Abstract
The DYRKs (dual specificity tyrosine phosphorylation-regulated kinases) are a conserved family of protein kinases that autophosphorylate a tyrosine residue in their activation loop by an intra-molecular mechanism and phosphorylate exogenous substrates on serine/threonine residues. Little is known about the identity of true substrates for DYRK family members and their binding partners. To address this question, we used full-length dDYRK2 (Drosophila DYRK2) as bait in a yeast two-hybrid screen of a Drosophila embryo cDNA library. Of 14 independent dDYRK2 interacting clones identified, three were derived from the chromatin remodelling factor, SNR1 (Snf5-related 1), and three from the essential chromatin component, TRX (trithorax). The association of dDYRK2 with SNR1 and TRX was confirmed by co-immunoprecipitation studies. Deletion analysis showed that the C-terminus of dDYRK2 modulated the interaction with SNR1 and TRX. DYRK family member MNB (Minibrain) was also found to co-precipitate with SNR1 and TRX, associations that did not require the C-terminus of the molecule. dDYRK2 and MNB were also found to phosphorylate SNR1 at Thr102 in vitro and in vivo. This phosphorylation required the highly conserved DH-box (DYRK homology box) of dDYRK2, whereas the DH-box was not essential for phosphorylation by MNB. This is the first instance of phosphorylation of SNR1 or any of its homologues and implicates the DYRK family of kinases with a role in chromatin remodelling.
Keywords: chromatin remodelling, dual-specificity kinase, dual specificity tyrosine phosphorylation-regulated kinase (DYRK), minibrain, Sf9 cell, yeast two-hybrid screen
Abbreviations: Brm, Brahma; CREB, cAMP-response-element-binding protein; Ct, C-terminal; DmcycE, Drosophila melanogaster cyclin E; DYRK, dual specificity tyrosine phosphorylation-regulated kinase; dDYRK2, Drosophila DYRK2; DH-box, DYRK homology box; FOXO, forkhead box O; GAL4, galactosidase 4 protein; GST, glutathione S-transferase; HA, haemagglutinin; HDAC, histone deacetylase; HNF1α, hepatocyte nuclear factor 1α; Kin, kinase; TRX, trithorax; HRX, human TRX; INI1, integrase interactor 1; LB/Amp, Luria–Bertani broth supplemented with 50 μg/ml ampicillin; LC-MS, liquid chromatography–MS; MAPK, mitogen-activated protein kinase; MBK-2, minibrain kinase 2; MBP, myelin basic protein; Mirk, minibrain related kinase; MNB, minibrain; NES, nuclear export signal; NFAT, nuclear factor of activated T-cells; Nt, N-terminal; OMA-1, oocyte maturation defective 1; SET domain, Su(var)3-9, Enhancer of Zeste, Trithorax domain; Sf9 cell, Spodoptera frugiperda 9 cell; SNR1, Snf5-related 1; STAT3, signal transducer and activator of transcription 3; WT, wild-type; X-α-Gal, 5-bromo-4-chloroindol-3-yl α-D-galactopyranoside; Yak1p, Saccharomyces cerevisiae DYRK family member encoding the YAK1 protein
INTRODUCTION
DYRKs (dual specificity tyrosine phosphorylation-regulated kinases) are an evolutionarily conserved family of protein kinases that autophosphorylate a critical tyrosine residue in the kinase domain activation loop, but phosphorylate exogenous substrates exclusively on serine and threonine residues [1–6]. DYRK family members share a conserved central kinase domain and adjacent N-terminal DH-box (DYRK homology box), but differ in their N- and C-terminal extensions. Two DYRK subclasses exist. Class 1 DYRKs, present only in multicellular organisms, contain an N-terminal nuclear localization signal and a C-terminal PEST (Pro-Glu-Ser-Thr) or GAS region (potential involvement in proteolytic breakdown of protein). Class 2 DYRKs, present in all eukaryotic organisms examined to date, do not contain any additional defined regions (Figure 1A) [6].
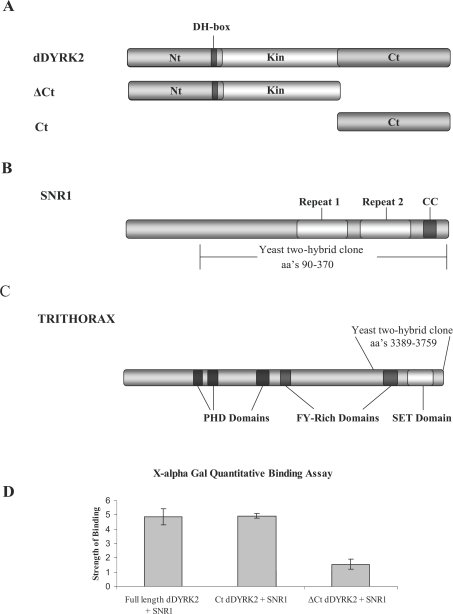
Figure 1. Yeast two-hybrid screen.
(A) Schematic representation of the dDYRK2 protein and deletion mutants showing the non-catalytic N-terminal (Nt) and C-terminal (Ct) regions flanking a central kinase (Kin) domain. The location of the highly conserved DH-box is also shown. (B) Schematic representation of SNR1 showing the conserved repeat domains 1 and 2, the coiled coil region (CC) and the portion of the molecule identified in the yeast two-hybrid screen. (C) Schematic representation of TRX showing the location of the PHD, FY-rich and SET domains as well as the portion of the molecule identified in the yeast two-hybrid screen. (D) Clones encoding full-length dDYRK2, Ct alone and ΔCt dDYRK2 proteins were co-transformed with SNR1 in yeast AH109 cells. X-α-Gal quantitative assays were then used to measure the relative binding strengths of different dDYRK2 molecules with SNR1. The results shown are the means±S.D. for four separate experiments.
Functional studies have been conducted on yeast, Caenorhabditis elegans, Drosophila and mammalian DYRK family members. In the yeast Saccharomyces cerevisiae, the sole DYRK family member, Yak1p (YAK1 protein), functions as an antagonist to the Ras/cAMP-dependent protein kinase (protein kinase A) pathway to negatively regulate cell growth [7]. The Dictyostelium counterpart, YakA, is essential for both a starvation-induced growth arrest and initiation of a developmental response, and has been suggested to function as a specific cell-cycle regulator to facilitate exit from the cell cycle and to mediate developmental events [8]. In Drosophila, mutation of the founding DYRK family member, mnb (minibrain), affects post-embryonic neurogenesis, resulting in specific reductions in the size of the optic lobes and central brain hemispheres [9]. The mammalian orthologue of the mnb gene, Dyrk1A, maps to the Down's syndrome critical region in humans, and mice with a single copy of the gene display region-specific reductions in the brain, indicating a conserved mode of action that determines normal growth and brain size in both mammals and flies [10]. The related family member Mirk (Minibrain related kinase)/Dyrk1B has been shown to play a critical role in skeletal-muscle development [11]. In C. elegans, the class 2 DYRK, MBK-2 (Minibrain kinase 2), phosphorylates OMA-1 (oocyte maturation defective 1) in a process that leads to proteolytic degradation of OMA-1, resulting in the transition of the oocyte to embryo [12–14].
