Abstract
Cellobiose phosphorylase, a member of the glycoside hydrolase family 94, catalyses the reversible phosphorolysis of cellobiose into α-D-glucose 1-phosphate and D-glucose with inversion of the anomeric configuration. The substrate specificity and reaction mechanism of cellobiose phosphorylase from Cellvibrio gilvus have been investigated in detail. We have determined the crystal structure of the glucose-sulphate and glucose-phosphate complexes of this enzyme at a maximal resolution of 2.0 Å (1 Å=0.1 nm). The phosphate ion is strongly held through several hydrogen bonds, and the configuration appears to be suitable for direct nucleophilic attack to an anomeric centre. Structural features around the sugar-donor and sugar-acceptor sites were consistent with the results of extensive kinetic studies. When we compared this structure with that of homologous chitobiose phosphorylase, we identified key residues for substrate discrimination between glucose and N-acetylglucosamine in both the sugar-donor and sugar-acceptor sites. We found that the active site pocket of cellobiose phosphorylase was covered by an additional loop, indicating that some conformational change is required upon substrate binding. Information on the three-dimensional structure of cellobiose phosphorylase will facilitate engineering of this enzyme, the application of which to practical oligosaccharide synthesis has already been established.
Keywords: carbohydrate-active enzyme, cellobiose phosphorylase, chitobiose phosphorylase, glycoside hydrolase family 94, substrate specificity, X-ray crystallography
Abbreviations: CBP, cellobiose phosphorylase; CDP, cellodextrin phosphorylase; CgCBP, CBP from Cellvibrio gilvus; CgCBP–SO4, CgCBP structure complexed with sulphate; CgCBP–PO4, CgCBP structure complexed with phosphate; ChBP, chitobiose phosphorylase; CuCBP, CBP from Cellulomonas uda; GH, glycoside hydrolase; Glc 1-P, glucose 1-phosphate; GT, glycosyl transferase; rmsd, rootmean square deviation; VpChBP, ChBP from Vibrio proteolyticus
INTRODUCTION
The enzymes involved in oligo- and poly-saccharide degradation and synthesis can be categorized into two major classes of Carbohydrate-Active enZymes, GH (glycoside hydrolases) and GT (glycosyl transferases) [1] (see CAZy website at http://www.cazy.org/CAZY). Enzymes of a third class, phosphorylases, catalyse phosphorolysis at glycosidic bonds, generating glycosylphosphates. This reaction is reversible, since the energy of a glycosyl–phosphate bond is not as high as that of a glycosyl–nucleotide bond, which acts as the sugar-donor for general GTs. Therefore, phosphorylases can be employed for both the synthesis and degradation of sugar chains.
Phosphorylases are found in both the GT and GH classes. GT-type phosphorylases have a similar fold and catalytic mechanism as GT enzymes. For example, glycogen phosphorylase (GT-35) has a GT-B fold [2–5] and also exhibits a strong resemblance in its active site to typical GT enzymes, such as OtsA (GT-20) [6] and glycogen synthase (GT-5) [7]. GH-type phosphorylases, such as GH-13, GH-65 and GH-94, have structures and catalytic mechanisms similar to those of GH enzymes. For example, maltose phosphorylase (GH-65) and ChBP [chitobiose (GlcNAcβ1–4GlcNAc) phosphorylase; GH-94] share a glucoamylase-like (α/α)6-barrel fold [8,9], and their proposed reaction mechanisms are similar to those of hydrolytic enzymes with the exception of molecules attacking the C1 atom (the hydroxyl ion in hydrolytic enzymes; the phosphate in phosphorylases).
CBP (cellobiose phosphorylase) catalyses the phosphorolysis of cellobiose into Glc 1-P (α-D-glucose 1-phosphate) and D-glucose, with inversion of the anomeric configuration. CBP also possesses cellobiose synthesis activity, using Glc 1-P as its sugar-donor and glucose as its sugar-acceptor. CBP has been classified into the GH-94 family, along with homologous ChBP, CDP (cello-dextrin phosphorylase), and cyclic-1,2-glucan synthase. Since none of these showed hydrolytic activity, GH-94 enzymes were formerly regarded as GT enzymes (GT-36). However, determination of the three-dimensional structure of VpChBP (chitobiose phosphorylase from Vibrio proteolyticus) revealed that it bore structural and mechanistic similarity to GH-15 glucoamylase and GH-65 maltose phosphorylase [8,10]. These findings led to the reclassification of GT-36 into a novel GH family, GH-94.
CgCBP (CBP from Cellvibrio gilvus) is the most studied enzyme in the GH-94 family [11–17]. CgCBP consists of 822 amino acid residues with a 33% identity with the amino acid structure of VpChBP. Its substrate specificity, especially its sugar-acceptor specificity in synthetic reactions, has been investigated in detail. Knowledge of the characteristics of this enzyme enabled us to synthesize several hetero-oligosaccharides {e.g. the branched trisaccharides, [Glcβ1–4(Glcα1–6)Glc)]} [17]. To better understand the function of CgCBP in practical oligosaccharide synthesis, it is necessary to determine its three-dimensional structure. In the present paper, we report the three-dimensional structure of two sugar–anion complexes (glucose–sulphate and glucose–phosphate) of CgCBP. The latter is the first such determination of a sugar–phosphate complex structure for an inverting phosphorylase. These structures have enabled us to examine the structural and functional differences between CgCBP and VpChBP.
