or 5.0 (lane 13) for 6 h and 4 h respectively. (B) Western blot analysis of siEGR-1-expressing clones (upper panel). Whole-cell lysates were prepared for immunoblot of EGR-1. (C) Northern blot analysis of siEGR1-expressing clones (upper panel). Total RNA was isolated, blotted and probed with labelled egr-1. (B and C) Lower panels: blots were re-probed with anti-total ERK1/2 antibody or with 18 S rRNA respectively as an internal control for protein or RNA loading. Blots are representative of at least three independent experiments with similar results.
EGR-1 plays a critical role in VV multiplication
In order to confirm that sustained expression of egr-1 mRNA followed by its continued translation upon VV infection were of biological relevance, the cell lines siEGR1-01 and -20 were infected with VV at an MOI of 10.0 and 12 hpi virus was collected and assayed for infectivity. As shown in Table 1, Egr-1 appears to be required for VV multiplication, since its silencing resulted in ∼10-fold reduction in virus yield (siEGR1-20). Of note, the decrease in virus production correlated well with the level of egr-1 silencing, and was less pronounced when the cells siEGR1-01 were used.
Table 1. Effects of siRNA targeted to egr-1 on VV multiplication.
Cell lines stably expressing siEGR1-01 and sIEGR1-20 or siIRNA-15 were infected with VV at an MOI of 10.0 and 12 hpi viruses were collected and titrated. Virus growth carried out with clone siIRNA-15 was taken as 100% (6.0×106 plaque-forming units/ml).
| Infection | Virus yield (%) | Inhibition (%) |
|---|---|---|
| siIRNA-15 | 100 | 0 |
| siEGR1-01 | 56 | 44 |
| siEGR1-20 | 13 | 87 |
Differential roles played by EGR-1 in orthopoxvirus biology
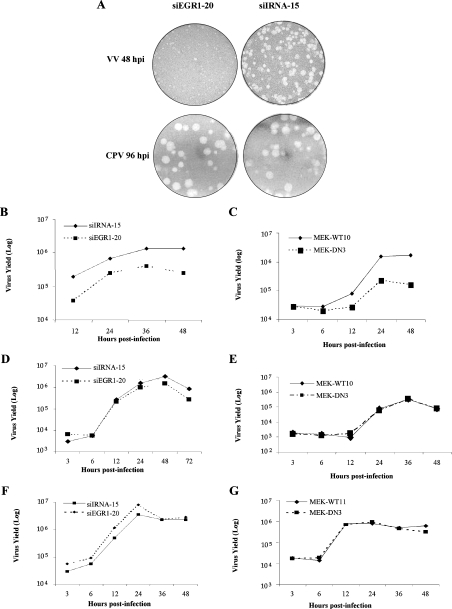
To gain some insights into whether egr-1 played differential roles in VV and CPV biology, the following set of experiments was carried out. First, as shown in Figure 7, VV multiplication performed with siEGR1-20, the cell clone expressing higher levels of gene silencing, resulted in reduction not only of viral plaque phenotype (Figure 7A, upper panel), but also of viral yield (Figure 7B). Secondly, and opposite, altered plaque phenotype was not verified (Figure 7A, lower panel), neither was CPV multiplication affected, when the experiments were performed with the same cell line, at different MOI (Figures 7D and 7F). Because VV- and CPV-stimulated EGR-1 expression is governed by the MEK/ERK pathway ([14], and the present study), finally, we tested whether or not viral multiplication performed with a cell line expressing MEK dominant-negative mutation (MEK-DN3) paralleled that carried out with the clone siEGR1-20. As shown in Figure 7(C), VV growth rates decreased approx. one log cycle in this line as compared with that expressing WT MEK (MEK-WT10). Once again, it is noteworthy that CPV multiplication did not rely on the MEK/ERK pathway, independently of the MOI used, as shown in Figures 7(E) and 7(G).
Figure 7. Effects of EGR-1 gene silencing and MEK dominant-negative mutation in viral biology.