Various other studies have identified a number of potential DYRK family substrates including the Gli1 (glioma-associated oncogene homologue 1) transcription factor, protein-synthesis initiation factor eIF2Bϵ (eukaryotic initiation factor 2Bϵ), the microtubule-associated protein tau, STAT3 (signal transducer and activator of transcription 3), the forkhead transcription factor FKHR (forkhead in rhabdosarcoma), CREB (cAMP-response-element-binding protein), cyclin L2, splicing factor SF3b (splicing factor 3b)/SAP155 (SIT4 associating protein 155), dynamin, the cyclin-dependent inhibitor p27Kip1, the cell-cycle inhibitor and survival molecule p21cip1, the cell-cycle regulator, cyclin D1, class II HDACs (histone deacetylases) (HDAC4, HDAC5, HDAC7 and HDAC9) and HNF1α (hepatocyte nuclear factor 1α) [2–4,15–18]. In vitro studies indicate that DYRKs are proline-directed and display a strong preference for the presence of an arginine residue at the −2 or −3 position N-terminal to the phosphorylated residue [2,19], producing a consensus phosphorylation recognition sequence R-X(X)-S/T-P. This sequence has proven useful in predicting potential phosphorylation sites but a number of exceptions have been reported [3,4,15,20]. Proteins shown to directly interact with DYRK family members include CREB, the brain-specific protein PAHX-AP1 (phytanoyl-CoA α-hydroxylase associated protein 1), Arip4, the dimerization cofactor of HNF1α [DCoHm (dimerization cofactor of HNF1α from muscle)], HNF1α, MKK3 [MAPK (mitogen-activated protein kinase) kinase 3], p38a/b MAPK and RanBPM (Ran binding protein M) [3,21–24].
In the present study using dDYRK2 (Drosophila DYRK2) as the bait in a yeast two-hybrid screen, we report the identification of SNR1 (Snf5-related 1) and TRX (trithorax) as dDYRK2-interacting proteins. Co-immunoprecipitation studies confirmed the association of dDYRK2 with these proteins and demonstrated that the DYRK family member MNB also co-precipitates with SNR1 and TRX. Both dDYRK2 and MNB were found to phosphorylate SNR1 on Thr102 in vitro and in vivo. For dDYRK2, the highly conserved DH-box was essential for this phosphorylation event.
MATERIALS AND METHODS
Yeast two-hybrid constructs
For the generation of full-length WT (wild-type) dDYRK2, an EcoRI site was cloned into the dDYRK2 cDNA immediately preceding the initiating ATG using the primer, 5′-ggacctgcatatggccatggaggccgaattcatgttggatcgatgcgaaatgccg-3′. The EcoRI fragment encoding the full-length dDYRK2 protein was then cloned in frame with the GAL4 (galactosidase 4 protein) binding domain of the yeast two-hybrid vector pGBKT7. For the generation of a construct containing the N-terminal and kinase domain of dDYRK2, a termination codon TGA was introduced immediately after the kinase domain of the construct pGBKT7/dDYRK2 full-length using the primer 5′-ggcggcccaccacgagtttctgtgagaattccagccatctgcctccagtcgcc-3′. For the generation of a construct containing only the C-terminal portion of dDYRK2, an EcoRI site was introduced immediately preceding the start of the C-terminal using the primers used for the N-terminus plus kinase domain construct. The EcoRI fragment encoding the C-terminus of dDYRK2 was then cloned in frame with the GAL4 binding domain of pGBKT7.
Yeast two-hybrid screen
Full-length and deletion mutants of dDYRK2 were cloned into the GAL4 binding domain vector, pGBKT7. Library DNA (Clontech MATCHMAKER Drosophila melanogaster Embryo cDNA library; catalogue no. 638839) was then mixed with pGBKT7/dDYRK2 plasmid DNA and used to transform competent AH109 yeast cells using a standard lithium acetate method (see the manufacturer's recommendations). After co-transformation, cells were collected by centrifugation at 1300 g for 5 min, washed in distilled water, and plated on to quadruple dropout (–Ade, –His, –Leu, –Trp) plates plus X-α-Gal (5-bromo-4-chloroindol-3-yl α-D-galactopyranoside), with or without 3-amino-1,2,4-triazole (5–25 mM; Sigma). Library plasmid DNA was isolated from positive yeast colonies using the YEASTMAKER kit (BD Biosciences/Clontech). Isolated DNA was then used to transform XL-10 gold ultracompetent Escherichia coli cells (Stratagene) for selection on LB/Amp (Luria–Bertani broth supplemented with 50 μg/ml ampicillin) plates to rescue library plasmids.
α-Gal quantification assay
The appropriate liquid SD (synthetic dropout; 3 ml) medium was inoculated with a fresh yeast colony expressing the pair of interacting proteins being analysed. The culture was incubated overnight at 30 °C with shaking at 250 rev./min. The culture (1 ml) was transferred to a microcentrifuge tube and centrifuged for 5 min at 16100 g, and the supernatant was removed for analysis. For the assay, 8 μl of cell-culture medium supernatant was added to 24 μl of Assay buffer {100 mM PNP-α-Gal (p-nitrophenyl α-D-galactopyranoside) solution and 1× NaOAc [1:2 (v/v) ratio]}. The reaction was incubated for 60 min at 30 °C before being stopped with 960 μl of 1× stop solution (0.1 M NaCO3). The absorbance at 410 nm was then recorded.
Baculovirus constructs
The generation of FLAG epitope-tagged WT- and kinase dead K227M-dDYRK2-expressing baculovirus and Sf9 cell (Spodoptera frugiperda 9 cell) cultures have been described previously [6]. For the generation of ΔCt dDYRK2, an EcoRI site was cloned into the dDYRK2 cDNA immediately preceding the C-terminus. The EcoRI fragment encoding the truncated dDYRK2 protein was then cloned into the pVL1393 baculoviral vector (BD Biosciences) for expression in Sf9 cells. ΔDH-dDYRK2 was generated by removing bases 562–582 (GACGATGATAATGGCAACTAC) of full-length dDYRK2 cDNA by site-directed mutagenesis using the QuikChange® site-directed mutagenesis kit (Stratagene). To generate HA (haemagglutinin)-tagged SNR1, a BglII site was introduced immediately upstream of the initiating ATG of the Snr1 cDNA (Invitrogen, clone GH08712) and an HA tag introduced immediately after the initiating ATG using the primer, 5′-accgtcccaccatcgggcgcggatcagatctcaacatgtacccatacgatgttccagattacgctgcactgcagacatacggggacaagc-3′. An EcoRI site was introduced immediately after the termination codon using the primer 5′-ggccaataccacaactggttggtgatcttcgaattccgccgatccagcaatgtgtcactaatgtaatc-3′ and the BglII/EcoRI fragment cloned into the pVL1392 baculoviral vector (BD Biosciences). To generate HA-tagged TRX-(3010–3759), a BamHI site was introduced immediately upstream of the initiating ATG of the TRX cDNA (Invitrogen, clone LD39445) and an HA tag was introduced immediately after the initiating ATG using the primer, 5′-gaaatcgcactactattggatccaacatgtacccatacgatgttccagattacgctggccgctccaagtttcccgg-3′. A NotI site was introduced immediately after the termination codon using the primer 5′-gtcggaagtacttaaattaataaaaggcggccgcgaagatcagtgtcagagataagaactaag-3′ and the BamHI/NotI fragment cloned into the pVL1393 baculoviral vector (BD Biosciences).