MATERIALS AND METHODS
Sample preparation, crystallization and data collection
Production and purification of the CgCBP protein were performed as described in [18]. CgCBP crystals were obtained at 25 °C using the sitting-drop vapour-diffusion method by mixing 2 μl of a protein solution (10 mg/ml) with 2 μl of a reservoir solution consisting of 1.5 M ammonium sulphate, 0.1 M Mes/NaOH (pH 6.5) and 5 mM glucose (for CgCBP–SO4), or 0.8 M sodium/potassium phosphate (pH 8.2) and 10 mM glucose at 4 °C (for CgCBP–PO4). The crystals were transferred to a reservoir solution containing glycerol [20% (v/v) for CgCBP–SO4, and 40% (v/v) for CgCBP–PO4] and flash-cooled in a stream of cold nitrogen gas at 100 K. X-ray-diffraction data sets were collected using synchrotron radiation [beam line BL-5A and NW12A, Photon Factory, High Energy Accelerator Research Organization (KEK), Tsukuba, Japan], and these data sets were processed and scaled using HKL2000 [19]. Data collection and processing statistics are shown in Table 1.
Table 1. Crystallographic data collection and refinement statistics.
| Crystal | ||
|---|---|---|
| Parameter | CgCBP–SO4 | CgCBP–PO4 |
| Data collection statistics | ||
| Beamline | PF BL-5A | PF-AR NW12A |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Space group | P21 | P21 |
| Unit-cell parameters | ||
| a (Å) | 84.8 | 85.3 |
| b (Å) | 98.4 | 98.8 |
| c (Å) | 104.0 | 105.1 |
| β (°) | 102.7 | 102.5 |
| Resolution (Å)* | 50.00–2.00 (2.07–2.00) | 50.00–2.10 (2.18–2.10) |
| Measured reflections | 392667 | 377058 |
| Unique reflections | 111886 | 99298 |
| Completeness (%) | 97.0 (92.1) | 100.0 (100.0) |
| Mean I/σ | 8.0 (2.8) | 8.1 (3.3) |
| Rmerge (%)† | 6.8 (30.7) | 9.4 (30.3) |
| Refinement statistics | ||
| Resolution range (Å) | 49.21–2.00 | 41.86–2.10 |
| Rfactor/Rfree‡ (%) | 17.6/21.3 | 18.0/22.0 |
| No. of protein atoms | 12856 | 12856 |
| No. of solvent atoms | 1074 | 931 |
| No. of heteroatoms | 34 | 46 |
| Average B-factors (Å2) | 23.7 | 22.0 |
| Protein (Chain A/B) | 22.8/23.7 | 20.7/22.7 |
| Glucose | 23.8/30.6 | 24.1/32.8 |
| Sulphate | 24.9/24.7 | |
| Phosphate | 19.5/18.0 | |
| Glycerol | 25.3/27.9 | |
| Water | 28.5 | 25.8 |
| Rmsd bond lengths (Å) | 0.005 | 0.006 |
| Rmsd bond angles (°) | 1.3 | 1.3 |
*Values in parentheses are for the highest resolution shell.
†Rmerge=ΣhΣi|I(h,i)−〈I(h)〉|/ΣhΣiI(h,i), where I(h,i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the average value over multiple measurements.
‡Calculated using a test data set; 5% of total data randomly selected from the observed reflections.
Phase calculation and refinement
Phase calculation was performed by molecular replacement with program MOLREP [20] in the CCP4 program suite [21] against the data set of CgCBP–SO4. The crystal structure of VpChBP (Protein Data Bank entry 1V7W) was used as a search model. Program ARP/wARP [22] was used for automatic model building. The structure of CgCBP–PO4 was refined starting from the refined CgCBP–SO4 structure. Visual inspection of the models was performed using XtalView [23]. Simulated annealing, energy minimization, and individual B factor refinement were carried out using CNS1.1 [24]. The Figures were prepared using MOLSCRIPT [25], RASTER3D [26], XtalView, SPOCK [27], and PyMol [http://www.pymol.org]. Superimposition of CgCBP and VpChBP structures were carried out with program TOPP [29] in the CCP4 program suite.
Enzyme assays and kinetic analysis
1,5-Anhydroglucitol, isomaltose, and melibiose were purchased from Sigma–Aldrich. Measurement of the synthetic reaction was carried out as described previously [11]. The initial rate was assayed at 37 °C in 50 mM Tris/HCl (pH 7.0) by measuring the amount of phosphate released from 10 mM Glc 1-P with a suitable concentration of a glucosyl acceptor [30]. The kinetic parameters were calculated by regressing the experimental data by the curve fit method using GraFit Version 4.0 (Erithacus Software, Surrey, U.K.).