Cell lines (4.0×105 cells) stably expressing either siEGR1-20 or siIRNA-15 (A, B, D and F) or WT MEK or dominant-negative MEK1 (MEK-DN3) (C, E and G), were infected with VV at 400 pfu (plaque-forming units)/well (A, upper panel) or with CPV at 150 pfu/well (A, lower panel) and 48 or 96 hpi respectively were fixed with formaldehyde. Cells were infected with VV at an MOI of 3.0 (B and C) for the times shown. Infections with CPV were performed at an MOI of 1.0 (D and E) or an MOI of 5.0 (F and G). Plaques were visualized after staining with Crystal Violet. Results are representative of at least three independent experiments with similar results.
Both CPV early gene expression and DNA replication do not depend on the MEK/ERK pathway, but sustained ERK1/2 phosphorylation is dependent on viral DNA replication
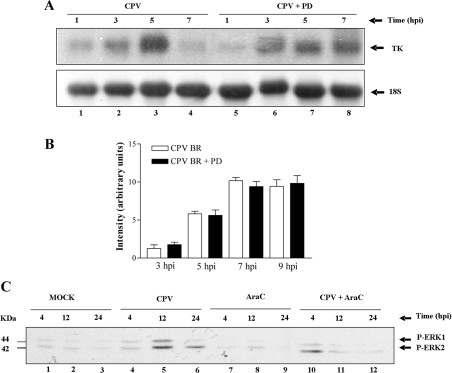
We have shown previously that pharmacological inhibition of MEK/ERK was associated with both delayed VV early viral gene (TK) expression and decreased DNA replication, in accordance with the reduction verified in virus yield [14]. In order to investigate whether both CPV TK gene expression and DNA replication also relied on signals transmitted by MEK/ERK, cells were infected with CPV at an MOI of 5.0 and viral RNA/DNA was isolated at the indicated times and blotted on to a nylon membrane. Figure 8(A) shows that TK gene expression was delayed when the infections were carried out in the presence of PD98059 (lanes 5–8). On the other hand, CPV viral DNA replication was not affected by MEK/ERK, since no statistically significant differences were found with or without inhibitor (Student non-parametric two-tailed t test) (Figure 8B), contrasting with the partial dependence on this pathway displayed by VV [14]. In an attempt to clarify whether the sustained ERK1/2 phosphorylation upon CPV infection was dependent on DNA replication or on a post-replicative protein synthesis event, experiments were carried out in either the presence or the absence of Ara C. As shown in Figure 8(C), inhibition of viral DNA synthesis was followed by a dramatic decrease in ERK1/2 phosphorylation.
Figure 8. Both CPV TK gene expression and DNA replication did not rely on MEK/ERK, but sustained ERK1/2 phosphorylation was dependent on viral DNA replication.
Cells were mock-infected or infected with CPV at an MOI of 5.0 in either the absence or the presence of 50 μM PD98059 (PD) (A and B) or Ara C (40 μg/ml) (C) for the times shown. (A) Viral RNA was isolated as described in the Experimental section, blotted and hybridized with 32P-labelled CPV TK probe. (B) Viral DNA was isolated, dot-blotted and hybridized with 32P-labelled CPV DNA. Results are means±S.D. of the quantification of duplicate samples for each time point. No statistically significant difference was seen between samples with or without PD98059 (Student's non-parametric two-tailed t test). (C) Whole-cell lysates were immunoblotted with anti-phospho-ERK1/2. Results were consistently confirmed in three independent experiments.
DISCUSSION
The large genome of VV codes for molecules necessary to promote self-expression of viral early genes, such as RNA polymerase [42], the VV early transcription factor (VETF) [43], and capping enzyme [44], among others. Consequently, early studies had suggested that VV transcription might rely entirely on virus-encoded products. However, it was quickly realized that the virus must also benefit from the interaction with its hosts, which provide not only the necessary energy to carry out essential metabolic functions, but also transcription factors needed for expression of intermediate and late virus genes [16,17,19].
Thus viral replication results from a complex network of virus–host interactions culminating with the most adequate intracellular environment for the generation of its progeny. As far as poxvirus replication is concerned, it has been shown that viral manipulation of signalling pathways is a fertile way of promoting replication [13]. MV illustrates this assumption. By activating PAK-1 and Raf-1, this rabbit-specific poxvirus makes mouse fibroblasts permissive for its replication [11].