Immunoprecipitation and immunoblotting
Immunoprecipitation using anti-dDYRK2, anti-FLAG M2 (Sigma) or anti-HA (Sigma) antibodies, and immunoblotting using 1:5000 anti-FLAG, 1:1000 anti-HA, 1:2000 anti-4G10 pTyr (Cell Signaling Technology), 1:1000 anti-phosphothreonine–proline (Cell Signaling Technology), 1:500 anti-C18-INI1 [where INI1 is integrase interactor 1 (human SNR1 homologue)] (Santa Cruz Biotechnology) or 1:5000 anti-dDYRK2 antibodies were performed as described previously [6].
Expression and purification of GST (glutathione S-transferase)–SNR1 from bacterial cells
The T102A mutation of SNR1 was introduced by site-directed mutagenesis using the QuikChange® kit. To create GST–SNR1 fusion proteins, WT and T102A-SNR1 genes were directionally cloned into BglII/EcoRI sites of pGEX 4T-1 (Amersham Biosciences) and transformed into BL21(DE3) bacterial cells (Stratagene). To express proteins, bacteria were grown in 250 ml of LB/Amp cultures at 37 °C to an A600 (absorbance) of 0.4–0.6, GST–SNR1 proteins were induced by the addition of isopropyl β-D-thiogalactoside (Sigma) to a final concentration of 1 mM, and the bacteria were incubated for an additional 2 h. Cells were then collected by centrifugation, resuspended in 10 ml of cold PBS and lysed by sonication. Samples were clarified by centrifugation, the supernatant was transferred to a fresh tube, and dithiothreitol was added to a final concentration of 1 mM. The sample was then added to a 50% (v/v) glutathione–Sepharose slurry and incubated with rotation at room temperature (25 °C) for 20 min. Sepharose beads were pelleted, washed three times in ice-cold PBS, and bound proteins were eluted by incubation in glutathione elution buffer [10 mM GSH in 50 mM Tris/HCl (pH 8.0)] at room temperature for 10 min. The eluted protein samples were collected and stored at −70 °C.
SNR1 and MBP (myelin basic protein) kinase reactions
Immunoprecipitated FLAG-tagged constructs of WT- and K227M-dDYRK2 or WT- and K193M-MNB were incubated with either bacterially expressed GST–SNR1 or MBP in a total volume of 50 μl containing 0.1 mM [γ-32P]ATP (2×106 c.p.m./nmol) and 10 mM MgCl2 in buffer A (50 mM Tris/HCl, pH 7.5, and 1 mM EGTA) for 30 min at 30 °C. The reaction was stopped by adding 15 μl of NuPAGE sample buffer (Invitrogen) and run on a 4–12% (w/v) NuPAGE gel (Invitrogen). 32P incorporation was visualized by autoradiography.
Unlabelled kinase assay of SNR1
Immunoprecipitated FLAG-tagged WT- or K227M-dDYRK2 was incubated with 2 μg of GST–SNR1 in buffer A with 0.1 mM ATP and 10 mM MgCl2 for 30 min with agitation at 30 °C. Proteins were subjected to SDS/PAGE and transferred on to nitrocellulose. Membranes were incubated in 1:2000 anti-phosphothreonine–proline antibody (Cell Signaling Technology) overnight at 4 °C. Horseradish peroxidase-conjugated anti-mouse antibody was used to detect anti-phosphothreonine–proline and then visualized using the ECL® (enhanced chemiluminescence) system (Amersham).
Identification of phosphorylated residues
HA-tagged SNR1 was expressed on its own or was co-expressed with WT- or K227M-dDYRK2, or with WT- or K193M-MNB in Sf9 cells. SNR1 was immunoprecipitated using anti-HA agarose beads (Sigma), isolated by SDS/PAGE (10% NuPAGE gel) and stained using Simply Blue Safe Stain (Invitrogen). The excised proteins were alkylated with 100 mM iodoacetamide and digested with 5 μg/ml trypsin. The tryptic digests were dissolved in 100 μl of 1% formic acid/4 mM EDTA in water and 20 μl was analysed by LC-MS (liquid chromatography–MS) with precursor 79 scanning on a 4000 Q-TRAP system as described previously [6,25]. Extracted ion chromatograms from precursor ion scans were generated using Analyst 1.4.1 software (MDS-Sciex, Canada). The sequences of the peptides and sites of phosphorylation were confirmed by database searching using Mascot v2.1 (Matrixscience, U.K.) run on a local server as well as by manual interpretation of the tandem MS spectra.
RESULTS
Identification of SNR1 and TRX as dDYRK2-interacting proteins
To identify interacting proteins, the full-length dDyrk2 gene was used as bait by cloning into the yeast pGBKT7 vector to express a GAL4 binding domain–dDYRK2 fusion protein. A pre-made Clontech MATCHMAKER Drosophila Embryo cDNA library was used to screen for binding partners. This screen, conducted using the highest stringency possible, identified 14 positive clones representing eight distinct genes as encoding dDYRK2-interacting proteins. Three of the 14 clones were independently derived from the chromatin remodelling factor SNR1 and three from the essential chromatin factor TRX.
The Snr1 gene encodes a protein product of 370 amino acids. SNR1 is a conserved counterpart of the yeast SWI/SNF complex (multiprotein chromatin remodelling complex) involved in ATP-dependent chromatin remodelling [26]. The protein contains two repeat domains (1 and 2) implicated in mediating critical protein–protein interactions, and a coiled coil region. All three clones obtained from the screen were approximately the same size (the largest encoding amino acids 90–370 and the smallest encoding amino acids 94–370) and encode proteins that contain both repeat domains and the coiled coil region (Figure 1B). The trx gene encodes a 3759-amino-acid protein which contains a number of domains including PHD fingers (plant homeodomain fingers domain), FY-rich regions and a C-terminal SET domain [Su(var)3-9, Enhancer of Zeste, Trithorax domain] (Figure 1C). SET domains are reported to mediate protein–protein interactions and the C-terminal SET domain of TRX has been shown to interact with SNR1 [27]. All TRX clones isolated in this screen contain only the C-terminal portion of the molecule (residues 3389–3759), which includes an FY-rich region and the SET domain.
The C-terminus of dDYRK2 interacts with SNR1 and TRX
DYRK proteins contain non-catalytic N-terminal (Nt) and C-terminal (Ct) regions flanking a central kinase domain (Kin) (Figure 1A). To identify the region of the protein responsible for the association with SNR1 and TRX, various dDYRK2 deletion constructs were prepared and interactions were tested by co-transformation of yeast cells. As shown in Table 1, when the Nt, Kin or Ct region of dDYRK2 was co-expressed with SNR1 or TRX, only the Ct construct supported growth in yeast cells. A construct containing both the Nt and kinase domain (ΔCt) did support growth; however, these ΔCt co-transformants grew much more slowly than did the full-length WT- or Ct-dDYRK2 plus SNR1 (or TRX) co-transformants. These interactions were explored further using the α-Gal quantitative assay, which is a sensitive colorimetric assay for the detection and quantification of α-galactosidase activity [28]. This analysis demonstrated that the interaction between SNR1 and the C-terminus of dDYRK2 is equal in strength to that of the full-length molecule and much stronger than the interaction between SNR1 and the ΔCt construct (Figure 1D). Similar results were obtained when the interaction of dDYRK2 with TRX was investigated (results not shown). These results suggest that the C-terminus of dDYRK2 plays a key role in binding to SNR1 and TRX but that other regions of the DYRK molecule are also involved in the association.
Table 1. Analysis of the interactions of SNR1 with dDYRK2 deletion constructs.
