Abstract
Isolation of bacterial mutants hypersusceptible to antibiotics can reveal novel targets for antibiotic potentiators. However, identification of such mutants is a difficult task which normally requires laborious replica plating of thousands of colonies. The technique proposed here allows for the positive selection of genetic knockout mutants leading to hypersusceptibility. This technique, designated SDR (selection for DNA release), involves introduction of random insertions of a marker gene into the chromosome of a highly transformable bacterial species, followed by treatment of the obtained library with an antibiotic at subinhibitory concentrations. DNA released by lysing bacteria is collected and used to transform fresh bacteria, selecting for insertion of the marker gene. These selection cycles are repeated until variants with a hypersusceptibility phenotype caused by insertion of the marker begin to dominate in the library. This approach allowed for isolation of a number of mutants of the gram-negative opportunistic pathogen Acinetobacter sp. susceptible to 4- to 16-times-lower concentrations of ampicillin than wild-type bacteria. The mutations affected proteins involved in peptidoglycan turnover and, surprisingly, proteins involved in exopolysaccharide production. A further modification of the SDR technique is described which allows for selecting mutants hypersensitive to agents that affect bacterial physiology but do not cause cell lysis, e.g., inhibitors of translation. This application of SDR is illustrated here by identification of several mutants of Acinetobacter sp. with increased susceptibility (two- to fivefold decrease in the MIC) to erythromycin. The same technique can be used to identify prospective targets for potentiators of many other antibacterial agents.
Gene knockout mutations leading to hypersusceptibility to antibiotics can help identify novel targets of antibiotic potentiators. Indeed, if bacteria become hypersensitive to a particular antibiotic upon disruption of a certain gene, an inhibitor of the protein product of this gene is likely to have the same effect and promote antibiotic action. Apart from genetic knockouts of known antibiotic resistance genes, only a limited number of hypersusceptibility mutations have been described to date, mostly due to the laboriousness of their isolation.
Almost by definition, such mutants that either die or stop growing in the presence of a low concentration of antibiotics cannot be selected directly. The standard approach to isolation of such mutants is replica plating of a library of mutagenized bacteria on a control plate and a plate with a subinhibitory concentration of an antibiotic, followed by identification of colonies that grow only on the control plate. Limited-size screens of this kind have revealed several hypersusceptibility mutations (3, 19, 29, 32, 34). However, this approach is very laborious. If mutagenesis is achieved by random chromosomal insertions of a marker genetic element, such as a transposon, an exhaustive screening of a typical bacterial genome would require replica plating of tens of thousands of colonies (14). To our knowledge, a work of this magnitude has never been performed to isolate hypersusceptibility mutants.
Potentially, identification of such mutants could also be conducted using a number of DNA-based techniques developed in the past several years. In these approaches, a library of insertional mutants that has been subjected to experimental conditions (e.g., a subinhibitory concentration of an antibiotic) is compared to the original library; clones that become extinct are identified using either PCR-based or hybridization-based methods (11, 12, 17). However, like replica plating, these DNA-based techniques require large-scale efforts and, to our knowledge, have not been used for isolation of hypersusceptibility mutants.
Here we describe a new genetic technique, selection for DNA release (SDR), which allows for positive selection of mutations leading to antibiotic hypersusceptibility. Instead of merely identifying mutant bacteria in the library of genetic knockouts, the SDR strategy directly selects for insertions of a marker gene that lead to hypersusceptibility. The DNA fragments containing such insertions are released into the medium by mutant bacteria exposed to a low antibiotic concentration. These fragments are rescued and used to transform a fresh batch of bacterial cells. Several cycles of such selection lead to dramatic enrichment of the library with the desired mutants.
The most immediate application of this strategy is the identification of genes whose disruption leads to hypersusceptibility to antibiotics causing bacterial lysis, such as ampicillin. Here, we used SDR to select several ampicillin-hypersusceptible mutants. We also demonstrate how the SDR strategy can be adapted for selecting bacterial mutants hypersusceptible to antibiotics that do not cause lysis, such as translational inhibitors. Specifically, we describe the selection of three mutants susceptible to low concentrations of erythromycin. This work demonstrates the promising potential of an SDR strategy in diverse areas of bacterial genetics and, specifically, for selection of mutants hypersusceptible to a wide variety of antimicrobial agents.
