Abstract
A Salmonella enterica serotype Cubana isolate exhibiting resistance to most β-lactam antibiotics, including oxyimino-cephalosporins and imipenem, was isolated from a 4-year-old boy with gastroenteritis in Maryland. β-Lactam resistance was mediated by a conjugative plasmid that encoded KPC-2, a class A carbapenemase previously found in a Klebsiella pneumoniae isolate from the Maryland area as well. Sequence analysis of the flanking regions indicated a potential association of blaKPC-2 with mobile structures.
Resistance to expanded-spectrum oxyimino-cephalosporins among Salmonella strains is mostly due to acquisition of plasmids encoding various class A extended-spectrum β-lactamases (1, 8, 13, 17, 19-21). Production of plasmid-mediated class C β-lactamases by Salmonella isolates has also been described previously (4, 22, 23). The emergence of such strains may have serious implications because of the limitation of therapeutic choices for patients with invasive Salmonella infections and by facilitation of the spread of bla genes in the community. We describe here an imipenem-resistant Salmonella enterica serotype Cubana isolate that produces the recently described KPC-2 β-lactamase (E. S. Moland, J. Johnson, J. A. Black, T. J. Lockhart, A. Hossain, V. L. Herrera, N. D. Hanson, and K. S. Thomson, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2226, 2001).
MATERIALS AND METHODS
Salmonella serotype Cubana was isolated in December 1998 from a stool specimen of a 4-year-old boy with diarrhea in a hospital in Maryland. The patient was chronically ill with Wiskott-Aldrich syndrome. In the 6 months before isolation of serotype Cubana, he had been hospitalized three times and had received intravenous antibiotics. During each of those hospitalizations he had received intravenous β-lactams, including ceftriaxone and ceftazidime, but not carbapenems. There was no history of recent travel. The isolate (AM04707) was submitted to the Centers for Disease Control and Prevention as part of the National Antimicrobial Resistance Monitoring System for enteric bacteria (http://www.cdc.gov/NARMS/) and was subsequently forwarded to the Hellenic Pasteur Institute.
Escherichia coli K-12 strain 14R525 (Nalr) was used as the recipient in conjugation experiments. E. coli DH5α (GIBCO-BRL, Carlsbad, Calif.) was used for transformation. Chloramphenicol-resistant plasmid pBCSK(+) (Stratagene, La Jolla, Calif.) was used for cloning and expression of bla genes.
The MICs of β-lactams were determined by an agar dilution technique (10). Susceptibilities to other antimicrobial agents were assessed by a disk diffusion method (11) and by a partial-range broth microdilution method (Sensititre; Trek Diagnostics; Westlake, Ohio), according to the instructions of the manufacturer.
Conjugation experiments were carried out in mixed broth cultures as described previously (5). Transconjugant clones were selected on Mueller-Hinton agar containing ampicillin (50 μg/ml) plus nalidixic acid (200 μg/ml). Plasmid DNA preparations were obtained by an alkaline lysis technique and resolved in 0.8% (wt/vol) agarose gels. Individual plasmids were excised as discrete bands from low-melting-point agarose (0.8%) and were subjected to partial digestion with various restriction enzymes including HindIII. Digests were ligated into the multicloning site of pBCSK(+). The resulting recombinant plasmids were used to transform E. coli DH5α competent cells. β-Lactam-resistant transformants were selected on Luria-Bertani agar containing chloramphenicol (20 μg/ml) and ampicillin (50 μg/ml). The nucleotide sequences of the cloned fragments were determined with an ABI Prism 377 DNA sequencer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). Sequence similarity searches were performed with the BLAST program (available at the website of the National Center for Biotechnology Information).