RESULTS AND DISCUSSION
Crystallography and overall structure
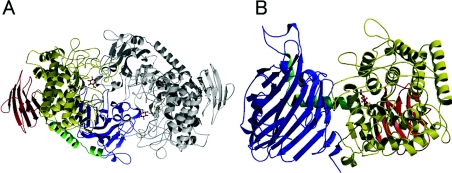
CgCBP crystals were obtained either complexed with sulphate (CgCBP–SO4) or complexed with phosphate (CgCBP–PO4). CgCBP–SO4 and CgCBP–PO4 structures were determined at 2.0 and 2.1 Å (1 Å=0.1 nm) resolution to R factor (Rfree) of 17.6% (21.3%) and 18.0% (22.0%) respectively (Table 1). The crystals belong to the space group P21 and contain two subunits per asymmetric unit (Figure 1A). The structures of the four subunits (CgCBP–SO4 chain A/B and CgCBP–PO4 chain A/B) are almost identical. The rmsd (root-mean square deviation) for Cα atoms between all pairs of these subunits was within 0.3 Å.
Figure 1. Ribbon diagrams of the CgCBP–PO4 structure.
(A) CgCBP dimer in an asymmetric unit. One subunit is shown in grey. (B) CgCBP–PO4 monomer (N-terminal domain, blue; linker helices, green; α-helix barrel domain, yellow; C-terminal domain, red). Bound glucose, glycerol and phosphate are shown as ball-and-stick models.
The two molecules in an asymmetric unit form a dimer (Figure 1A), which seems to correspond to the dimeric form in solution [31]. The rmsd for Cα atoms between the dimer was 0.27 Å, both in CgCBP–SO4 and CgCBP–PO4. The two subunits are tightly connected through hydrophobic interactions and a number of hydrogen bonds. Upon dimer formation, 3200 Å2 of solvent-accessible surface area (12% of the total monomer surface) is buried. This dimer form is similar to that of VpChBP. Since the residues involved in dimer interaction are conserved, as well as contributing to the formation of the active-site pocket (see below), it is likely that GH-94 CBPs from other sources also form dimers, although some CBPs have been reported to be monomeric in solution [32,33].
Figure 1(B) shows a ribbon diagram of the monomeric structure of CgCBP–PO4, consisting of 822 amino acid residues from the original N-terminal methionine residue; an additional sequence, carrying a His6-tag at the N-terminal end, is disordered. Structurally, the CgCBP monomer exhibits a complex architecture, consisting of four distinct regions, similar to that of VpChBP: an N-terminal domain (residues 1–289), a helical linker region (residues 290–320), an α-helix barrel domain (residues 329–745), and a C-terminal domain (residues 321–328 and 746–822). The rmsd between CgCBP and VpChBP is 1.58 Å for the Cα atoms of 766 residues.
Complex structures with SO4 and PO4
We have previously reported the ternary (protein–sugar–anion) structure of VpChBP, complexed with two GlcNAc, at subsites −1 and +1, and an SO4 anion [8]. The SO4-binding site was located in the vicinity of the N-termini of two helices, making it also suitable for other anions, such as chloride or PO4. Due to the similar shapes and electrostatic properties of phosphate and sulphate ions, it is likely that the SO4-binding site is also the PO4-binding site.
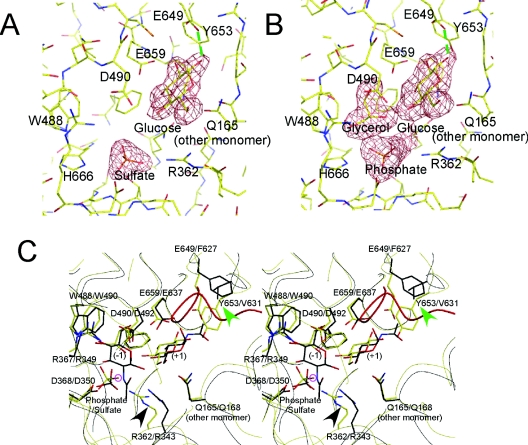
We were able to obtain two protein–sugar–anion complex structures of CgCBP, CgCBP–SO4 and CgCBP–PO4, in which the SO4 and PO4 were derived from the precipitants used for crystallization. The SO4 or PO4 was located at the anion-binding site, and a glucose moiety was located at subsite +1 (Figures 2A and B). Kinetic analysis has shown that the binding affinity of subsite −1 for glucose is much weaker; the Ki of glucose to subsites −1 and +1 has been estimated to be 170 mM and 3.0 mM respectively [34]. The glucose molecule at subsite +1 takes on a chair (4C1) conformation in both of the CgCBP complex structures. This glucose overlaps with the GlcNAc molecule at subsite +1 in the VpChBP structure (Figure 2C), enabling us to compare sugar recognition at this site in detail (see below). Interestingly, the CgCBP–PO4 contained a glycerol at subsite −1 (Figure 2B; see below), whereas subsite −1 of CgCBP–SO4 was empty.
Figure 2. Active site structure of CgCBP.