MATERIALS AND METHODS
Strains, media, and transformation.
Escherichia coli strain JM109 and Acinetobacter sp. strain ADP1, alternatively designated as Acinetobacter calcoaceticus strain BD413 (ATCC 33305), were grown in Luria-Bertani (LB) medium or on LB agar at 37°C. Antibiotics were used at the following concentrations (in micrograms per milliliter): for E. coli, ampicillin at 100, kanamycin (KAN) at 50, and spectinomycin-streptomycin (SPT-STR) at 50 and 10; and for Acinetobacter sp. strain ADP1, KAN at 12.5 and SPT-STR at 50 and 10. For transformation of Acinetobacter sp., an overnight culture was diluted 50-fold into fresh LB medium and grown for 2 h, at which point DNA was added for 1 h and the cells were plated on antibiotic-containing agar plates (22). DNA released into the medium by lysed bacteria was isolated as follows. Cells were removed from a 5-ml culture by centrifugation, and the medium was supplemented with 3 g of guanidinium chloride and 1 ml of Wizard PCR-purification silica resin (Promega). After 1 min, the resin was collected in a Promega mini-column and washed with washing solution (Promega), and DNA bound to the resin was eluted with 65°C water. The amount of DNA was measured by using Picogreen dsDNA quantitation reagent (Molecular Probes). MICs of antibiotics for Acinetobacter sp. were determined by broth microdilution test according to the methods of the National Committee for Clinical Laboratory Standards (20) or by replica plating logarithmically growing cells on LB agar plates supplemented with different concentrations of antibiotics.
Genetic constructs.
To create pTnLysKm, a lysis cassette consisting of the S, R, and Rz genes was amplified from λ DNA without their own promoter sequence with the primers LysDir (GACTATCGATTGGGGGTAAGACTAGAAGATGC), containing a ClaI site (underlined), and LysRev (ATCGATATGGTCGACTCTATCTG), containing a SalI site. nptII (Kmr) was amplified from transposon Tn5 with the primers KmDir (ATAAGAACGCGTCGACAGCAAGCGAACCGGAATTG), containing a SalI site, and KmRev (GAGAATCGATGAGTCCCGCTCAGAAGAAC), containing a ClaI site. The lysis cassette and nptII were ligated after digesting with SalI, followed by PCR amplification with the primers LysDir and KmRev. The resulting PCR product was cloned into the ClaI site of pMOD (Epicentre Technologies) to flank it with 19-bp repeats recognizable by Tn5 transposase. To create pAlkMLysKm, alkM was amplified from Acinetobacter genomic DNA with the insertion of a MluI site in the middle of the gene and cloned into pUC18, yielding pAlkM. A lysis cassette with a Kmr marker was amplified from pTnLysKm with the primers LysMlu (ATCCACGCGTTGGGGGTAAGACTAGAAGATGC) and KmMlu (GAGAACGCGTGAGTCCCGCTCAGAAGAAC) and cloned into the MluI site of pAlkM. The resulting plasmid, in which lysis cassette genes were oriented in the same direction as alkM, was linearized by digestion with ScaI within the vector region and used for natural transformation of Acinetobacter strain ADP1, thus placing the lysis cassette under the control of the chromosomal alkM promoter. To create pTnΩ, a Ω cassette containing a Sm-Spec resistance gene (24) was released from pUI1638 (kindly provided by Ellen Neidle, University of Georgia, Athens) by digesting with ClaI and XbaI and cloned between the ClaI and XbaI sites of pMOD. To construct strain LIT1acrBAc::Ω, acrBAc, a homolog of acrB of E. coli, was amplified from Acinetobacter genomic DNA with the primers AcrBDir (GCGCAATATCCGACGATT) and AcrBRev (CCAGACTCACGTTCTTCA) and the Ω cassette was inserted into the obtained PCR product by in vitro transposition of TnΩ, using the procedure recommended by Epicentre Technologies (www.epicentre.com/item.asp?ID=283&CatID=11&SubCatID=45). The resulting DNA was used to disrupt acrBAc in strain LIT1 by homologous recombination. The insertion of the Ω cassette into chromosomal acrBAc was confirmed by PCR analysis.
Construction of insertional mutant libraries in Acinetobacter strain ADP1.