AH109 yeast cells were co-transformed with Snr1 and WT-, Ct-, Nt-, Kin-, or Nt+Kin-dDYRK2 constructs. An interaction between proteins was assessed by plating samples on quadruple dropout media. Growth of colonies containing Nt+Kin dDYRK2 was much slower and blue colour appeared much later than those containing WT- or Ct-dDYRK2.
| dDYRK2 construct | Growth on selective media |
|---|---|
| Full-length DD2 Nt+Kin+Ct | Yes |
| Nt+Kin | Yes (slow growth) |
| Ct | Yes (fast growth) |
| Nt | No |
| Kin | No |
dDYRK2 interacts with SNR1 and TRX in vitro
To determine if SNR1 and TRX interact with dDYRK2 in an independent system, we conducted a series of co-immunoprecipitation studies. For these experiments, full-length SNR1 and the C-terminus of TRX (residues 3010–3759) were tagged with the HA epitope for expression in Sf9 cells. The proteins were then expressed alone or were co-expressed with WT or kinase-inactive (ATP-binding Lys227 altered to methionine) versions of dDYRK2. dDYRK2 proteins were then immunoprecipitated from cell lysates and immunocomplexes were probed for the presence of SNR1. As shown, SNR1 did co-precipitate with dDYRK2 (Figure 2A). An interaction between these two molecules was also observed when SNR1 immunoprecipitates were probed for the presence of dDYRK2 (Figure 2B). These interactions did not depend on dDYRK2 kinase activity as kinase-inactive K227M-dDYRK2 also co-immunoprecipitated with SNR1 (Figures 2A and 2B).
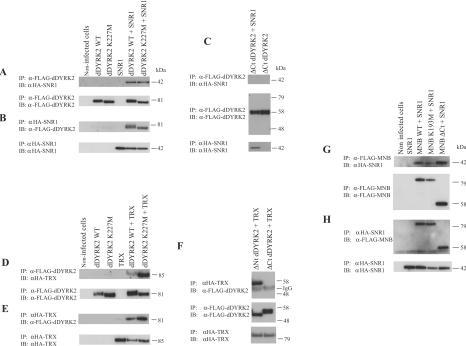
Figure 2. SNR1 and TRX co-immunoprecipitate with dDYRK2 and MNB.
FLAG-tagged WT-, kinase-inactive and truncated forms of dDYRK2 or MNB were expressed alone or were co-expressed with HA–SNR1 or HA–TRX (residues 3010–3759) in Sf9 cells. dDYRK2 or MNB proteins were then immunoprecipitated (IP) from cell lysates with FLAG antibody; or SNR1 or TRX proteins were immunoprecipitated with anti-HA (α-HA) antibody. Immunocomplexes were probed (immunoblotted; IB) for the presence of DYRK (dDYRK2/MNB), SNR1 or TRX as indicated. (A) dDYRK2 immunoprecipitates were probed for the presence of SNR1 (upper) or dDYRK2 (lower). (B) SNR1 immunoprecipitates were probed for the presence of dDYRK2 (upper) or SNR1 (lower). (C) ΔCt dDYRK2 immunoprecipitates were probed for the presence of SNR1 (top) or ΔCt dDYRK2 (middle). The presence of SNR1 in lysates was confirmed (bottom). (D) dDYRK2 immunoprecipitates were probed for presence of TRX (upper) or dDYRK2 (lower). (E) TRX immunoprecipitates were probed for the presence of dDYRK2 (upper) or TRX (lower). (F) ΔCt dDYRK2 immunoprecipitates were probed for the presence of TRX (top) or ΔCt dDYRK2 (middle). Presence of TRX in lysates was confirmed (bottom). (G) FLAG-tagged WT-, K193M- or ΔCt MNB immunoprecipitates were analysed for the presence of SNR1 (upper) or MNB (lower). (H) SNR1 immunoprecipitates were analysed for the presence of MNB (upper) or SNR1 (lower). Molecular masses in kDa and proteins visualized are shown on the right of the Figure. Results are representative of three replicate experiments.
A series of deletion mutants were used to localize the region of dDYRK2 responsible for the association with SNR1 in vitro. The DH-box, a small conserved region of charged amino acids thought to be involved in substrate recognition and/or protein binding [29], was included in our analysis. As above, dDYRK2 deletion proteins were expressed individually or were co-expressed with SNR1 or TRX, and an interaction between the two proteins was assessed by co-immunoprecipitation. Removal of any portion (including the DH-box) or the entire non-catalytic Nt of dDYRK2 did not eliminate the interaction with SNR1. However, consistent with evidence obtained from the yeast two-hybrid system, when the non-catalytic Ct was deleted, the dDYRK2 mutant protein failed to associate with SNR1 in co-immunoprecipitation reactions (Figure 2C). These same dDYRK2 constructs were then used to assess TRX binding. As observed for SNR1, TRX co-precipitated with WT- and kinase-inactive-dDYRK2 (Figure 2D) and dDYRK2 co-precipitated with TRX (Figure 2E). In addition, while removal of the N-terminus of dDYRK2 did not abolish the interaction, deletion of the C-terminus dramatically reduced the association with TRX (Figure 2F).
Next, we used co-immunoprecipitation experiments to ask if another DYRK family member, MNB, would also interact with SNR1 and TRX. WT- or kinase-inactive MNB (K193M-MNB) was expressed singly or co-expressed with HA–SNR1 (or HA–TRX) in Sf9 cells. SNR1, TRX or MNB proteins were then immunoprecipitated from cell lysates, fractionated by SDS/PAGE and subjected to immunoblot analysis. As shown, SNR1 co-precipitated with both WT and kinase-inactive MNB (Figure 2G), and both forms of the MNB protein co-precipitated with SNR1 (Figure 2H). However, in contrast with dDYRK2, both SNR1 and TRX co-precipitated with ΔCt MNB (Figures 2G and 2H and results not shown).
dDYRK2 and MNB phosphorylate SNR1 on Thr102
These studies were extended to determine if dDYRK2 or MNB could phosphorylate SNR1. For in vitro assays, full-length SNR1 was cloned into pGEX 4T-1 to create a GST–SNR1 fusion protein. WT or kinase-inactive dDYRK2 and MNB were then incubated with GST–SNR1 in the presence of [γ-32P]ATP. The samples were separated by SDS/PAGE and phosphorylated proteins were visualized by autoradiography. Both WT-dDYRK2 (Figure 3A) and WT-MNB (results not shown) were found to efficiently phosphorylate SNR1 compared with the kinase-inactive mutants.
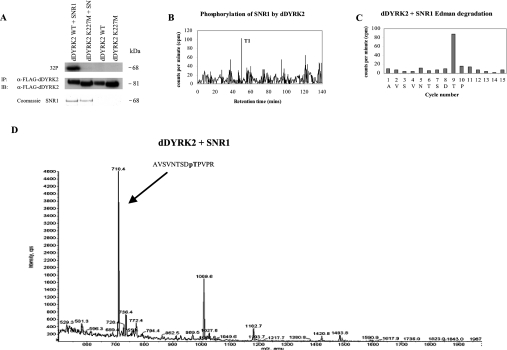
Figure 3. dDYRK2 and MNB phosphorylate SNR1 on Thr102.