(A) Wireframe model of CgCBP–SO4, and the unbiased |Fobs| − |Fcalc| electron-density map of the bound glucose and phosphate (3.0σ). The residues (single-letter amino acid codes are used) involved in subsite formation (R362, W488, E649, Y653, E659, and Q165 from the adjacent subunit), sulphate recognition (H666), and catalysis (D490) are labelled. Glucose takes on the β-anomer configuration and forms a hydrogen bond with E649 (green broken line). (B) Wireframe model of CgCBP–PO4, and the unbiased |Fobs| − |Fcalc| electron-density map of the bound glucose, glycerol, and phosphate (3.0σ). (C) Stereo view of CgCBP–PO4 and VpChBP superimposed at the active site. The backbone traces of CgCBP and VpChBP are coloured yellow and black, respectively. The bound glucose, glycerol, and phosphate ion in CgCBP (yellow), and the GlcNAc molecules and sulphate ion in VpChBP (black) are shown as wireframe models. Subsites −1 and +1 are labelled (−1) and (+1), respectively. The structurally conserved residues are shown as a wireframe model and labelled in the order corresponding to CgCBP and VpChBP. The 495–513 loop in CgCBP is coloured red. The residues related to N-acetyl/hydroxy group recognition at the C-2 position in each subsite are indicated by arrowheads. The oxygen atom suitable for direct attack on the C-1 atom of glucose at subsite −1 is circled in pink. Single-letter amino acid codes are used.
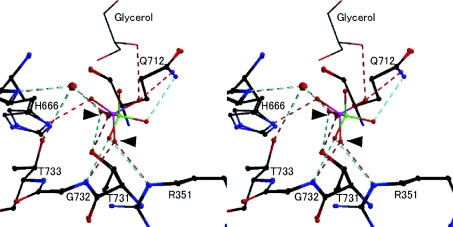
The bound forms of SO4 and PO4 were almost identical. Among the highly conserved residues involved in anion recognition are Arg351, His666, Thr731, Gly732, and Thr733 (Figure 3). Interestingly, the orientations of the bound SO4 and PO4 are different, in that one P–O bond points to His666, whereas one S–O bond points in the opposite direction (Figure 3). The pH values for crystallization (pH 8.2 for CgCBP–PO4 and pH 6.5 for CgCBP–SO4) suggest that the anions are probably in states with two negative charges (HPO42− and SO42−). Therefore, the different orientations of the bound anions may be caused by an interaction with glycerol, which was present only in CgCBP–PO4, not by different charge states.
Figure 3. Stereo view of superimposition around the anion binding site of CgCBP–PO4 and CgCBP–SO4.
The phosphate in CgCBP- PO4 and the sulphate in CgCBP–SO4 are coloured pink and green respectively. Broken lines indicate hydrogen bonds between CgCBP and the anion (red for phosphate and cyan for sulphate). The two oxygen atoms held by T731, G732 and R351 are indicated by arrowheads. Single-letter amino acid codes are used.
In PO4 recognition, three hydrogen bonds, i.e. those with the side-chain hydroxy group of Thr731, the main-chain NH group of Gly732, and the NεH group of Arg351, hold two of the four oxygen atoms of PO4 (indicated by arrowheads in Figure 3). The other two oxygen atoms, which are positioned differently from those of SO4, form two additional hydrogen bonds, one with the Nδ atom of His666 and the other, which is held by Gln712 and glycerol (circled in Figure 2C), appears to be suitable for the direct attack on the C-1 atom of glucose at subsite −1. The structure of the CgCBP–PO4 complex supports our hypothesis that the reaction mechanism of inverting phosphorylases begins with direct nucleophilic attack by phosphate on the anomeric centre [8].
Sugar-donor specificity (subsite −1)
Subsites −1 and +1 in the phosphorolytic reaction of CBP act as sugar-donor and sugar-acceptor sites in the synthetic reaction, respectively. Although we could not obtain a complex structure of CgCBP with glucose at subsite −1 (sugar-donor site), a glycerol, which was derived from the cryoprotectant, was bound to subsite −1 in CgCBP–PO4. Glycerol occasionally occupies sugar-binding subsites [35]. The carbon and oxygen atoms of glycerol in CgCBP almost completely overlap the C-4 to C-6 and O-4 to O-6 atoms of GlcNAc in VpChBP (Figure 2C). The average distance between these atoms was 0.42 Å (maximum 0.65 Å) when the polypeptide chains of CgCBP and VpChBP were superimposed by the least-squares method. Although the sugar ring at subsite −1 may form a reaction intermediate with an oxocarbonium ion-like conformation during phosphorolysis [8], the shift of the C-4, C-5, and C-6 positions is likely to be relatively small. Therefore, the structure of the complex with glycerol does not conflict with the reaction mechanism.
Residues surrounding subsite −1 are basically conserved between CgCBP and VpChBP. At the C-2 position of the substrate sugar, however, they carry a different group, hydroxy in CBP and N-acetyl in ChBP. In the VpChBP structure, Arg343 and Asp350 form hydrogen bonds with the N-acetyl group (Figure 2C). These residues are conserved in CgCBP (Arg362 and Asp368), but the arginine side chain is significantly close to the sugar substrate when the two structures are superimposed. This arginine side chain sterically hinders an N-acetyl group at this site (indicated by a black arrowhead in Figure 2C). These structural features at this subsite are consistent with the substrate specificities of these enzymes, in that CBP cannot accommodate GlcNAc 1-P, whereas ChBP can accommodate both GlcNAc 1-P and Glc 1-P [36].