A library of Ω cassette insertional mutants (∼34,000 clones) in Acinetobacter strain ADP1 was generated by using a transposome technique (7). In brief, transposomes were formed by incubating TnΩ (200 ng/μl), released by PvuII digestion from pTnΩ, with 1 U of modified Tn5 transposase (Epicentre Technologies) for 30 min at 37°C in 20 μl of 50 mM Tris-acetate-200 mM potassium glutamate (pH 7.5), transferred by gel filtration into 5 mM Tris-acetate (pH 7.5), and electroporated into Acinetobacter strain ADP1. To generate an insertional library of lysis cassette flanked with a Kmr marker, TnLysKm was amplified from pTnLysKm with the primers PMODDir (GGGCCTCTTCGCTATTAC) and PMODRev (CGCGTTGGCCGATTCATT), combined with transposase, and used for in vivo transposome mutagenesis as described above, yielding ∼11,000 KAN-resistant clones. Strain LIT1 selected from this TnLysKm library (see Results) served as a host for construction of a double insertion mutant library, which was obtained by natural transformation of strain LIT1 with genomic DNA isolated from the library of TnΩ insertional mutants.
Northern blotting and sequence analysis.
Total RNA was isolated from Acinetobacter strain ADP1 by the standard procedure developed for E. coli (1), separated by 1% agarose gel electrophoresis with formaldehyde (27), and transferred to nylon membrane. DNA probe labeled with 32P was prepared with the 431-bp PCR product of the Rz gene amplified from λ DNA by using the Prime-a-Gene labeling kit (Promega). In order to determine the sites of transposome insertions, genomic DNA isolated from mutant cells by the standard procedure developed for E. coli (1) was sequenced directly, without additional subcloning, with primers complementary to the ends of transposomes by using the fmol DNA cycle sequencing system (Promega). Obtained 30- to 80-bp-long sequences were searched in the Acinetobacter genome database that was made available to us by Michael Fonstein (Integrated Genomics Inc., Chicago, Ill.), using the ERGO database interface (http://wit.integratedgenomics.com). An alternative version of the genome of the same strain is available at the website of Institut Pasteur (http://www.genoscope.cns.fr/externe/English/Projets/Projet_DY/DY.html).
RESULTS
Simple SDR strategy: basic principles.
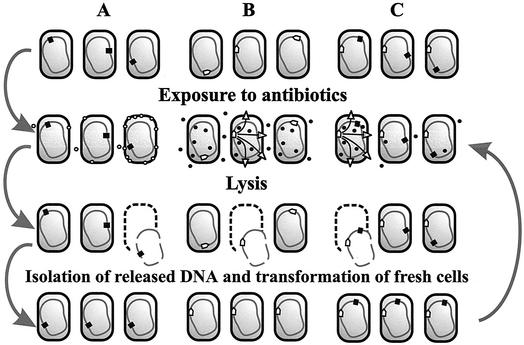
In its simplest form, the SDR strategy consists of the following steps (Fig. 1A). A library of mutants is obtained in which mutations are caused by random insertions of an antibiotic resistance marker. This library is subjected to experimental conditions causing the lysis of desired mutant cells but not of wild-type cells. As shown in Fig. 1A, this condition is an exposure to a low concentration of a bacteriolytic antibiotic. DNA released by bacteria dying under these conditions are collected from the medium (hence the name of the strategy, selection for DNA release). This DNA is then used to transform fresh bacteria with the selection for insertion of the marker gene. In transformants, the marker inserts by homologous recombination into the same position of the chromosome as in the original library and disrupts the same genetic locus, generating the same hypersusceptibility phenotype. Marker insertions that cause cell lysis under experimental conditions are significantly enriched in the obtained library of transformants, compared to the original library. To continue the process of enrichment, the new library is subjected to the same experimental treatment, released DNA is collected, and selection cycles are repeated until desired mutants begin to dominate in the library.
FIG. 1.
SDR strategy. (A) The simplest form of SDR, in which bacteria mutagenized by random insertion of a marker gene (black square) are selected for hypersusceptibility to agents causing bacterial lysis, such as β-lactam antibiotics (open circles). For the ease of presentation, hypersusceptible mutants are presumed to accumulate more of the lytic agent, although the exact mechanism of hypersusceptibility can be different. (B) Selection of a strain that lyses in response to a drug (filled circles). A promoterless lysis cassette of bacteriophage λ fused with the selectable marker (white arrow) is randomly inserted into the chromosome. In clones where it is inserted under the control of a drug-activated promoter, the presence of the drug causes cell lysis. The SDR procedure selects such clones. (C) Use of SDR for isolation of mutants hypersusceptible to a drug that does not cause bacterial lysis. The procedure is essentially identical to the one shown in panel A, except that a lysis reporter strain, obtained as shown in panel B, is used as the host strain instead of wild-type bacteria.