β-Lactamases were extracted in one of two ways. One method involved ultrasonic treatment of bacterial cells, followed by suspension in phosphate buffer (100 mM; pH 7.0) and clarification by centrifugation (Fig. 1A). A small-scale freeze-thaw method with modifications provided by J. W. Biddle and J. K. Rasheed was used for comparison of KPC-1 and KPC-2 (3) (Fig. 1B). Detection of carbapenemase activity was performed by bioassay as described previously (24). Analytical isoelectric focusing was performed in polyacrylamide gels containing ampholytes (pH range, 3.5 to 9.5; APBiotech, Piscataway, N.J.). β-Lactamase activity was visualized with nitrocefin (Oxoid Ltd., Basingstoke, United Kingdom, or Becton-Dickinson, Lexington, Ky.). In situ inhibition of β-lactamase activity was performed by soaking the gels in solutions containing clavulanate (0.5 mM), tazobactam (0.5 mM), or EDTA (5 mM). Maximum hydrolysis rates for various β-lactam substrates were estimated by UV spectrophotometry, and inhibition by clavulanate and tazobactam was assessed by using nitrocefin as the reporter substrate. The respective procedures were as described previously (5).
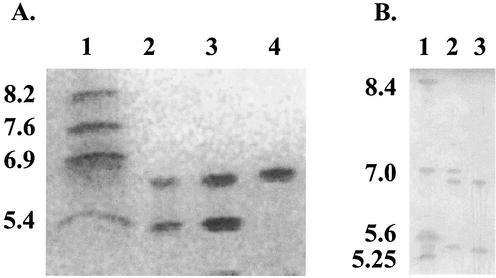
FIG. 1.
(A) Isoelectric focusing of β-lactamase preparations from Salmonella serotype Cubana 4707, E. coli(pST4707), and E. coli(pST-H1) (lanes 2, 3, and 4, respectively). β-Lactamases with known pIs are in lane 1 (TEM-1, pI 5.4; PSE-3, pI 6.9; SHV-1, pI 7.6; SHV-5, pI 8.2). (B) Comparison of preparations from K. pneumoniae 1534 (lane 2) and Salmonella serotype Cubana 4707 (lane 3). β-Lactamases with known pIs are in lane 1 (TEM-12, pI 5.25; TEM-2, pI 5.6; SHV-3, pI 7.0; MIR-1, pI 8.4).
Nucleotide sequence accession number.
The sequence of the 5.2-kb fragment of pST-H1 containing blaKPC-2 has been submitted to the GenBank database and assigned accession number AF481906.
RESULTS
Salmonella serotype Cubana 4707 exhibited either resistance or decreased susceptibility to all β-lactams tested including β-lactam-β-lactamase inhibitor combinations, oxyimino-cephalosporins, aztreonam, and carbapenems. In the presence of clavulanic acid, the MICs of ceftazidime and imipenem were decreased by 4 and 2 doubling dilutions, respectively (Table 1). The isolate was also resistant to streptomycin, trimethoprim, and sulfamethoxazole. The isolate was susceptible to nalidixic acid (MIC, ≤4 μg/ml) and ciprofloxacin (MIC, ≤0.015 μg/ml).
TABLE 1.
Susceptibilities to antibiotics of Salmonella serotype Cubana 4707, an E. coli transconjugant containing wild-type plasmid pST4707, and an E. coli DH5α transconjugant containing plasmid pSTH-1
| Antibiotic | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Serotype Cubana 4707 | E. coli(pST4707) | E. coli DH5α(pSTH-1) | E. coli DH5α | |
| Ampicillin | >256 | >256 | >256 | 2 |
| Amoxicillin | >256 | >256 | >256 | 4 |
| Amoxicillin-CLAa | 64 | 32 | 8 | 2 |
| Ticarcillin | >128 | >128 | >128 | 0.5 |
| Ticarcillin-CLAb | >128 | >128 | >128 | 0.5 |
| Piperacillin | >128 | >128 | >128 | 1 |
| Piperacillin-TAZc | 128 | >128 | 32 | 1 |
| Cefamandole | >64 | >64 | >64 | 1 |
| Cefoxitin | 8 | 16 | 8 | 2 |
| Ceftazidime | 64 | 32 | 8 | ≤0.5 |
| Ceftazidime-CLAc | 4 | 2 | 2 | ≤0.5 |
| Cefotaxime | 32 | 16 | 8 | ≤0.5 |
| Ceftriaxone | 64 | 64 | 16 | ≤0.5 |
| Cefepime | 8 | 8 | 4 | ≤0.5 |
| Aztreonam | 64 | 64 | 32 | ≤0.5 |
| Imipenem | 16 | 16 | 8 | ≤0.5 |
| Imipenem-CLAc | 4 | 2 | 1 | ≤0.5 |
| Meropenem | 8 | 8 | 2 | ≤0.5 |
CLA, clavulanic acid. Penicillin/inhibitor ratio, 2:1.
