Abstract
Inhibition of human cytomegalovirus (HCMV) by 1263W94 was additive dosewise in combination with ganciclovir, acyclovir, and foscarnet. None of the commonly used anti-human immunodeficiency virus agents antagonized the inhibition of HCMV by 1263W94. The data were analyzed by a modified isobologram procedure that measures the strength and statistical significance of drug interactions.
1263W94 (maribavir; 5,6-dichloro-2-isopropylamino-1-β-l-ribofuranosyl-1H-benzimidazole) is a benzimidazole l-riboside that is a potent and selective inhibitor of human cytomegalovirus (HCMV) replication with a novel mechanism of action (2). It is under development for the treatment of HCMV infection. Because patients with HCMV infection frequently receive concomitant antiviral medication, assessment of the antiviral activity of 1263W94 in combination with other antiviral agents would be useful in evaluating 1263W94 as a potential therapeutic agent. For this reason we examined inhibition of HCMV replication by 1263W94 in combination with various antiherpetic or anti-human immunodeficiency virus (anti-HIV) agents in vitro. We describe a modification of fractional inhibitory concentration (FIC) analysis that provides a measure of the strength of any drug interaction and also indicates whether the interaction is statistically significant. Use of the method is not limited to combinations of antivirals. It could be used to analyze interactions of inhibitors of a wide variety of biological processes. The investigational drug 1263W94 was synthesized at GlaxoSmithKline (Research Triangle Park, N.C.); the remaining antiviral drugs were obtained from the manufacturers. 1263W94 was tested in combination with antiherpetic agents (acyclovir [GlaxoSmithKline], ganciclovir [Roche Global Development], cidofovir [Gilead Sciences, Inc.], foscarnet [Astra USA, Inc.]) and anti-HIV agents (zidovudine, lamivudine, amprenavir, and abacavir [all from GlaxoSmithKline], indinavir [Merck Research Laboratories], didanosine [Bristol-Myers Squibb Co.], and zalcitabine [Hoffmann-La Roche Inc.]).
MRC-5 cells (BioWhittaker, Walkersville, Md.) were infected with HCMV laboratory strain AD169 (American Type Culture Collection, Manassas, Va.) as described previously (13). MRC-5 cells were cultured in minimal essential medium (MEM) with 8% (MEM 8-1-1) or 2% (MEM 2-1-1) fetal bovine serum (HyClone, Logan, Utah), 2 mM l-glutamine, 100 U of penicillin G per ml, and 100 μg of streptomycin sulfate per ml. Except as noted, reagents and culture media were obtained from Gibco BRL (Grand Island, N.Y.).
Replication of HCMV was measured by either a multicycle DNA hybridization assay or a single-cycle yield reduction assay as described previously (2). For multicycle DNA hybridization assays, each 96-well plate had two columns of eight wells that were mock infected and that served as uninfected controls. Two other columns of eight wells contained no drug and served as infected controls. The remaining eight columns of eight wells contained a matrix of concentrations of the two drugs such that each concentration of each drug was present in combination with each concentration of the other drug, including no drug. For antiherpetic drugs, the highest concentration was 5 to 20 times the 50% inhibitory concentration (IC50). For the anti-HIV drugs, the highest concentrations were chosen on the basis of the clinically therapeutic levels in serum. From the highest to the lowest concentrations, the serial dilutions decreased by a factor of √10. For each drug combination, three or four replicate plates of infected cells were set up, with one plate of mock-infected cells used for microscopic estimation of cytotoxicity. At the concentrations used, none of the drugs produced any overt cytotoxic effects, either alone or in combination. Single-cycle yield reduction assays were carried out on 24-well plates by using a layout similar to that described above.
The interaction of each pair of compounds was analyzed by use of the model first proposed by Gaddum (5) and later independently by Elion et al. (4). In this model, synergy and antagonism are defined as deviations from dosewise additivity, which results when two drugs interact as if they were the same drug. These concepts have been most clearly elaborated by Cornfield (3), Harvey (7), and Berenbaum (1). We extended the analysis by calculating a new parameter, the average deviation from dosewise additivity, which permits estimation of the nature and the intensity of any interaction between a combination of drugs and of the statistical significance of the interaction.
If a combination of n inhibitors shows dosewise additivity, then for a particular level of inhibition
 |
(1) |
where the FIC for inhibitor I (FICI) is the ratio of xI, the concentration required to produce the particular level of inhibition when the drug is acting in combination, to XI, the concentration required to produce this level of inhibition when the drug is acting alone:
 |
(1a) |
In our study, analysis was based on the concentrations of inhibitory compounds that resulted in a 50% reduction in HCMV replication compared to the amount of viral replication observed in the absence of any inhibitory compound; that is, those concentrations at which
 |
(1b) |
where v(I1, I2) is the yield of virus in the presence of the combination of concentrations I1 and I2 of inhibitors 1 and 2, respectively, and v0 is the yield in the absence of inhibitors. The data were analyzed to calculate the concentration of each compound that produced a 50% reduction in viral replication (i.e., the IC50) alone and in the presence of each concentration of the second drug.