(A) WT- or K227M-dDYRK2 immunoprecipitates as described in the text were incubated for 30 min at 30 °C with or without recombinant GST–SNR1 in the presence of [γ-32P]ATP, as indicated. Following SDS/PAGE, radiolabelled SNR1 was visualized by autoradiography (top panel). The blot was then probed with anti-FLAG antibody to visualize dDYRK2 (middle panel). GST–SNR1 levels were monitored by Coomassie stain (bottom panel). IP, immunoprecipitation; IB, immunoblotting. (B) GST–SNR1 was radiolabelled with WT-dDYRK2 as described in the text. SNR1 was then isolated, digested with trypsin, and the resulting phosphopeptides were separated on a Vydac C18 column. 32P radioactivity present in each fraction was analysed by Cerenkov counting. The position of the major phosphopeptide T1 is shown. (C) The T1 phosphopeptide was analysed by N-terminal Edman degradation and the 32P radioactivity released at each cycle is shown (filled bars). Amino acids corresponding to the putative SNR1 peptide sequence are displayed. (D) SNR1 was phosphorylated by WT-dDYRK2 using unlabelled ATP as described in the Materials and methods section. SNR1 was then isolated, treated with trypsin, and tryptic phosphopeptides were separated and analysed on a 4000 Q-Trap mass spectrometer. Using the electrospray ionization MS in precursor ion scanning mode, the precursors of scans obtained over the 45 min peptide separation were summed and the major phosphopeptide ions were annotated. The arrow shows the position and amino acid sequence of the major SNR1 phosphopeptide isolated (pT, phosphothreonine).
To identify individual sites of phosphorylation, SNR1 was first radiolabelled in the presence of dDYRK2 or MNB, then digested with trypsin, and the resulting tryptic phosphopeptides were separated by HPLC. When the amount of radioactivity was determined for each fraction, one significant peak eluting at approx. 49 min was observed for WT-SNR1, following 30 min incubation at 30 °C with WT-dDYRK2 (Figure 3B) or WT-MNB (results not shown), suggesting that both DYRKs phosphorylate SNR1 at a single site in vitro. The peak HPLC fraction was then analysed by N-terminal Edman degradation. Significant counts were released from dDYRK2- and MNB-radiolabelled SNR1 only after 9 cycles of N-terminal degradation (Figure 3C). Previous studies have shown that a proline residue in the +1 position to the DYRK phosphorylation site is important for optimal substrate recognition [2,19]. Three tryptic peptides from SNR1 contain threonine residues followed by a proline residue, Thr102, Thr133 and Thr198. The only tryptic peptide to contain one of these at position 9 is the one containing Thr102, AVSVNTSDTPVPR, suggesting that Thr102 may be the site phosphorylated on SNR1 by dDYRK2 and MNB.
We then used MS analysis with precursor ion scanning to monitor SNR1 phosphorylation in vivo. WT-dDYRK2 or MNB was co-expressed with HA–SNR1 in Sf9 cells. SNR1 was immunoprecipitated with anti-HA antibody, fractionated by SDS/PAGE, and the isolated protein was digested with trypsin. The digests were then analysed by LC-MS with precursor ion scanning [25] using a 4000 QTRAP mass spectrometer. The fragmentation and analysis of the tryptic phosphopeptides revealed that Thr102 of SNR1 was the major site of phosphorylation by dDYRK2 (Figure 3D) and by MNB (results not shown).
The DH-box of dDYRK2 is essential for SNR1 phosphorylation
Phospho-specific antibodies can provide a powerful reagent for monitoring the phosphorylation state of individual sites. As SNR1 is phosphorylated on a Thr-Pro motif, we tested a commercially available antibody that reacts with phosphorylated threonine followed by proline (phospho-T-P) to determine if the antibody could be used to monitor SNR1 phosphorylation. Site-directed mutagenesis was used to mutate SNR1-Thr102 to non-phosphorylatable alanine (T102A). GST–WT-SNR1 or GST–T102A-SNR1 was then incubated with dDYRK2 or MNB proteins in the presence of unlabelled ATP for 30 min at 30 °C. The proteins were resolved by SDS/PAGE and immunoblotted using the phospho-T-P antibody. SNR1 was efficiently phosphorylated on threonine residue when incubated for 30 min at 30 °C with WT-MNB (Figure 4A) or dDYRK2 (results not shown) but was not phosphorylated when incubated as above with kinase-inactive mutants. As expected, the phospho-T-P signal was lost when SNR1-Thr102 was altered to alanine. These results are consistent with the phosphorylation of Thr102 of SNR1 by dDYRK2 and MNB, and demonstrate that the phospho-T-P antibody can be used to monitor this event.
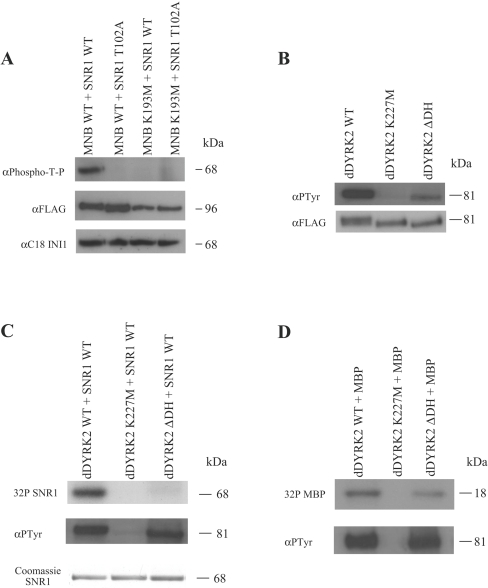
Figure 4. The DH-box of dDYRK2 is critical for SNR1 phosphorylation in vitro.
(A) WT- or K193M-MNB immunoprecipitates (described in the text) were incubated with recombinant WT- or T102A-GST–SNR1 in the presence of unlabelled ATP for 30 min at 30 °C. Samples were immunoblotted and probed for the presence of phospho-Thr102-SNR1 (α-phospho-T-P), MNB (α-FLAG) or SNR1 (α-C18-INI1) as indicated. (B) As above except, WT-, K227M- or ΔDH-dDYRK2 immunoprecipitates were probed for the presence of tyrosine-phosphorylated dDYRK2 (α-pTyr) or total dDYRK2 (α-FLAG). (C) Immunoprecipitates containing WT-, K227M- or ΔDH-dDYRK2 proteins were incubated with recombinant GST-SNR1 in the presence of [γ-32P]ATP for 30 min at 30 °C. Following SDS/PAGE, SNR1 phosphorylation was visualized by autoradiography (top), or dDYRK2 tyrosine autophosphorylation was visualized by probing with anti-pTyr antibody (middle). GST–SNR1 was visualized by Coomassie stain (bottom). (D) Immunoprecipitates of WT-, K227M- or ΔDH-dDYRK2 were incubated with MBP in the presence of [γ-32P]ATP for 30 min at 30 °C, and analysed as above. Phosphorylated MBP was visualized by autoradiography (upper panel) and dDYRK2 tyrosine autophosphorylation by anti-pTyr antibody (lower panel). Results are representative of two to three replicate experiments.