By its nature, the SDR strategy selects for cosegregation of a marker and hypersusceptibility phenotype, thus ensuring that only those mutants that acquired this phenotype due to the marker insertion are selected. This allows for easy identification of the genetic cause of the phenotype: direct sequencing of the regions flanking the marker gene in selected mutants identifies the gene involved. Obviously, the SDR strategy is applicable only to bacterial species with a high efficiency of transformation and homologous recombination. In this work we used one such species, Acinetobacter sp. (strain ADP1).
The simple SDR strategy presented in Fig. 1A is directly applicable to antimicrobial agents that cause cell lysis and the release of DNA. Below, we describe the use of this strategy for selecting mutants susceptible to low concentrations of one such agent, the β-lactam antibiotic ampicillin.
Selection of mutants hypersusceptible to ampicillin.
The library of genetic knockouts of Acinetobacter strain ADP1 was obtained by random insertion of a selectable marker, an Ω cassette carrying a Spec-Sm resistance gene (24), into the chromosome. This was achieved by electrotransformation of bacteria with transposomes (7), complexes of the marker DNA with Tn5 transposase. To enable the Ω cassette to form these complexes, it was flanked with 19-bp inverted repeats that are recognized by the transposase. When transposomes enter the cytoplasm, the transposase activated by intracellular Mg2+ ions inserts the marker DNA into a random position of the chromosome (7). The entire library contained ∼34,000 independent SPT-STR-resistant clones, so that a marker was inserted on average every ∼110 bp of the ∼3.6-Mb genome.
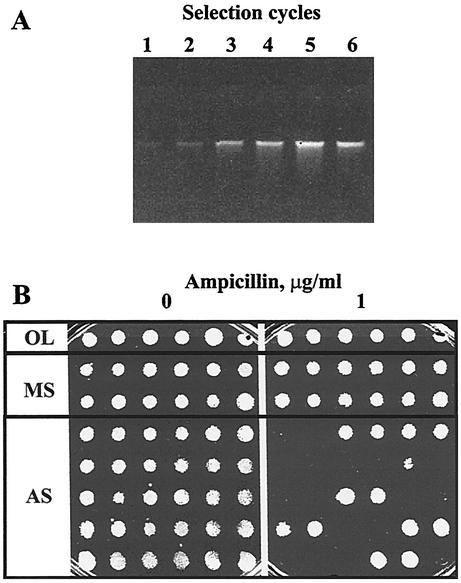
The library was grown to an optical density at 600 nm (OD600) of 0.4 and subjected to a 2-h incubation with a low concentration of ampicillin (3 μg/ml). At this concentration, wild-type bacteria continued to grow and the amount of DNA released into the medium did not differ from the amount released without antibiotic (results not shown). After collecting DNA from the medium by binding to silica resin and transforming fresh Acinetobacter cells, with selection for SPT-STR resistance, a new library of approximately 50,000 clones was obtained, and the selection was repeated. With each cycle of selection, the amount of DNA recovered from the medium was increasing, indicating that selection was indeed happening, until it reached a plateau level at cycle 5 (Fig. 2A). In a mock selection, performed without ampicillin, the amount of released DNA did not rise significantly (data not shown).
FIG. 2.
SDR selection of mutants hypersusceptible to ampicillin. (A) Gel electrophoresis of DNA released in each cycle of selection. (B) Replica plating of randomly chosen colonies from the original library (OL), cycle 5 transformants of the mock selection (MS), and cycle 5 transformants of the ampicillin selection (AS). Colonies were inoculated into 96-well plates, and logarithmic cultures were replica plated, using a 48-pin replicator, on an LB agar plate and an LB agar plate containing ampicillin at 1 μg/ml.