To calculate the IC50s, the data were fitted to the Hill equation (8, 10), shown below:
 |
(2) |
where v(I1, I2) is the yield of virus in the presence of the combination of a variable concentration I1 and a fixed concentration I2 of inhibitors 1 and 2, respectively; v(I2) is the yield in the presence of fixed concentration I2 of inhibitor 2 with inhibitor 1 absent; K is the value of I1 that gives v(I1, I2) equal to v(I2)/2, and N is the Hill constant. Unweighted nonlinear regression (NLIN; SAS Institute) was used to obtain estimates of the three parameters v(I2), K, and N of equation 2.
The IC50s were calculated from these estimates by using the equation
 |
(3) |
where IC50(I1, I2) is the concentration of inhibitor 1 that reduces HCMV replication to 50% of the replication observed in the absence of any inhibitors and in the presence of the fixed concentration I2 of inhibitor 2. Equation 3 is obtained by substituting equation 1b into equation 2. The analysis thus generates a set of IC50s of inhibitor 1 as a function of the concentration of inhibitor 2. Repeating the analysis after switching of the subscripts in equations 2 and 3 generates an equivalent set of IC50s for inhibitor 2. The analysis was repeated for each of the replicate plates, allowing an estimate of the mean and standard error for each IC50. It must be emphasized that the use of the Hill equation is not intended to imply any particular mechanism of drug action or interaction. It is used here only because it gives a robust fit to curves of a variety of forms and thus provides a convenient determination of IC50. The use of another function or of nonparametric estimation of IC50 could be equally valid.
The FICs at which viral replication was inhibited by 50% (FIC50s) were calculated from the IC50 of each component by equation 1a. The resulting calculations can be represented graphically as isobolograms (5). Dosewise additivity results when two drugs interact as if they were the same drug (3, 7) and is indicated by a straight line of slope −1 which intercepts both FIC axes at a value of 1. Datum points that fall significantly above the line representing dosewise additivity result from antagonism between the two drugs. Datum points that fall significantly below the line indicate synergy.
Since the isobologram is simply a graphical representation of equation 1, observed deviations from dosewise additivity (D) can be quantitated and the statistical significance tested. If we make M observations of the effects of a given combination of n inhibitors, equation 1 states that the sum of the FICs for the M observations will be equal to M if the combination is additive. The value of D can thus be calculated as the average deviation of the data from equation 1:
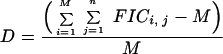 |
(4) |
and the standard error can be calculated from the standard errors of the individual IC50 measurements involved in the calculations of the FIC50s. The statistical significance of differences from dosewise additivity may be tested by a one-sample t test with the hypothesis that D is equal to 0; P values <0.05 (determined by the t test) would be considered significant.
D is a parameter that represents both the nature and the intensity of the interaction of a combination. The value of D is negative when the combination is synergistic, and very strong synergistic interactions give values approaching −0.5 for experiments with a reasonably uniform range of FICs. Values in the range of −0.2 to −0.1 could be interpreted as indicating weak synergy. Antagonistic combinations give a positive value of D, and the possible range of values is not limited, as is the case for synergism. However, a D value of 0.5 would represent strong antagonism.
To demonstrate the validity of the methodology, we examined the interactions of two different concentrations of ganciclovir, using the multicycle DNA hybridization assay. As expected for dosewise additivity, the ganciclovir isobologram is linear with a slope of −0.99 ± 0.1 and an intercept of 0.99 ± 0.05 (Fig. 1). The average D (−0.06 ± 0.11) is clearly not significantly different from zero (P = 0.29 by the t test). The interactions of two solutions of 1263W94 were examined by the yield reduction assay and again produced the expected result of dosewise additivity (Table 1).
FIG. 1.
Isobologram for inhibition of HCMV replication when drugs 1 and 2 are both ganciclovir (GCV), as determined by the multicycle DNA hybridization assay. Error bars show the standard errors of the two FICs plotted.
TABLE 1.