We next asked if the DYRK DH-box was important for SNR1 phosphorylation. As dDYRK2 and MNB serine/threonine kinase activity requires prior autophosphorylation of a tyrosine residue in the activation loop of the molecule [6], we assessed tyrosine phosphorylation levels of Sf9-expressed WT-, kinase-inactive- and ΔDH-versions of dDYRK2 and MNB. As shown by anti-pTyr immunoblot analysis, removal of the DH-box from dDYRK2 reduced autophosphorylation of the mutant protein when compared with the WT molecule (Figure 4B). Removal of the DH-box from MNB affected tyrosine autophosphorylation to a far lesser extent (results not shown). WT and mutant forms of dDYRK2 and MNB were then incubated for 30 min at 30 °C with GST–WT-SNR1 in the presence of [γ-32P]ATP. Autoradiography of samples demonstrated that while WT-dDYRK2 efficiently phosphorylated SNR1, removal of the DH-box of dDYRK2 almost completely abolished in vitro phosphorylation even after normalization of tyrosine autophosphorylation levels (Figure 4C). This is not a general property of dDYRK2 substrate phosphorylation as evidenced by differences in MBP (Figure 4D) and dFOXO (where FOXO is forkhead box O) (R. Kinstrie and V. Cleghon, unpublished work) phosphorylation where removal of the DH-box has much less effect. In contrast, removal of the DH-box from MNB had little effect on SNR1 phosphorylation.
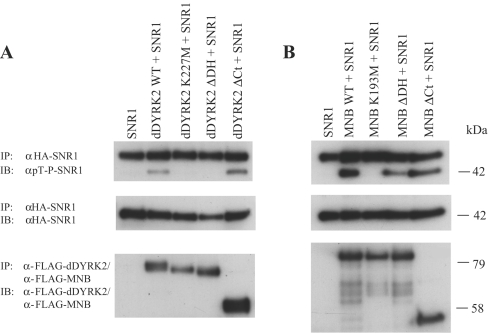
To determine if dDYRK2 or MNB was able to phosphorylate SNR1 in intact cells, HA–SNR1 was expressed on its own or was co-expressed with various WT or mutant DYRK proteins in Sf9 cells. Using the phospho-T-P antibody to monitor SNR1-Thr102 phosphorylation, we found that SNR1 was efficiently phosphorylated in vivo only when co-expressed with WT-dDYRK2 (Figure 5A) or WT-MNB (Figure 5B). However, bacterially expressed SNR1, SNR1 expressed on its own in Sf9 cells or coexpressed with kinase-inactive versions of dDYRK2 or MNB was not phosphorylated. The requirement for the C-terminus (Ct) of dDYRK2 in this phosphorylation event was investigated by co-expressing SNR1 with ΔCt-dDYRK2 in Sf9 cells and monitoring the phosphorylation of Thr102. This analysis revealed that SNR1 was phosphorylated on Thr102 when co-expressed with ΔCt-dDYRK2 (Figure 5A). The importance of the DH-box was assessed by co-expression of SNR1 with ΔDH-dDYRK2 or ΔDH-MNB in Sf9 cells and immunoblot analysis using the anti-phospho-T-P antibody. As seen with in vitro phosphorylation, removal of the DH-box greatly reduced dDYRK2-mediated phosphorylation of SNR1 (Figure 5A) but affected MNB phosphorylation to a much lesser extent (Figure 5B). Taken together, these results suggest that dDYRK2 and MNB can phosphorylate SNR1 on Thr102 in vitro or in an intact cell.
Figure 5. dDYRK2 and MNB phosphorylate SNR1 in Sf9 cells.
HA–SNR1 was expressed alone in Sf9 cells or was co-expressed with various FLAG-tagged versions of dDYRK2 (A) or MNB (B), as described in Figure 2. HA–SNR1 immunoprecipitates were then probed for the presence of phospho-Thr102 (α-phospho-T-P; top panel), or SNR1 (α-HA, middle panel). The presence of DYRK proteins was confirmed (α-FLAG; bottom panel). (A) HA–SNR1 co-expressed with WT, K227M-, ΔDH- or ΔC-dDYRK2. IP, immunoprecipitation; IB, immunoblotting. (B) HA–SNR1 co-expressed with WT-, K193M-, ΔDH- or ΔC-MNB. Results are representative of three replicate experiments.
DISCUSSION
Using a yeast two-hybrid screen and co-immunoprecipitation studies, we identified the chromatin remodelling factors SNR1 and TRX as dDYRK2- and MNB-interacting proteins. SNR1, an essential subunit of the Drosophila Brm (Brahma) ATP-dependent chromatin remodelling complex, has counterparts in yeast (SNF5) and in mammals (INI1) [23]. The mammalian homologue, INI1, is a tumour suppressor gene mutated in most of the AT/RT (atypical teratoid and malignant rhabdoid tumours) [30]. TRX, a highly conserved chromatin component, is part of the TRX group of epigenetic regulators that together with Polycomb group genes act to maintain homeotic gene expression. TRX is the fly homologue of the ALL-1 (human Trithorax homologue) or hrx (human TRX) gene which is involved in acute leukaemia, in particular infant and secondary leukaemia [31]. SNR1 and TRX have been shown to interact physically and appear to co-operate in establishing open and repressive chromatin states to activate/inactivate transcription [24]. The regions of these proteins involved in their interaction, repeat region 2 of SNR1 and the SET domain of TRX, are contained in the clones identified in our screen. In addition, these same regions are highly conserved in their mammalian counterparts, suggesting that INI1 and HRX/ALL-1 may also interact with mammalian DYRKs.
The association lead us to ask if SNR1 or TRX was also a DYRK substrate. Studies have proposed a consensus recognition sequence for phosphorylation by DYRKs of R-X(X)-S/T-P [2,19], and recent work on C. elegans MBK-2 demonstrates that this sequence is useful in identifying sites on a suspected substrate [14]. However, more than one-third of reported DYRK substrates (i.e. STAT3, p27Kip1, p21Cip1, cyclin D1 and splicing factor), including SNR1 (the present study), do not contain the R-X(X)-S/T-P motif [4,15,32]. The site phosphorylated, Thr102, lies in a Thr-Pro motif (similar to other exceptions), consistent with reports of the importance of a proline residue in the +1 position [2,19]. Although SNR1 does not contain a consensus recognition sequence, it was phosphorylated by dDYRK2 and MNB as well or better than any DYRK family substrate we have tested. This is the first instance of SNR1 or any of its mammalian homologues being shown to be a phosphoprotein [33]. Owing to the size of the protein (3759 amino acids), we were unable to determine whether TRX was a DYRK substrate at this time. However, TRX contains 38 Ser/Thr-Pro motifs (most within the N-terminal third of the molecule) including two consensus DYRK recognition sequences (Ser378 and Ser552), and interacts with DYRK proteins, suggesting that TRX should also be considered a potential substrate.
The ability of dDYRK2 and MNB to associate with and phosphorylate SNR1 indicates that the proteins co-localize within the cell. SNR1 localizes predominantly in the nucleus of cells [34]. Class 1 DYRKs (i.e. MNB/DYRK1A) contain a nuclear localization sequence and have been observed to be predominantly nuclear as well [35–37], whereas class 2 DYRKs (i.e. dDYRK2) appear to be mainly cytosolic [6,36]. There are two extant explanations for this seeming anomaly: re-localization of the kinase and/or re-localization of the substrate. Both class 1 and 2 DYRKs have been shown to alter their ‘normal’ subcellular distribution [38,39], and in the case of class 2 DYRKs the change in intracellular localization in response to environmental or developmental signals allowed phosphorylation of a substrate in a time- and location-specific manner [13–15,40].