We have shown previously that the MEK/ERK/RSK2/Elk-1 pathway is required for maximal VV replication [14]. By combining both pharmacological and genetic inhibition of MEK, we show now that viral stimulation of this pathway leads to the expression of egr-1 (Figures 1C, 1F and 3B). Consistent with the viral requirement for activation of the signalling cascade, induction of egr-1 is also dependent on viral multiplication, de novo protein synthesis and active gene transcription, since UV-irradiated viruses are unable to stimulate the gene expression and the same is verified when infection is carried out in the presence of the respective inhibitor (Figures 1C and 1D). Whether host or viral (or both) protein and RNA synthesis are required for VV-stimulated egr-1 expression, under our experimental conditions, it is not possible to discriminate between them, and it remains to be demonstrated further. Stimulation of the MEK/ERK/EGR-1 pathway is also a CPV-regulated process (Figures 2A–2E, 3A and 3C). Of note, the kinetics of egr-1 mRNA/protein accumulation is prolonged until times where viral morphogenesis had already come to completion (Figures 1F, 2E, 7B, 7D and 7F).
Also remarkable is the observation that, upon VV infection, EGR-1 translocates into the nucleus and binds to the regulatory sequence EBS, a cis-acting element found at the promoter region of EGR-1-regulated genes, suggesting a regulatory role played by EGR-1. Once again, both processes rely upon MEK/ERK pathway (Figure 4, bottom row; Figure 5, lane 11). Nonetheless, other genes under EGR-1 regulation and the roles they may play on VV biology, remain to be investigated further. CPV was also able to stimulate EGR-1 nuclear translocation (results not shown).
Collectively, however, our data suggest that VV- and CPV-regulated induction of egr-1 should be of biological relevance. The beneficial role played by egr-1 on the HSV replication cycle, was reported previously [30]. Thus, through its binding to the HSV LAT (latency-associated transcript) promoter, EGR-1 prevents TBP from binding and RNA polymerase II from initiating transcription of LAT, and thus reactivates HSV from latency. In our study, the roles played by EGR-1 in VV biology were confirmed by knocking-down this gene (Figure 6). This host factor not only affects the generation of virus progeny (decrease of about one log cycle), but also alters the virus plaque phenotype. Virus replication carried out with siEGR1-20 cells presents a smaller plaque size than that observed with the control siIRNA cells (Figures 7A, upper panel, and 7B). Gene silencing, however, does not appear to affect the expression of the viral early gene (TK), because comparable levels of mRNA accumulation are verified in both knocked-down and control cells (results not shown). On the other hand, the knocking-down process does seem to affect late VV gene expression as revealed by immunoblot assay carried out using the anti-viral H3L antibody (results not shown). Moreover, the dominant-negative approach carried out with MEK/ERK, the pathway that ultimately leads to VV-stimulated egr-1 expression, also shows about one log cycle decrease in virus yield (Figure 7C), accompanied by a reduction in plaque size (results not shown), confirming the virus-dependence on MEK/ERK/EGR-1. It is worth noting that the levels of egr-1 silencing are in accordance with the decrease observed in virus yield (Table 1), which appears to point to a threshold level of gene product expression required to be of biological significance. Contribution of egr-1 to virus replication could be even more pronounced considering that the knocking-down process hardly reaches 90% of gene silencing, as observed with other viruses [45–47]. Even though EGR-1 appears to have multiple implications for VV biology and, in addition, it also appears to be regulated by the modified VV Ankara [48]. In remarkable contrast, however, this host factor does not seem to play a relevant role for the orthopoxvirus CPV, as far as virus yield (Figures 7D and 7F) and virus plaque phenotype (Figure 7A, lower panel) are concerned, since none of them is affected when assayed either with the siEGR1-20 or dominant-negative mutant MEK cell lines. In addition, CPV DNA replication is not affected at all by disruption of the MEK/ERK pathway, and, even though TK expression is delayed, it does not affect CPV multiplication, which is in contrast with what we have observed previously with VV [14]. Whether intermediate or late CPV genes are affected by EGR-1 or MEK/ERK, it remains to be investigated further. Also remarkable is the observation that CPV-stimulated ERK1/2 phosphorylation is dependent on virus DNA replication (Figure 8C), once again in a remarkable contrast with what has been reported for VV [14]. Thus, despite their closely related evolution, and their ability to infect common hosts, evidence is provided that virus-stimulated MEK/ERK/EGR-1 pathway play distinct and yet overlapping roles in orthopoxvirus biology.