Sugar-acceptor specificity (subsite +1)
The electron density of glucose bound to subsite +1 (sugar-acceptor site) was clear in shape. Therefore, we could confidently determine the conformation of the glucose. When we compared the structures of CgCBP and VpChBP, we found that the Asp490/492, Lys658/636, Glu659/637, and Gln712/690 residues were conserved. In addition, the Gln165/168 residue, in the N-terminal domain of the other monomer, was conserved to form the active-site pocket (Figure 2C). As with the O-1 atom at the reducing end of cellobiose, which shows strict β-anomer specificity [37], the reducing end O-1 atom of glucose formed a hydrogen bond with Glu649 of CgCBP (green broken line in Figures 2A and 2B), and electron density showed that the glucose moiety was exclusively in the β-anomer configuration. In contrast, VpChBP, which shows no anomer specificity at the reducing-end GlcNAc, did not form hydrogen bonds with the O-1 atom of GlcNAc, and electron density showed a mixture of the α- and β-anomer configurations [8]. CgCBP and VpChBP also show distinct specificities for β-substituted sugar-acceptors. Unlike VpChBP, which can phosphorolyse β-substituted chitobioses such as o-nitrophenyl chitobiose [38], CgCBP cannot [34]; this is possibly due to the existence of a loop, consisting of residues 495–513, in CgCBP (see below).
Differences in C-2 group specificity at subsite +1 can be clearly explained by the respective structural features of the two enzymes. The substrate-binding pocket of VpChBP is locally hydrophobic (indicated by a green arrowhead in Figure 2C). The Val631 residue is involved in the formation of this small hydrophobic pocket, which contains the methyl portion of the N-acetyl group of GlcNAc. In CgCBP, the area corresponding to the small hydrophobic pocket of VpChBP is filled by a bulky residue, Tyr653, which forms a hydrogen bond with the bound glucose. These residues, valine in ChBP and tyrosine in CBP, are fully conserved in each enzyme group.
At subsite +1 of CgCBP, the kinetic parameters for several analogues of glucose have been determined (Table 2). The Gibbs energy of activation difference (the ΔΔG values) and the net strengths of hydrogen-bonding interactions can be estimated from the differences in catalytic efficiency of enzyme for each analogue compared with that for glucose [32,39]. The ΔΔG values of 1,5-anhydroglucitol (4.4 kcal/mol; 1 kcal=4.184 kJ) and 2-deoxyglucose (3.3 kcal/mol), relative to D-glucose, suggest that the O-1 and O-2 hydroxy groups form strong interactions (probably via hydrogen bonds) with CgCBP. As with some reducing-end-specific carbohydrate-acting enzymes, recognition of the anomeric hydroxy group of CgCBP seems to be necessary to increase recognition position for fixing the reducing end unit [40]. In contrast, the ΔΔG value of 6-deoxyglucose (1.3 kcal/mol) indicates that the recognition of the O-6 hydroxy group was relatively weak. In addition, removal of the hydroxymethyl group at C-6 decreased kcat/Km 125-fold, with a loss of binding energy of 3.0 kcal/mol. Therefore we estimate that C-6 stabilizes substrate binding by 1.7 (3.0–;1.3) kcal/mol, probably via a hydrophobic interaction. Similar kinetic results have been reported for CuCBP (CBP from Cellulomonas uda) [32]. By determining the kinetic parameters for the binding of the 3-deoxy and deoxyfluoro analogues of glucose, the interaction between glucose and CuCBP was predicted to occur through the formation of hydrogen bonds between each hydroxy group and a negatively charged (at O-1, O-3, and O-4) or non-charged (at O-2 and O-6) group, as well as by a hydrophobic interaction at C-6.
Table 2. Kinetic parameters for the synthesis reactions of CgCBP with different glucosyl acceptors, and interactions in the complex structure.
The ΔΔG is the Gibbs energy of activation for kcat/Km for the enzymatic glucosyl transfer to an acceptor, relative to transfer to D-glucose, was calculated according to ΔΔG=RTln[(kcat/Km)D-glucose/(kcat/Km)acceptor], whereby R is 1.987 cal·mol−1·K−1 and T is 310.15 K [39]. Values in parentheses are taken from the results of CuCBP (T is 303.15 K) [32]. Values for 1,5-anhydroglucitol, isomaltose, gentiobiose and melibiose were determined in the present study.