Re-localization of the kinase need not be invoked as a mechanism. In a recent study, both class 1 and 2 DYRKs co-operated in maintaining an appropriate NFAT (nuclear factor of activated T-cells)-mediated response by phosphorylating and thereby altering the cytoplasmic/nuclear distribution of the NFAT signalling molecule [20]. Consistent with this premise, SNR1 contains a conserved NES (nuclear export signal) within its repeat 2 region, indicating that the protein spends time outside the nucleus. Furthermore, a mutant form of SNR1 that removes a portion of the protein immediately adjacent to the NES was found at increased levels in the cytoplasm [34]. Thr102, the residue identified in the present study, lies just outside of the NES region and it is possible that DYRKs regulate the nuclear/cytoplasmic distribution of SNR1. While the NES region is conserved in the mammalian counterpart, Thr102 is not. At this time, we do not know if phosphorylation is specific to Drosophila SNR1, or like the circadian rhythm protein PER (Drosophila PERIOD), the same family of kinases phosphorylate the fly and mammalian protein, but the residues phosphorylated are in different regions of the molecule [41].
The present study demonstrates several shared properties between dDYRK2 and MNB (i.e. both associate with TRX and SNR1 and phosphorylate SNR1), but it also provides evidence for fundamental structural/functional differences between the molecules. First, an interaction with SNR1 or TRX is mediated in large part through the C-terminus of dDYRK2, whereas MNB does not require this portion of the molecule. Secondly, loss of the DH-box from MNB had little effect on SNR1 phosphorylation, but removal of this conserved region from dDYRK2 abolished phosphorylation of SNR1, in vitro and in vivo (see Figure 4C). Interestingly, loss of the DH-box did not appear to reduce significantly dDYRK2 phosphorylation of other substrates such as dFOXO (R. Kinstrie and V. Cleghon, unpublished work) or MBP. The DH-box, a highly conserved region found in the N-terminus of the DYRK family, has been suggested to play a role in substrate binding [29], but the present study is the first to provide experimental evidence for a functional role for this conserved region.
At this time, we can only speculate on the possible functional role for the association of DYRKs with SNR1 and TRX. All three proteins are implicated in regulating permanent developmental events and in cell-cycle progression, and indeed chromatin is remodelled in both a developmental and cell-cycle-dependent manner [33,42]. SNR1 genetically interacts with a number of cell-cycle control genes including HDACs, DmcycE (D. melanogaster cyclin E) and CDK2 (cyclin-dependent kinase 2) [33], and human INI1 exerts tumour suppression function through the direct recruitment of HDAC activity to the cyclin D1 promoter, resulting in G0/G1 arrest [43]. SNR1 and TRX also interact with DmcycE to regulate negatively the G1 to S transition [44]. DYRK family members Yak1p, Pom1 (S. pombe DYRK family member encoding POM1 protein) and MIRK/DYRK1B are all active G0/G1 kinases capable of arresting cell growth in the G1 phase of the cell cycle [1,8,11], and have been shown to interact with many of the same cell-cycle control genes as SNR1 and TRX, including phosphorylation of HDACs and cyclin D1 [16–18,45]. With regard to phosphorylation, TRX has been shown to interact with the type 1 serine/threonine phosphatase PP1β9C (protein phosphatase 1β9C) [46] and with the antiphosphatase Sbf1 (SET binding factor 1) [47] on polytene chromosomes. Phosphorylation has also been suggested as a control mechanism for components of the mammalian Brm complexes for the onset of mitosis [48,49]. In addition, yeast SFH1p, a close relative of SNR1, appears to be phosphorylated during the G1 phase [50]. These observations suggest that cell-cycle chromatin remodelling may be regulated in part by phosphorylation. Our results suggest that the DYRK family of kinases may play a role in this process.
Acknowledgments
We thank J. Wyke, D. Gillespie and A. Dhillon at The Beatson Institute for Cancer Research (Glasgow, Scotland, U.K.) for critically reading this paper. We also thank M. Rylatt and T. Rawjee (Beatson Institute for Cancer Research) for useful discussions, and A. Dingwall (Cardinal Bernardin Cancer Center, Loyola University of Chicago) for reagents and helpful comments. This work was funded by Cancer Research U.K.
References
- 1.Bahler J., Nurse P. Fission yeast Pom1p kinase activity is cell cycle regulated and essential for cellular symmetry during growth and division. EMBO J. 2001;20:1064–1073. doi: 10.1093/emboj/20.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell L. E., Proud C. G. Differing substrate specificities of members of the DYRK family of arginine-directed protein kinases. FEBS Lett. 2002;510:31–36. doi: 10.1016/s0014-5793(01)03221-5. [DOI] [PubMed] [Google Scholar]
- 3.Galceran J., de Graaf K., Tejedor F. J., Becker W. The MNB/DYRK1A protein kinase: genetic and biochemical properties. J. Neural. Transm. Suppl. 2003;67:139–148. doi: 10.1007/978-3-7091-6721-2_12. [DOI] [PubMed] [Google Scholar]
- 4.Mercer S. E., Ewton D. Z., Deng X., Lim S., Mazur T. R., Friedman E. Mirk/Dyrk1B mediates survival during the differentiation of C2C12 myoblasts. J. Biol. Chem. 2005;280:25788–25801. doi: 10.1074/jbc.M413594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lochhead P. A., Sibbet G., Kinstrie R., Cleghon T., Rylatt M., Morrison D. K., Cleghon V. dDYRK2: a novel dual-specificity tyrosine-phosphorylation-regulated kinase in Drosophila. Biochem. J. 2003;374:381–391. doi: 10.1042/BJ20030500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lochhead P. A., Sibbet G., Morrice N., Cleghon V. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell. 2005;121:925–936. doi: 10.1016/j.cell.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 7.Smith A., Ward M. P., Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Souza G. M., Lu S., Kuspa A. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development. 1998;125:2291–2302. doi: 10.1242/dev.125.12.2291. [DOI] [PubMed] [Google Scholar]
- 9.Tejedor F., Zhu X. R., Kaltenbach E., Ackermann A., Baumann A., Canal I., Heisenberg M., Fischbach K. F., Pongs O. Minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14:287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- 10.Fotaki V., Dierssen M., Alcantara S., Martinez S., Marti E., Casas C., Visa J., Soriano E., Estivill X., Arbones M. L. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol. Cell. Biol. 2002;22:6636–6647. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng X., Ewton D. Z., Pawlikowski B., Maimone M., Friedman E. Mirk/dyrk1B is a Rho-induced kinase active in skeletal muscle differentiation. J. Biol. Chem. 2003;278:41347–41354. doi: 10.1074/jbc.M306780200. [DOI] [PubMed] [Google Scholar]
- 12.Shirayama M., Soto M. C., Ishidate T., Kim S., Nakamura K., Bei Y., van den Heuvel S., Mello C. C. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr. Biol. 2006;16:47–55. doi: 10.1016/j.cub.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 13.Stitzel M. L., Pellettieri J., Seydoux G. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr. Biol. 2006;16:56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 14.Nishi Y., Lin R. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans. Dev. Biol. 2005;288:139–149. doi: 10.1016/j.ydbio.2005.09.053. [DOI] [PubMed] [Google Scholar]