Since both viruses regulate the expression of EGR-1 until late times in their infective cycles and a number of reports support the conclusion that VV infection shuts down host protein synthesis and decreases mRNA half life [1,49], how virus deals with the selective up-regulation of some host genes, as demonstrated in the present study and in [18,48] and yet down-regulates others, remains largely a matter of speculation.
We believe that the effect caused by knocking-down egr-1 is specific because (i) both the mRNA and the gene product are down-regulated, thus ruling out the possibility of a translation attenuation effect mediated by miRNA (micro RNA) [50], (ii) translation attenuation associated with IFN production seems to be unlikely because these viruses present unequivocal mechanisms to counteract IFN production and actions [7,10,51,52]. Moreover, we were not able to detect induction of IFN-stimulated genes in the VV-infected cells expressing siEGR1 (results not shown).
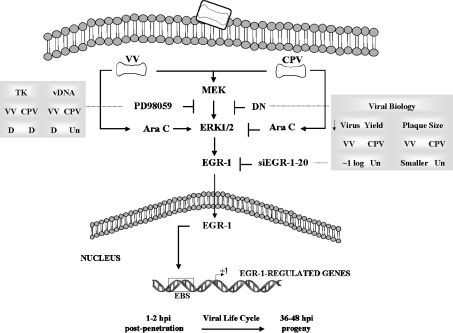
On the basis of the findings in the present study and our previous one [14], we propose a model for the orthopoxvirus–host cell interaction and the roles played by the MEK/ERK/EGR-1 pathway in viral biology. Although both VV and CPV stimulate the same transduction pathway, distinct biological outcomes, nonetheless, are verified (Figure 9).
Figure 9. Model of the orthopoxvirus–host cell interaction.
Both VV and CPV stimulate the MEK/ERK/EGR-1 pathway from early until late times of viral infective cycle. Upon stimulation, EGR-1 translocates into the nucleus where it binds to the regulatory region (EBS) of EGR-1-regulated genes to control their expression. Pharmacological (PD98059) and genetic (dominant-negative mutation, DN) inhibition of MEK/ERK in conjunction with gene silencing (siEGR1-20) approaches demonstrate a distinct requirement of the pathway for viral biology (see right-hand box; Un, unaffected). While viral TK gene expression upon pharmacological inhibition was equally delayed (D) for both viruses (see left-hand box), viral DNA (vDNA) replication was unaffected (Un) only for CPV. A post-replicative event is required (Ara C) for CPV- but not for VV-stimulated ERK1/2 phosphorylation.
To the best of our knowledge, this is the first report to describe a novel category of host factors required for efficient VV replication, although the role played in CPV biology remains to be demonstrated further. While others have reported and characterized host factors associated with viral intermediate and late transcription termination or host factors necessary for in vitro virus replication, for example, VITF-2 [47], VLTF [17,18], HSP-90 [20], cyclophilin A [21], RNA polymerase II, and the transcription factors YY1, SP1 and TBP [22], although these data await confirmation from in vivo experimentation, the characterization of EGR-1 is novel in the sense that this host factor, in contrast with the others, is virus-regulated throughout its entire life cycle, and relies upon a signalling pathway. Thus we believe that this study has contributed to lay one more brick to building the picture of the interaction of the brick-shaped VV with host cells.