| Km (mM) | kcat (s−1) | kcat/Km (s−1·M−1) | ΔΔG (kcal/mol) | Hydrogen-bond pair | Distance (Å) | |||
|---|---|---|---|---|---|---|---|---|
| Glucose [11] | 2.1 | 98 | 46800 | |||||
| O-1 | 1,5-Anhydroglucitol | 89 | 5.3 | 59 | 4.11 | (>7) | Glu649 Oϵ | 2.6 |
| O-2 | Mannose [11] | 115 | 39 | 342 | 3.03 | (2.65) | Tyr653 Oη | 2.8 |
| 2-Deoxyglucose [11] | 168 | 37 | 220 | 3.30 | (2.24) | Glu659 Oϵ1 | 3.0 | |
| Glucosamine [11] | 13 | 15 | 1160 | 2.28 | (2.19) | |||
| O-3 | (3-Deoxyglucose) | (4.01) | Lys658 Nζ | 3.0 | ||||
| Glu659 Oϵ2 | 2.7 | |||||||
| O-4 | Asp490 Oδ | 2.6 | ||||||
| O-5 | 5a-Carba-β-D-glucopyranose [12] | 55 | 93 | 1680 | 2.05 | Gln165 | ||
| (other monomer) Nϵ | 3.2 | |||||||
| O-6 | 6-Deoxyglucose [11] | 24 | 139 | 5780 | 1.29 | (0.77) | None | |
| Xylose [11] | 84 | 31 | 372 | 2.98 | (2.15) | |||
| Isomaltose | 71 | 1.9 | 27 | |||||
| Gentiobiose | 83 | 7.2 | 87 | |||||
| Melibiose | 93 | 2.3 | 24 | |||||
These estimates of the strength of interaction were consistent with the structural features of the enzyme. For example, the O-1 and O-2 hydroxy groups of glucose form one strong and two relatively weak hydrogen bonds with CgCBP, respectively, consistent with the estimated ΔΔG values (Table 2). The hydrogen bond between O-2 and Tyr653 appears to be weaker than the interactions at O-1 and O-3, probably because Tyr653 possesses a non-charged side chain. The O-3 hydroxy group of glucose in CgCBP forms a direct hydrogen bond with Glu659, and the short distance (2.7 Å) indicates that CgCBP strictly recognizes the C-3 equatorial hydroxy group of glucose. Lys658 also contributes to the interaction at O-3. The O-4 hydroxy group interacts with Asp490, which acts as a general acid in phosphorolysis. In the synthesis reaction, Asp490 would assist deprotonation of the O-4 hydroxy group of the sugar-acceptor molecule, as predicted for CuCBP. The O-6 hydroxy group does not form any direct hydrogen bonds with CgCBP, but there is a hydrophobic interaction with the guanidium plane of Arg362 (Figure 2C). Consistently, the ΔΔG value for O-6 is smaller than that of any other hydroxy groups, and replacement with hydrogen increases Km. The O-6 atom in CgCBP is exposed to the solvent, and there is a large open space ahead of it. This structural feature accounts for the ability of CgCBP to synthesize branched trisaccharides by accepting O-6 substituted disaccharides, such as isomaltose (Glcα1–6Glc), gentiobiose (Glcβ1–6Glc), and melibiose (Galα1–6Glc), at this site (Table 2) [17].
Active-site-pocket formation
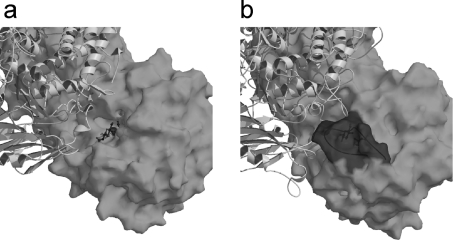
The active sites of VpChBP and CgCBP are located at the dimer interface between the α-helix barrel domain and the N-terminal domain of the adjacent subunit (Figure 1A). The dimer interaction forms a pocket-type active site, which is appropriate for short disaccharide substrates such as chitobiose and cellobiose [8]. Therefore, the substrate specificity of GH-94 enzymes with regard to the degree of polymerization, e.g. the difference between CBP and CDP, seems to be determined by subunit interaction. Figures 4(A) and 4(B) show the molecular surfaces of VpChBP and CgCBP respectively. Although both enzymes phosphorolyse disaccharide substrates, the shape of the entrance to their active sites is different. In VpChBP, the active site pocket is relatively ‘open’, and the substrates are able to gain access without any structural change of the enzyme. In contrast, the active-site entrance of CgCBP is ‘closed’, in that it is covered by a long loop consisting of residues 495–513 (loop coloured red in Figure 2C). This loop, which is found only in CBPs, but not in ChBPs or CDPs, in GH-94, exhibits a relatively high B-factor (30 Å2 in average), indicating that it is flexible. Therefore, this loop may undergo some conformational change on substrate binding. This structural feature may provide a solution to the problem of discrepancy in the proposed substrate binding order of four very similar CBPs from Cellvibrio gilvus, Clostridium thermocellum, Cellulomonas uda, and Thermotoga maritima (sequence identity >61% for each pair) [31,32,41,42]. All of these enzymes are likely to exhibit a random-ordered Bi Bi mechanism, since the movement of the loop covering the active site may have affected inhibition patterns.
Figure 4. Active-site-pocket formation in VpChBP (a) and CgCBP (b).
The molecular surface of one subunit and a ribbon diagram of the adjacent subunit are shown. (a) The bound GlcNAc molecules in VpChBP are shown as a wireframe model. (b) The molecular surface is rendered transparent to show the bound glucose beneath loop 495–513 covering the active-site-pocket.