Interactions of 1263W94 with anti-HCMV agents
| Type of assay | Drug 2 | No. of expts | HCMV IC50 (μM) |
D
|
Interaction with 1263W94 | ||
|---|---|---|---|---|---|---|---|
| Mean | SEM | P (t test) | |||||
| Multicycle DNA hybridization | Acyclovir | 2 | 41 ± 8 | −0.09 | 0.10 | 0.19 | Additive |
| Ganciclovir | 2 | 0.83 ± 0.23 | −0.02 | 0.11 | 0.44 | Additive | |
| Foscarnet | 2 | 14.3 ± 2.6 | −0.11 | 0.12 | 0.19 | Additive | |
| Cidofovir | 3 | 0.12 ± 0.025 | −0.45 | 0.06 | <0.001 | Synergistic | |
| 1263W94 | 9 | 0.07 ± 0.01 | NDa | ND | ND | ND | |
| Single-cycle yield reduction | 1263W94 | 2 | 0.02 | 0.046 | 0.14 | 0.37 | Additive |
| Ganciclovir | 1 | 0.06 | 0.16 | 0.14 | 0.15 | Additive | |
| Foscarnet | 1 | 7 | −0.09 | 0.35 | 0.41 | Additive | |
| Cidofovir | 1 | 0.06 | −0.07 | 0.16 | 0.34 | Additive | |
ND, not done.
The interactions of 1263W94 with other anti-HCMV agents were measured by both the multicycle HCMV DNA hybridization assay and the single-cycle yield reduction assay. The results are presented in Table 1. The mean D values show that the interactions of 1263W94 with ganciclovir, acyclovir, and foscarnet were not significantly different from dosewise additivity when measured by either assay. The isobologram for the interaction of 1263W94 and ganciclovir is shown in Fig. 2. The interaction of 1263W94 with cidofovir was synergistic in the multicycle HCMV DNA hybridization assay but additive in the single-cycle yield reduction assay (Table 1; Fig. 3). We have no explanation for this discrepancy.
FIG. 2.
Isobologram for inhibition of HCMV replication by the combination of 1263W94 and ganciclovir (GCV). The results pooled from two experiments performed by the multicycle DNA hybridization assay are shown.
FIG. 3.
Isobolograms for inhibition of HCMV replication by the combination of 1263W94 and cidofovir. The results pooled from three experiments performed by the multicycle DNA hybridization assay (A) and the yield reduction assay (B) are shown.
Table 2 shows the results of experiments assessing the inhibition of HCMV replication in the presence of pairwise combinations of 1263W94 and HIV type 1 inhibitors. All but one (abacavir) of the anti-HIV agents tested showed little or no inhibition of HCMV replication even at the highest concentrations tested. If one member of a combination is not an inhibitor, data analysis is simpler, being reduced to determining whether the IC50 of the active drug is affected by the concentration of the second, noninhibitor compound. If drug 1 is the active inhibitor, the IC50 is determined by the calculation of K in equation 2. The effect of drug 2 on drug 1 can then be tested by linear regression of the IC50 of drug 1 versus the logarithm of the concentration of drug 2. A slope significantly greater than zero indicates antagonism, whereas a slope significantly less than zero indicates potentiation. Potentiation is similar to synergy and is defined as improvement of the potency of an inhibitory compound by addition of a drug that does not by itself act as an inhibitor.
TABLE 2.
Interactions of 1263W94 with anti-HIV agents as determined by multicycle DNA hybridization assay
| Drug 2 | No. of expts | HCMV IC50 (μM) | Effect on 1263W94 HCMV IC50
|
||
|---|---|---|---|---|---|
| Interaction | Correlation | P (t test) | |||
| Zidovudine | 3 | >200 | Slight potentiation | −0.74 | 0.003 |
| Lamivudine | 3 | >250 | None | −0.07 | 0.76 |
| Didanosine | 3 | >100 | None | 0.1 | 0.7 |
| Zalcitabine | 3 | >10 | None | −0.47 | 0.36 |
| Indinavir | 2 | >30 | Slight potentiation | −0.83 | 0.002 |
| Amprenavir | 2 | >100 | None | −0.5 | 0.08 |
| Abacavir | 3 | 78 ± 13 | Synergya | NAb | <0.001 |
D = −0.27 ± 0.06.
NA, not applicable.
Except for zidovudine and abacavir, the nucleoside reverse transcriptase inhibitors showed no effect on the anti-HCMV activity of 1263W94; however, as shown in Fig. 4, zidovudine interacted with 1263W94 to increase the anti-HCMV potency of 1263W94. The two protease inhibitors tested (amprenavir and indinavir) also produced a slight but significant potentiation of the anti-HCMV effect of 1263W94 (Table 2). The reverse transcriptase inhibitor abacavir showed slight anti-HCMV activity in the absence of 1263W94 (Table 2) and moderate synergy in the presence of 1263W94.