Acknowledgments
We are grateful to Angela S. Lopes, Hilda M.V. Gama, Maria A. Souza and João R. dos Santos for their secretarial/technical assistance. We also thank Dr H. A. Armelin and Dr M. C. Sogayar (Departmento de Bioquímica-Universidade de São Paulo, São Paulo, Brazil), who kindly provided us with the A31 cell line. WT VV, VV strain WR and CPV BR were obtained from Dr C. Jungwirth (Universität Würzburg, Germany) and the chimaeric VV vF13L–GFP was a gift from Dr B. Moss (National Institute of Allergy and Infectious Diseases, Bethesda, MD, U.S.A.). Plasmids carrying the WT and MEK1 dominant-negative mutation were a gift from Dr P. J. S. Stork (Vollum Institute for Advanced Biomedical Research, Oregon Health & Science University, Portland, OR, U.S.A.). egr-1 probe was generously provided by Dr T. Curran (St. Jude Children's Research Hospital, Memphis, TE, U.S.A.). This work was supported by grants from Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). P.N.G.S., J.A.P.S. and B.S.A.F.B. are recipients of pre-doctoral fellowships from CNPq, CAPES (Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior; Brazilian Ministry of Culture, Science and Technology), and FAPEMIG respectively. C.A.B., E.G.K. and P.C.P.F. are recipients of research fellowships from CNPq. We are also grateful to Dr B. Moss (NIH) and Dr P. Traktman (Department of Microbiology and Molecular Genetics, Medical College of Wisconsin, Milwaukee, WI, U.S.A.) for their valuable comments on the manuscript.
References
- 1.Moss B. Poxviridae viruses and their replication. In: Fields B. N., Knipe D. M., Howley P. M., editors. Fields Virology, vol. 2. 3rd edn. New York: Lippincott-Raven Publishers; 1996. pp. 2637–2672. [Google Scholar]
- 2.Smith G. L., McFadden G. Smallpox: anything to declare? Nat. Rev. Immunol. 2002;2:521–527. doi: 10.1038/nri845. [DOI] [PubMed] [Google Scholar]
- 3.Lalani A. S., Masters J., Zeng W., Barrett J., Pannu R., Everett H., Arendt C. W., McFadden G. Use of chemokine receptors by poxviruses. Science. 1999;286:1968–1971. doi: 10.1126/science.286.5446.1968. [DOI] [PubMed] [Google Scholar]
- 4.Seet B. T., Johnston J. B., Brunetti C. R., Barrett J. W., Everett H., Cameron C., Sypula J., Nazarian S. H., Lucas A., McFadden G. Poxviruses and immune evasion. Annu. Rev. Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 5.Smith G. L., Symons J. A., Khanna A., Vanderplasschen A., Alcami A. Vaccinia virus immune evasion. Immunol. Rev. 1997;159:137–154. doi: 10.1111/j.1600-065x.1997.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 6.Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 7.Beattie E., Denzler K. L., Tartaglia J., Perkus M. E., Paoletti E., Jacobs B. L. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith E. J., Marie I., Prakash A., Garcia-Sastre A., Levy D. E. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 2001;276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- 9.Bowie A., Kiss-Toth E., Symons J. A., Smith G. L., Dower S. K., O'Neill L. A. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10162–10167. doi: 10.1073/pnas.160027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonjardim C. A. Interferons (IFNs) are key cytokines in both innate and adaptive antiviral immune responses – and viruses counteract IFN action. Microbes. Infect. 2005;7:569–578. doi: 10.1016/j.micinf.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Johnston J. B., Barrett J. W., Chang W., Chung C. S., Zeng W., Masters J., Mann M., Wang F., Cao J., McFadden G. Role of the serine-threonine kinase PAK-1 in myxoma virus replication. J. Virol. 2003;77:5877–5888. doi: 10.1128/JVI.77.10.5877-5888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F., Ma Y., Barrett J. W., Gao X., Loh J., Barton E., Virgin H. W., McFadden G. Disruption of ERK-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 2004;5:1266–1274. doi: 10.1038/ni1132. [DOI] [PubMed] [Google Scholar]