- 15.de Graaf K., Czajkowska H., Rottmann S., Packman L. C., Lilischkis R., Luscher B., Becker W. The protein kinase DYRK1A phosphorylates the splicing factor SF3b1/SAP155 at Thr434, a novel in vivo phosphorylation site. BMC Biochem. 2006;7:7. doi: 10.1186/1471-2091-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X., Mercer S. E., Shah S., Ewton D. Z., Friedman E. The cyclin-dependent kinase inhibitor p27Kip1 is stabilized in G(0) by Mirk/dyrk1B kinase. J. Biol. Chem. 2004;279:22498–22504. doi: 10.1074/jbc.M400479200. [DOI] [PubMed] [Google Scholar]
- 17.Zou Y., Ewton D. Z., Deng X., Mercer S. E., Friedman E. Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288. J. Biol. Chem. 2004;279:27790–27798. doi: 10.1074/jbc.M403042200. [DOI] [PubMed] [Google Scholar]
- 18.Deng X., Ewton D. Z., Mercer S. E., Friedman E. Mirk/dyrk1B decreases the nuclear accumulation of class II histone deacetylases during skeletal muscle differentiation. J. Biol. Chem. 2005;280:4894–4905. doi: 10.1074/jbc.M411894200. [DOI] [PubMed] [Google Scholar]
- 19.Himpel S., Tegge W., Frank R., Leder S., Joost H. G., Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 20.Gwack Y., Sharma S., Nardone J., Tanasa B., Iuga A., Srikanth S., Okamura H., Bolton D., Feske S., Hogan P. G., Rao A. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature (London) 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- 21.Bescond M., Rahmani Z. Dual-specificity tyrosine-phosphorylated and regulated kinase 1A (DYRK1A) interacts with the phytanoyl-CoA alpha-hydroxylase associated protein 1 (PAHX-AP1), a brain specific protein. Int. J. Biochem. Cell Biol. 2005;37:775–783. doi: 10.1016/j.biocel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Sitz J. H., Tigges M., Baumgartel K., Khaspekov L. G., Lutz B. Dyrk1A potentiates steroid hormone-induced transcription via the chromatin remodeling factor Arip4. Mol. Cell. Biol. 2004;24:5821–5834. doi: 10.1128/MCB.24.13.5821-5834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim S., Jin K., Friedman E. Mirk protein kinase is activated by MKK3 and functions as a transcriptional activator of HNF1alpha. J. Biol. Chem. 2002;277:25040–25046. doi: 10.1074/jbc.M203257200. [DOI] [PubMed] [Google Scholar]
- 24.Lim S., Zou Y., Friedman E. The transcriptional activator Mirk/Dyrk1B is sequestered by p38alpha/beta MAP kinase. J. Biol. Chem. 2002;277:49438–49445. doi: 10.1074/jbc.M206840200. [DOI] [PubMed] [Google Scholar]
- 25.Williamson B. L., Marchese J., Morrice N. A. Automated identification and quantification of protein phosphorylation sites by LC/MS on a hybrid triple quadrupole linear ion trap mass spectrometer. Mol. Cell Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Dingwall A. K., Beek S. J., McCallum C. M., Tamkun J. W., Kalpana G. V., Goff S. P., Scott M. P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozenblatt-Rosen O., Rozovskaia T., Burakov D., Sedkov Y., Tillib S., Blechman J., Nakamura T., Croce C. M., Mazo A., Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aho S., Arffman A., Pummi T., Uitto J. A novel reporter gene MEL1 for the yeast two-hybrid system. Anal. Biochem. 1997;253:270–272. doi: 10.1006/abio.1997.2394. [DOI] [PubMed] [Google Scholar]
- 29.Becker W., Joost H. G. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- 30.Biegel J. A., Zhou J. Y., Rorke L. B., Stenstrom C., Wainwright L. M., Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 31.Tkachuk D. C., Kohler S., Cleary M. L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo R., Ochiai W., Nakashima K., Taga T. A new expression cloning strategy for isolation of substrate-specific kinases by using phosphorylation site-specific antibody. J. Immunol. Methods. 2001;247:141–151. doi: 10.1016/s0022-1759(00)00313-6. [DOI] [PubMed] [Google Scholar]
- 33.Zraly C. B., Marenda D. R., Dingwall A. K. SNR1 (INI1/SNF5) mediates important cell growth functions of the Drosophila Brahma (SWI/SNF) chromatin remodeling complex. Genetics. 2004;168:199–214. doi: 10.1534/genetics.104.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zraly C. B., Marenda D. R., Nanchal R., Cavalli G., Muchardt C., Dingwall A. K. SNR1 is an essential subunit in a subset of Drosophila brm complexes, targeting specific functions during development. Dev. Biol. 2003;253:291–308. doi: 10.1016/s0012-1606(02)00011-8. [DOI] [PubMed] [Google Scholar]
- 35.Hammerle B., Carnicero A., Elizalde C., Ceron J., Martinez S., Tejedor F. J. Expression patterns and subcellular localization of the Down syndrome candidate protein MNB/DYRK1A suggest a role in late neuronal differentiation. Eur. J. Neurosci. 2003;17:2277–2286. doi: 10.1046/j.1460-9568.2003.02665.x. [DOI] [PubMed] [Google Scholar]
- 36.Becker W., Weber Y., Wetzel K., Eirmbter K., Tejedor F. J., Joost H. G. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J. Biol. Chem. 1998;273:25893–25902. doi: 10.1074/jbc.273.40.25893. [DOI] [PubMed] [Google Scholar]
- 37.Alvarez M., Estivill X., de la Luna S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 2003;116:3099–3107. doi: 10.1242/jcs.00618. [DOI] [PubMed] [Google Scholar]
- 38.Wegiel J., Kuchna I., Nowicki K., Frackowiak J., Dowjat K., Silverman W. P., Reisberg B., DeLeon M., Wisniewski T., Adayev T., et al. Cell type- and brain structure-specific patterns of distribution of minibrain kinase in human brain. Brain Res. 2004;1010:69–80. doi: 10.1016/j.brainres.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Chen-Hwang M. C., Chen H. R., Elzinga M., Hwang Y. W. Dynamin is a minibrain kinase/dual specificity Yak1-related kinase 1A substrate. J. Biol. Chem. 2002;277:17597–17604. doi: 10.1074/jbc.M111101200. [DOI] [PubMed] [Google Scholar]
- 40.Moriya H., Shimizu-Yoshida Y., Omori A., Iwashita S., Katoh M., Sakai A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 2001;15:1217–1228. doi: 10.1101/gad.884001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J.-M., Schroeder A., Allada R. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J. Neurosci. 2005;25:11175–11183. doi: 10.1523/JNEUROSCI.2159-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H. S., Gavin M., Dahiya A., Postigo A. A., Ma D., Luo R. X., Harbour J. W., Dean D. C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z. K., Davies K. P., Allen J., Zhu L., Pestell R. G., Zagzag D., Kalpana G. V. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol. Cell. Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brumby A. M., Zraly C. B., Horsfield J. A., Secombe J., Saint R., Dingwall A. K., Richardson H. Drosophila cyclin E interacts with components of the Brahma complex. EMBO J. 2002;21:3377–3389. doi: 10.1093/emboj/cdf334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanyon C. A., Liu G., Mangiola B. A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R. L., Jr A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudenko A., Bennett D., Alphey L. PP1beta9C interacts with trithorax in Drosophila wing development. Dev. Dyn. 2004;231:336–341. doi: 10.1002/dvdy.20146. [DOI] [PubMed] [Google Scholar]
- 47.Petruk S., Sedkov Y., Smith S., Tillib S., Kraevski V., Nakamura T., Canaani E., Croce C. M., Mazo A. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 48.Muchardt C., Reyes J. C., Bourachot B., Leguoy E., Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 49.Sif S., Stukenberg P. T., Kirschner M. W., Kingston R. E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao Y., Cairns B. R., Kornberg R. D., Laurent B. C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol. Cell. Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]