Acknowledgments
We thank Dr N. Igarashi, Dr N. Matsugaki, Dr M. Suzuki and Dr S. Wakatsuki for data collection at the Photon Factory, KEK, Tsukuba, Japan. This work was supported by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research 17780079 to S.F.), Research Fellowships of the Japan Society for the Promotion of Science for Young Scientist (15-11327 and 17·182 to M.H.), the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and in part by the National Project on Protein Structural and Functional Analyses funded by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Coutinho P., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H. J., Davies G. J., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 2.Geremia S., Campagnolo M., Schinzel R., Johnson L. N. Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase. J. Mol. Biol. 2002;322:413–423. doi: 10.1016/s0022-2836(02)00771-4. [DOI] [PubMed] [Google Scholar]
- 3.O'Reilly M., Watson K. A., Johnson L. N. The crystal structure of the Escherichia coli maltodextrin phosphorylase–acarbose complex. Biochemistry. 1999;38:5337–5345. doi: 10.1021/bi9828573. [DOI] [PubMed] [Google Scholar]
- 4.O'Reilly M., Watson K. A., Schinzel R., Palm D., Johnson L. N. Oligosaccharide substrate binding in Escherichia coli maltodextrin phosphorylase. Nat. Struct. Biol. 1997;4:405–412. doi: 10.1038/nsb0597-405. [DOI] [PubMed] [Google Scholar]
- 5.Watson K. A., McCleverty C., Geremia S., Cottaz S., Driguez H., Johnson L. N. Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question. EMBO J. 1999;18:4619–4632. doi: 10.1093/emboj/18.17.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson R. P., Tarling C. A., Roberts S., Withers S. G., Davies G. J. The donor subsite of trehalose-6-phosphate synthase–binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 Å resolution. J. Biol. Chem. 2004;279:1950–1955. doi: 10.1074/jbc.M307643200. [DOI] [PubMed] [Google Scholar]
- 7.Buschiazzo A., Ugalde J. E., Guerin M. E., Shepard W., Ugalde R. A., Alzari P. M. Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 2004;23:3196–3205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hidaka M., Honda Y., Kitaoka M., Nirasawa S., Hayashi K., Wakagi T., Shoun H., Fushinobu S. Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (α/α)6 barrel fold. Structure. 2004;12:937–947. doi: 10.1016/j.str.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Egloff M. P., Uppenberg J., Haalck L., van Tilbeurgh H. Crystal structure of maltose phosphorylase from Lactobacillus brevis: unexpected evolutionary relationship with glucoamylases. Structure. 2001;9:689–697. doi: 10.1016/s0969-2126(01)00626-8. [DOI] [PubMed] [Google Scholar]
- 10.Stam M. R., Blanc E., Coutinho P. M., Henrissat B. Evolutionary and mechanistic relationships between glycosidases acting on α- and β-bonds. Carbohydr. Res. 2005;340:2728–2734. doi: 10.1016/j.carres.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Kitaoka M., Sasaki T., Taniguchi H. Synthetic reaction of Cellvibrio gilvus cellobiose phosphorylase. J. Biochem. (Tokyo) 1992;112:40–44. doi: 10.1093/oxfordjournals.jbchem.a123862. [DOI] [PubMed] [Google Scholar]
- 12.Kitaoka M., Sasaki T., Taniguchi H. Conversion of sucrose into cellobiose using sucrose phosphorylase, xylose isomerase and cellobiose phosphorylase. Denpun Kagaku. 1992;39:281–283. [Google Scholar]
- 13.Kitaoka M., Sasaki T., Taniguchi H. Purification and properties of laminaribiose phosphorylase (EC 2.4.1.31) from Euglena gracilis Z. Arch. Biochem. Biophys. 1993;304:508–514. doi: 10.1006/abbi.1993.1383. [DOI] [PubMed] [Google Scholar]
- 14.Kitaoka M., Aoyagi C., Hayashi K. Colorimetric quantification of cellobiose employing cellobiose phosphorylase. Anal. Biochem. 2001;292:163–166. doi: 10.1006/abio.2001.5049. [DOI] [PubMed] [Google Scholar]
- 15.Liu A., Tomita H., Li H., Miyaki H., Aoyagi C., Kaneko S., Hayashi K. Cloning, sequencing and expression of the cellobiose phosphorylase gene of Cellvibrio gilvus. J. Ferment. Bioeng. 1998;85:511–513. [Google Scholar]
- 16.Percy A., Ono H., Watt D., Hayashi K. Synthesis of β-D-glucopyranosyl-(1-4)-D-arabinose, β-D-glucopyranosyl-(1-4)-L-fucose, and β-D-glucopyranosyl-(1-4)-D-altrose catalysed by cellobiose phosphorylase from Cellvibrio gilvus. Carbohydr. Res. 1998;305:543–548. [Google Scholar]
- 17.Percy A., Ono H., Hayashi K. Acceptor specificity of cellobiose phosphorylase from Cellvibrio gilvus: synthesis of three branched trisaccharides. Carbohydr. Res. 1998;308:423–429. doi: 10.1016/s0008-6215(98)00109-8. [DOI] [PubMed] [Google Scholar]
- 18.Hidaka M., Kitaoka M., Hayashi K., Wakagi T., Shoun H., Fushinobu S. Crystallization and preliminary X-ray analysis of cellobiose phosphorylase from Cellvibrio gilvus. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1877–1878. doi: 10.1107/S0907444904017767. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 21.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Perrakis A., Morris R., Lamzin V. S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 23.McRee D. E. XtalView Xfit–a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 24.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Kraulis P. J. Molscript – a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 26.Merritt E. A., Bacon D. J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 27.Christopher J. A., Baldwin T. O. SPOCK: Real-time collaborative molecular modelling. J. Mol. Graphics Model. 1998;16:285. [Google Scholar]
- 28. Reference deleted.