- 13.McFadden G. Poxvirus tropism. Nat. Rev. Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrade A. A., Silva P. N., Pereira A. C., De Sousa L. P., Ferreira P. C., Gazzinelli R. T., Kroon E. G., Ropert C., Bonjardim C. A. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. Biochem. J. 2004;381:437–446. doi: 10.1042/BJ20031375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Magalhaes J. C., Andrade A. A., Silva P. N., Sousa L. P., Ropert C., Ferreira P. C., Kroon E. G., Gazzinelli R. T., Bonjardim C. A. A mitogenic signal triggered at an early stage of vaccinia virus infection: implication of MEK/ERK and protein kinase A in virus multiplication. J. Biol. Chem. 2001;276:38353–38360. doi: 10.1074/jbc.M100183200. [DOI] [PubMed] [Google Scholar]
- 16.Rosales R., Sutter G., Moss B. A cellular factor is required for transcription of vaccinia viral intermediate-stage genes. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3794–3798. doi: 10.1073/pnas.91.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broyles S. S., Liu X., Zhu M., Kremer M. Transcription factor YY1 is a vaccinia virus late promoter activator. J. Biol. Chem. 1999;274:35662–35667. doi: 10.1074/jbc.274.50.35662. [DOI] [PubMed] [Google Scholar]
- 18.Guerra S., Lopez-Fernandez L. A., Pascual-Montano A., Munoz M., Harshman K., Esteban M. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 2003;77:6493–6506. doi: 10.1128/JVI.77.11.6493-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright C. F., Hubbs A. E., Gunasinghe S. K., Oswald B. W. A vaccinia virus late transcription factor copurifies with a factor that binds to a viral late promoter and is complemented by extracts from uninfected HeLa cells. J. Virol. 1998;72:1446–1451. doi: 10.1128/jvi.72.2.1446-1451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung J. J., Chung C. S., Chang W. Molecular chaperone Hsp90 is important for vaccinia virus growth in cells. J. Virol. 2002;76:1379–1390. doi: 10.1128/JVI.76.3.1379-1390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro A. P., Carvalho T. M., Moussatche N., Damaso C. R. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 2003;77:9052–9068. doi: 10.1128/JVI.77.16.9052-9068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh J., Broyles S. S. Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J. Virol. 2005;79:12852–12860. doi: 10.1128/JVI.79.20.12852-12860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 24.Silverman E. S., Collins T. Pathways of Egr-1-mediated gene transcription in vascular biology. Am. J. Pathol. 1999;154:665–670. doi: 10.1016/S0002-9440(10)65312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C., Rangnekar V. M., Adamson E., Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 1998;5:3–28. [PubMed] [Google Scholar]
- 26.Sukhatme V. P., Cao X. M., Chang L. C., Tsai-Morris C. H., Stamenkovich D., Ferreira P. C., Cohen D. R., Edwards S. A., Shows T. B., Curran T., et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 27.Khachigian L. M., Lindner V., Williams A. J., Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 28.Feng W. H., Hong G., Delecluse H. J., Kenney S. C. Lytic induction therapy for Epstein–Barr virus-positive B-cell lymphomas. J. Virol. 2004;78:1893–1902. doi: 10.1128/JVI.78.4.1893-1902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson T., Zetterberg H., Wang Y. C., Rymo L. Promoter-proximal regulatory elements involved in oriP-EBNA1-independent and -dependent activation of the Epstein–Barr virus C promoter in B-lymphoid cell lines. J. Virol. 2001;75:5796–5811. doi: 10.1128/JVI.75.13.5796-5811.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatarowicz W. A., Martin C. E., Pekosz A. S., Madden S. L., Rauscher F. J., 3rd, Chiang S. Y., Beerman T. A., Fraser N. W. Repression of the HSV-1 latency-associated transcript (LAT) promoter by the early growth response (EGR) proteins: involvement of a binding site immediately downstream of the TATA box. J. Neurovirol. 1997;3:212–224. doi: 10.3109/13550289709018296. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y., Dong B., Mittelstadt P. R., Xiao H., Ashwell J. D. HIV Tat binds Egr proteins and enhances Egr-dependent transactivation of the Fas ligand promoter. J. Biol. Chem. 2002;277:19482–19487. doi: 10.1074/jbc.M201687200. [DOI] [PubMed] [Google Scholar]