The inhibitor concentration was fixed at 2 μg/ml.
TAZ, tazobactam. The inhibitor concentration was fixed at 4 μg/ml.
Transfer of β-lactam resistance to E. coli by conjugation was successful. Transconjugants exhibited a phenotype of resistance to β-lactams similar to that of Salmonella serotype Cubana 4707 (Table 1). They were also resistant to streptomycin, trimethoprim, and sulfonamides. Analysis of plasmid DNA indicated transfer of a plasmid (pST4707) that, according to conventional gel electrophoresis in a 0.8% gel, was between 24.5 and 42 MDa (data not shown).
β-Lactamase extracts from Salmonella serotype Cubana 4707 and E. coli(pST4707) were positive in a carbapenemase bioassay performed as described by Yigit et al. (24; data not shown). Isoelectric focusing demonstrated that the extracts contained two β-lactamases with apparent pIs of 5.4 and 6.7, respectively (Fig. 1). Both enzymes were inhibited in situ by clavulanic acid and tazobactam but not by EDTA. The β-lactamase with a pI of 5.4 is consistent with a TEM-1 enzyme, the presence of which was supported by sequencing of 95% of the blaTEM-1-coding region (data not shown).
Cloning of a 5.2-kb HindIII fragment of pST4707 yielded a pBCSK(+) derivative (pST-H1) which mediated resistance only to β-lactams. E. coli(pST-H1) exhibited resistance or decreased susceptibility to all β-lactam antibiotics tested, including carbapenems (Table 1). Also, it was positive in the carbapenemase bioassay and produced a single β-lactamase with a pI of 6.7 (Fig. 1). Hydrolysis experiments performed with extracts from E. coli(pST-H1) showed that the rate of imipenem hydrolysis relative to that of penicillin G, which was set at 100, was 45 ± 5. The value for cefotaxime was 58 ± 3. Hydrolysis rates for ceftazidime and cefoxitin were too low to obtain reliable values, although the slightly elevated cefoxitin MIC indicates some degree of hydrolysis (Table 1). The hydrolysis rates for nitrocefin and cephalothin were significantly higher than that for penicillin G (2.4- and 3.1-fold, respectively). The 50% inhibitory concentrations of clavulanate and tazobactam were 1.4 and 0.08 μM, respectively.
The 5.2-kb HindIII fragment included an 882-bp open reading frame (ORF) that differed by only 1 bp (nucleotide 520) from blaKPC-1 found in Klebsiella pneumoniae (24). An identical sequence (blaKPC-2), also from K. pneumoniae, has recently appeared in GenBank (accession number AY034847; Moland et al., 41st ICAAC). The deduced protein, KPC-2, differs from KPC-1 by one amino acid residue (KPC-2 contains Gly instead of Ser at position 175). Gly-175 is also found in SME-1, NMC-A, and IMI-1 (9, 12, 16). KPC-2 contained the motifs typical for class A β-lactamases (7) as well as residues characteristic for the carbapenem-hydrolyzing enzymes of this molecular class, including Cys at positions 69 and 238, His at position 105, and a hydroxylated residue (Thr) at position 237 (14, 15, 18). The putative secretory signal sequence comprises 24 residues. The mature KPC-2 has a predicted molecular weight of 28,480. The calculated pI (6.4) differs slightly from the apparent pI of the native β-lactamase. The pI reported for KPC-1 was 6.7 (24). When isoelectric focusing was done with both β-lactamases, the pI of K. pneumoniae KPC-1 was very similar to that of KPC-2 from Salmonella serotype Cubana (Fig. 1).