- 29.Lu G. G. TOP: a new method for protein structure comparisons and similarity searches. J. Appl. Crystallogr. 2000;33:176–183. [Google Scholar]
- 30.Lowry O. H., Lopez J. A. The determination of inorganic phosphate in the presence of labile phosphate esters. J. Biol. Chem. 1946;162:421–428. [PubMed] [Google Scholar]
- 31.Kitaoka M., Sasaki T., Taniguchi H. Phosphorolytic reaction of Cellvibrio gilvus cellobiose phosphorylase. Biosci. Biotechnol. Biochem. 1992;56:652–655. doi: 10.1271/bbb.56.652. [DOI] [PubMed] [Google Scholar]
- 32.Nidetzky B., Eis C., Albert M. Role of non-covalent enzyme–substrate interactions in the reaction catalysed by cellobiose phosphorylase from Cellulomonas uda. Biochem. J. 2000;351:649–659. [PMC free article] [PubMed] [Google Scholar]
- 33.Reichenbecher M., Lottspeich F., Bronnenmeier K. Purification and properties of a cellobiose phosphorylase (CepA) and a cellodextrin phosphorylase (CepB) from the cellulolytic thermophile Clostridium stercorarium. Eur. J. Biochem. 1997;247:262–267. doi: 10.1111/j.1432-1033.1997.00262.x. [DOI] [PubMed] [Google Scholar]
- 34.Kitaoka M., Hayashi K. Carbohydrate-processing phosphorolytic enzymes. Trends Glycosci. Glycotechnol. 2002;14:35–50. [Google Scholar]
- 35.Fushinobu S., Hidaka M., Honda Y., Wakagi T., Shoun H., Kitaoka M. Structural basis for the specificity of the reducing end xylose-releasing exooligoxylanase from Bacillus halodurans C-125. J. Biol. Chem. 2005;280:17180–17186. doi: 10.1074/jbc.M413693200. [DOI] [PubMed] [Google Scholar]
- 36.Honda Y., Kitaoka M., Hayashi K. Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 glycosyltransferase in Vibrio. Biochem J. 2004;377:225–232. doi: 10.1042/BJ20031171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitaoka M., Ogawa S., Taniguchi H. A cellobiose phosphorylase from Cellvibrio gilvus recognizes only the β-D-form of 5a-carba-glucopyranose. Carbohydr. Res. 1993;247:355–359. doi: 10.1016/0008-6215(93)84271-7. [DOI] [PubMed] [Google Scholar]
- 38.Honda Y., Kitaoka M., Hayashi K. Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus. In: Ohmiya K., Sakka K., Karita S., Kimura T., Sakka M., Onishi Y., editors. Biotechnology of Lignocellulose Degradation and Biomass Utilization. Tokyo: Uni Publishers; 2003. pp. 484–493. [Google Scholar]
- 39.Street I. P., Armstrong C. R., Withers S. G. Hydrogen bonding and specificity. Fluorodeoxy sugars as probes of hydrogen bonding in the glycogen phosphorylase–glucose complex. Biochemistry. 1986;25:6021–6027. doi: 10.1021/bi00368a028. [DOI] [PubMed] [Google Scholar]
- 40.Honda Y., Kitaoka M. A family 8 glycoside hydrolase from Bacillus halodurans C-125 (BH2105) is a reducing end xylose-releasing exo-oligoxylanase. J. Biol. Chem. 2004;279:55097–55103. doi: 10.1074/jbc.M409832200. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y.-K., Kitaoka M., Krishnareddy M., Mori Y., Hayashi K. Kinetic studies of a recombinant cellobiose phosphorylase (CBP) of the Clostridium thermocellum YM4 strain expressed in Escherichia coli. J. Biochem. (Tokyo) 2002;132:197–203. doi: 10.1093/oxfordjournals.jbchem.a003210. [DOI] [PubMed] [Google Scholar]
- 42.Rajashekhara E., Kitaoka M., Kim Y.-K., Hayashi K. Characterization of a cellobiose phosphorylase from a hyperthermophilic eubacterium, Thermotoga maritima MSB8. Biosci. Biotechnol. Biochem. 2002;66:2578–2586. doi: 10.1271/bbb.66.2578. [DOI] [PubMed] [Google Scholar]