- 32.da Fonseca F. G., Trindade G. S., Silva R. L., Bonjardim C. A., Ferreira P. C., Kroon E. G. Characterization of a vaccinia-like virus isolated in a Brazilian forest. J. Gen. Virol. 2002;83:223–228. doi: 10.1099/0022-1317-83-1-223. [DOI] [PubMed] [Google Scholar]
- 33.Husain M., Moss B. Intracellular trafficking of a palmitoylated membrane-associated protein component of enveloped vaccinia virus. J. Virol. 2003;77:9008–9019. doi: 10.1128/JVI.77.16.9008-9019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joklik W. K. The purification of four strains of poxvirus. Virology. 1962;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- 35.Locker J. K., Kuehn A., Schleich S., Rutter G., Hohenberg H., Wepf R., Griffiths G. Entry of the two infectious forms of vaccinia virus at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell. 2000;11:2497–2511. doi: 10.1091/mbc.11.7.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Sousa L. P., Brasil B. S., Silva B. M., Freitas M. H., Nogueira S. V., Ferreira P. C., Kroon E. G., Bonjardim C. A. Plasminogen/plasmin regulates c-fos and egr-1 expression via the MEK/ERK pathway. Biochem. Biophys. Res. Commun. 2005;329:237–245. doi: 10.1016/j.bbrc.2005.01.123. [DOI] [PubMed] [Google Scholar]
- 37.Bonjardim C. A. A mutant cell line partially responsive to both IFN-α and IFN-γ. Braz. J. Med. Biol. Res. 1997;30:41–50. doi: 10.1590/s0100-879x1997000100007. [DOI] [PubMed] [Google Scholar]
- 38.Zimarino V., Wu C. Induction of sequence-specific binding of Drosophila heat shock activator protein without protein synthesis. Nature (London) 1987;327:727–730. doi: 10.1038/327727a0. [DOI] [PubMed] [Google Scholar]
- 39.Horgan A. M., Stork P. J. Examining the mechanism of ERK nuclear translocation using green fluorescent protein. Exp. Cell Res. 2003;285:208–220. doi: 10.1016/s0014-4827(03)00037-5. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J., Fritsch E. F., Maniatis T. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 41.Dykxhoorn D. M., Novina C. D., Sharp P. A. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 42.Baroudy B. M., Moss B. Purification and characterization of a DNA-dependent RNA polymerase from vaccinia virions. J. Biol. Chem. 1980;255:4372–4380. [PubMed] [Google Scholar]
- 43.Broyles S. S., Yuen L., Shuman S., Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J. Biol. Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- 44.Shuman S., Broyles S. S., Moss B. Purification and characterization of a transcription termination factor from vaccinia virions. J. Biol. Chem. 1987;262:12372–12380. [PubMed] [Google Scholar]
- 45.Gitlin L., Karelsky S., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature (London) 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 46.Qin X. F., An D. S., Chen I. S., Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. U.S.A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randall G., Grakoui A., Rice C. M. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludwig H., Mages J., Staib C., Lehmann M. H., Lang R., Sutter G. Role of viral factor E3L in modified vaccinia virus ankara infection of human HeLa cells: regulation of the virus life cycle and identification of differentially expressed host genes. J. Virol. 2005;79:2584–2596. doi: 10.1128/JVI.79.4.2584-2596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Condit R. C., Niles E. G. Regulation of viral transcription elongation and termination during vaccinia virus infection. Biochim. Biophys. Acta. 2002;1577:325–336. doi: 10.1016/s0167-4781(02)00461-x. [DOI] [PubMed] [Google Scholar]
- 50.Ruvkun G., Wightman B., Ha I. The 20 years it took to recognize the importance of tiny RNAs. Cell. 2004;116:S93–96. doi: 10.1016/s0092-8674(04)00034-0. [DOI] [PubMed] [Google Scholar]
- 51.Takeda K., Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 52.Harte M. T., Haga I. R., Maloney G., Gray P., Reading P. C., Bartlett N. W., Smith G. L., Bowie A., O'Neill L. A. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 2003;197:343–351. doi: 10.1084/jem.20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]