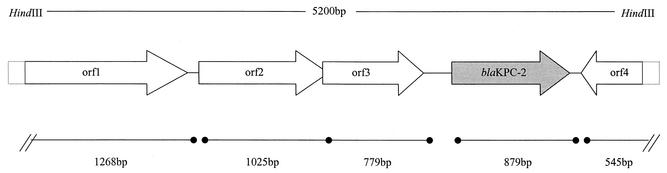
The flanking sequences of blaKPC-2 (120 bp upstream, including the ribosome binding site and the −10 and −35 regions, and 300 bp downstream) were identical to those reported for blaKPC-1 (24). Analysis of the rest of the sequence of the 5.2-kb fragment showed at least four additional ORFs (orf1 to orf4) (Fig. 2). The respective putative peptides (P1 through P4) were significantly similar to proteins associated with transposable elements (Table 2). P1 was highly similar to the C terminus of a transposase A (TnpA) found in the Tn3 family of transposons from Pseudomonas putida (6). P2 and P3 displayed similarity to a TnpA and a transposition helper protein, respectively, found in Ralstonia solanacearum. P4 shared a moderate degree of similarity with putative transposases described in Burkholderia cepacia and Ralstonia spp.
FIG. 2.
Schematic representation of the cloned HindIII fragment (GenBank accession number AF481906). Note that an additional HindIII site is located within the blaKPC-2 gene.
TABLE 2.
ORFs, including blaKPC-2, of the 5.2-kb HindIII fragment of plasmid pST4707
| ORF | Length (no. of amino acids) | Position | G + C content (%) | BLASTP scores (% identity-% similarity) | GenBank accession no. | Remarks |
|---|---|---|---|---|---|---|
| orf1 | 422 | 1-1269 | 60.5 | 1833 (83-89) | T28654 | Transposase from P. putida |
| orf2 | 341 | 1376-2401 | 63.6 | 829 (55-68) | AAL47163 | Putative transposase from R. solanacearum |
| orf3 | 259 | 2398-3177 | 61.9 | 931 (72-85) | AAL47164 | Putative transposition helper protein from R. solanacearum |
| blaKPC-2 | 293 | 3564-4445 | 61.7 | Identical | AY034847 | Class A carbapenemase KPC-2 |
| orf4 | 177 | 5228-4964 | 61.2 | 259 (35-52) | AAL50017 | Putative transposase from Ralstonia metalidurans |
DISCUSSION
KPC-2, along with KPC-1 (24), SME-1 (9), NMC-A (12), and IMI-1 (16), comprise a small group of class A β-lactamases (functional group 2f [2, 15]) with potent carbapenemase activities. The KPC enzymes seem to differ from the rest of the group in that they hydrolyze oxyimino-cephalosporins more efficiently. KPC-2 conferred levels of resistance to β-lactams that were comparable to those conferred by KPC-1 (24). The two enzymes, however, were expressed in different systems, and they also differed by an amino acid residue of the putative omega loop. The latter structure is significant in determining the catalytic properties of class A β-lactamases (7). Hence, differences in the substrate specificity cannot be ruled out.
The high degree of homology between blaKPC-1 and blaKPC-2 and the identity of their flanking sequences indicate that these variant bla genes could be parts of a single structure. However, KPC-2-encoding plasmid pST4707 was self-transferable, while the blaKPC-1-encoding plasmid could not be conjugated into E. coli (24). Also, the blaKPC-1-encoding plasmid, unlike pST4707, neither encoded TEM nor mediated resistance to antibiotics other than β-lactams. Therefore, it can be speculated that the blaKPC variants resulted from insertion of a similar or common mobile element into different plasmid backgrounds. Sequencing data are compatible with this notion, although the existence of a transposable element was not proven.
KPC-1 was found in a K. pneumoniae isolate from a hospital in North Carolina. KPC-producing K. pneumoniae strains have also been found in a Maryland hospital (Moland et al., 41st ICAAC). The isolation of Salmonella serotype Cubana 4707, also in Maryland, indicates a certain degree of spread of blaKPC genes in the United States. The plasmid locations of these genes and their association with possibly mobile structures may facilitate their spread. Continued surveillance is essential to monitor for the potential spread of blaKPC genes among Salmonella strains.
Acknowledgments
We thank Jennifer Nelson, David Blythe, Dipti Shah, and David I. Wheeler for contributions to this work.