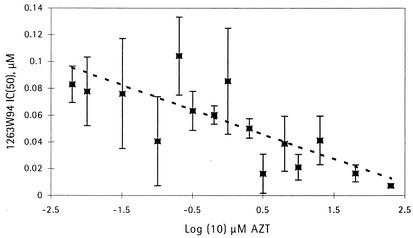
FIG. 4.
Effect of zidovudine (AZT) on the IC50 for the inhibition of HCMV replication by 1263W94. The results pooled from three experiments are shown.
The method described here enhances the traditional analysis of drug interactions by the FIC method (4, 5). The new parameter D provides an estimation of the nature and the intensity of any interaction between a combination of drugs, without requiring any assumptions regarding the underlying mechanisms of any observed interactions. More importantly, it allows evaluation of the statistical significance of the interaction. It is arguably a less sophisticated method than the response surface analysis methods that have been described (6, 11, 12). However, we have found these methods to be impractical. Where the analysis yields results, it is often because of oversimplification of the underlying models, as in the case of Prichard and Shipman (11), who made the simplifying assumption of linear dose-response curves for inhibition. Where simplifying assumptions are not made, as in the case of Greco et al. (6), the resulting equations describing the dose-response surfaces require so many parameters that their estimation is far from robust and impossible with most data sets.
From the definitions given above, additivity clearly results when two drugs have identical mechanisms of action. However, the converse is not necessarily true, and our observation that the interactions of 1263W94 with ganciclovir, acyclovir, and foscarnet were additive does not necessarily imply that the drugs have identical mechanisms of action. For example, 1263W94 and ganciclovir inhibit viral replication through different mechanisms. Ganciclovir inhibits HCMV DNA polymerase through its triphosphorylated derivative (9), whereas 1263W94 does not directly inhibit HCMV DNA polymerase and is not phosphorylated (2).
In terms of clinical application, the results presented here should be interpreted with caution. However, the absence of strong antagonism in vitro is encouraging and suggests that it will be feasible to administer 1263W94 in combination with these anti-HCMV agents should there be a need to do so. However, in the case of the additive combinations, there would be an advantage to combination anti-HCMV therapy only if the combination resulted in lower levels of toxicity or if the emergence of drug-resistant mutants was a problem.
The absence of antagonism with commonly used anti-HIV agents is also encouraging since it indicates that treatment of HCMV infection with 1263W94 in HIV-infected patients will not be compromised by concurrent HIV therapy. It is even possible that synergy or potentiation could result from the use of certain combinations.
REFERENCES
- 1.Berenbaum, M. C. 1985. The expected effect of a combination of agents: the general solution. J. Theor. Biol. 114:413-431. [DOI] [PubMed] [Google Scholar]
- 2.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith III, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornfield, J. 1975. A statistician's apology. J. Am. Stat. Assoc. 70:7-14. [Google Scholar]
- 4.Elion, G., S. Singer, and G. Hitchings. 1954. Antagonists of nucleic acid derivatives. VII. Synergism in combinations of biochemically related antimetabolites. J. Biol. Chem. 208:477-488. [PubMed] [Google Scholar]
- 5.Gaddum, J. 1949. Pharmacology. Oxford University Press, London, United Kingdom.
- 6.Greco, W., H. Park, and Y. Rustum. 1990. Application of a new approach for the quantitation of drug synergism to the combination of cis-diamminedichloroplatinum and 1-beta-d-arabinofuranosylcytosine. Cancer Res. 50:5318-5327. [PubMed] [Google Scholar]
- 7.Harvey, R. 1982. Synergism in the folate pathway. Rev. Infect. Dis. 42:255-260. [DOI] [PubMed] [Google Scholar]
- 8.Hill, A. 1910. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. (London) 40:4-8. [Google Scholar]
- 9.Martin, J., C. Dvorak, D. Smee, T. Matthews, and J. Verheyden. 1983. 9-[(1,3-Dihydroxy-2-propoxy)methyl]guanine: a new potent and selective antiherpes agent. J. Med. Chem. 26:759-761. [DOI] [PubMed] [Google Scholar]
- 10.Monod, J., J.-P. Changeux, and F. Jacob. 1963. Allosteric proteins and cellular control systems. J. Mol. Biol. 6:306-329. [DOI] [PubMed] [Google Scholar]
- 11.Prichard, M., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 12.Suhnel, J. 1990. Evaluation of synergism or antagonism for the combined action of antiviral agents. Antivir. Res. 13:23-39. [DOI] [PubMed] [Google Scholar]
- 13.Underwood, M., R. Harvey, S. Stanat, M. Hemphill, T. Miller, J. Drach, L. Townsend, and K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]