REFERENCES
- 1.Bradford, P. A., Y. Yang, D. Sahm, I. Grop, D. Gardovska, and G. Storch. 1998. CTX-M5, a novel cefotaxime hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K., and S. B. Singer. 1989. Effective cooling allows sonication to be used for liberation of β-lactamases from gram-negative bacteria. J. Antimicrob. Chemother. 24:82-84. [DOI] [PubMed] [Google Scholar]
- 4.Fey, P., P. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 5.Giakkoupi, P., L. S. Tzouvelekis, A. Tsakris, V. Loukova, D. Sofianou, and E. Tzelepi. 2000. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 44:2247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauf, U., C. Muller, and H. Herrmann. 1998. The transposable elements resident on the plasmids of Pseudomonas putida strain H, Tn5501 and Tn5502, are cryptic transposons of the Tn3 family. Mol. Gen. Genet. 259:674-678. [DOI] [PubMed] [Google Scholar]
- 7.Matagne, A., J. Lamotte-Brasseur, and J.-M. Frere. 1998. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem. J. 330:581-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morosini, M. I., R. Canton, J. Martinez-Beltran, M. C. Negri, J. C. Perez-Diaz, F. Baquero, and J. Blazquez. 1995. New extended-spectrum TEM-type β-lactamase from Salmonella enterica subsp. enterica isolated in a nosocomial outbreak. Antimicrob. Agents Chemother. 39:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naas, T., L. Wandel, W. Sougakoff, D. M. Livermore, and P. Nordmann. 1994. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 38:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4 (M100-S7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6 (M100-S7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Nordmann, P., S. Mariotte, T. Naas, R. Labia, and M.-H. Nicolas. 1993. Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 37:939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poupart, M. C., C. Chanal, D. Sirot, R. Labia, and J. Sirot. 1991. Identification of CTX-2, a novel cefotaximase from a Salmonella mbandaka isolate. Antimicrob. Agents Chemother. 35:1498-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raquet, X., J. Lamotte-Brasseur, F. Bouillenne, and J.-M. Frere. 1997. A disulfide bridge near the active site of carbapenem-hydrolyzing class A β-lactamases might explain their unusual substrate profile. Proteins 27:47-58. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen, B. A., K. Bush, D. Keeney, Y. Yang, R. Hare, C. O'Gara, and A. A. Medeiros. 1996. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 40:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Revathi, G., K. P. Shannon, P. D. Stapleton, B. K. Jain, and G. L. French. 1998. An outbreak of extended-spectrum beta-lactamase-producing Salmonella senftenberg in a burns ward. J. Hosp. Infect. 40:295-302. [DOI] [PubMed] [Google Scholar]
- 18.Sougakoff, W., T. Naas, P. Nordmann, E. Collatz, and V. Jarlier. 1999. Role of Ser-237 in the substrate specificity of the carbapenem-hydrolyzing class A β-lactamase Sme-1. Biochim. Biophys. Acta 1433:153-158. [DOI] [PubMed] [Google Scholar]
- 19.Tassios, P. T., M. Gazouli, E. Tzelepi, H. Milch, N. Kozlova, S. V. Sidorenko, N. J. Legakis, and L. S. Tzouvelekis. 1999. Spread of a Salmonella typhimurium clone resistant to expanded-spectrum cephalosporins in three European countries. J. Clin. Microbiol. 37:3774-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vahaboglu, H., L. M. Hall, L. Mulazimoglu, S. Dodanli, I. Yildirim, and D. M. Livermore. 1995. Resistance to extended-spectrum cephalosporins, caused by PER-1 beta-lactamase, in Salmonella typhimurium from Istanbul, Turkey. J. Med. Microbiol. 43:294-299. [DOI] [PubMed] [Google Scholar]
- 21.Villa, L., C. Pezzella, F. Tosini, P. Visca, A. Petrucca, and A. Carattoli. 2000. Multiple-antibiotic resistance mediated by structurally related IncL/M plasmids carrying an extended-spectrum β-lactamase gene and a class 1 integron. Antimicrob. Agents Chemother. 44:2911-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffman, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]