Individual transformants randomly chosen from cycle 5 of ampicillin selection were tested for their sensitivity to ampicillin by replica plating on LB plates and LB plates containing ampicillin at 1 μg/ml. As Fig. 2B demonstrates, approximately half of the transformants failed to grow in the presence of this low concentration of the antibiotic. For comparison, wild-type Acinetobacter under the same conditions grew in up to 16 μg/ml of the drug. As also shown in Fig. 2B, clones randomly picked from cycle 5 of the mock selection did not demonstrate increased sensitivity to ampicillin.
Although the SDR procedure directly selects for genetic linkage between the insertion of the marker gene and the hypersusceptibility phenotype, we tested this linkage for five hypersusceptible mutants. Chromosomal DNA was isolated from these mutants and used to transform wild-type Acinetobacter. As expected, all SPT-STR-resistant transformants demonstrated increased sensitivity to ampicillin. The sites of insertion of the marker gene were determined by direct sequencing of chromosomal DNA of the selected mutants, using outward-oriented primers corresponding to the termini of the Ω cassette. Some of the clones yielded the same sequences, indicating that they represented subclones of the same knockout mutants. Nevertheless, we were able to identify a number of distinct sites of insertion leading to a hypersusceptibility phenotype.
The genes whose disruption caused hypersusceptibility to ampicillin in our experiments included a homolog of the E. coli gene encoding the murein transglycosylase Slt70 protein (13); a homolog of the tail-specific protease Prc, involved in the cleavage of the C terminus of penicillin-binding protein PBP-3 (9); and a number of genes involved in the biosynthesis of exopolysaccharides. These included a homolog of the Serratia marcescens gene wbbL, which encodes rhamnosyl transferase involved in the production of O-antigen (26); a homolog of the E. coli gene galU encoding UTP-glucose-1-phosphate uridylyltransferase (three independent insertions in different positions within the gene); a homolog of a gene encoding glucose-6-phosphate isomerase; and a homolog of the E. coli gene encoding transporter Wza. The latter two enzymes and the transporter are involved in biosynthesis of polysaccharide capsule (4, 6, 10). Three other mutants carried insertions of the Ω cassette in hypothetical genes showing no clear homology to known genes in other bacteria.
The presented data demonstrate that the SDR strategy can successfully select for ampicillin hypersusceptibility mutations and probably for mutations leading to hypersusceptibility to any other agent causing bacterial lysis. Detailed characterization of the obtained mutants is ongoing and is beyond the scope of this publication.
Application of the SDR strategy for selecting mutants hypersusceptible to other antibiotics: basic principles.
Only a limited group of antimicrobial agents cause bacterial lysis. Below, we describe a modification of the SDR approach for selecting mutants hypersusceptible to other antibacterial agents. This modification is based on the work of Kloos et al. (15), who reported that expression of a lysis gene cassette of bacteriophage λ in Acinetobacter causes cell lysis and release of DNA into the medium. We selected a special lysis reporter strain of Acinetobacter in which the lysis cassette of λ was placed under the control of a chromosomal promoter activated by inhibition of protein biosynthesis. The lysis reporter strain served as a host for generating a library of insertional mutants. This library was then used for selecting mutant variants hypersusceptible to a translational inhibitor, the antibiotic erythromycin. Both the creation of a lysis reporter strain (Fig. 1B) and selection of erythromycin-hypersusceptible mutants (Fig. 1C) were performed using SDR, as described below.
Selection of a strain lysing in response to inhibitors of translation.
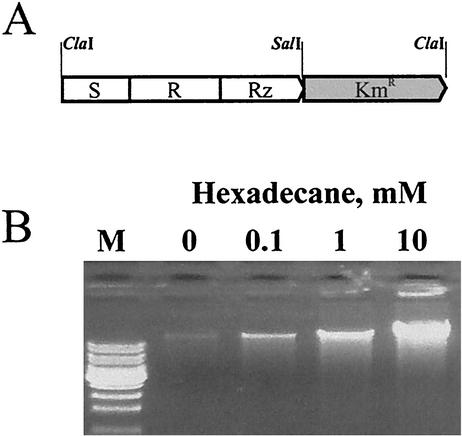
A construct was created in which a cassette of genes of bacteriophage λ causing bacterial lysis was linked to a selectable marker, the Km resistance gene (Fig. 3A). The goal was to insert this cassette randomly into the genome of Acinetobacter and, using SDR, select clones that would lyse in the presence of translational inhibitors. The lysis cassette, containing bacteriophage genes S, R, and Rz (8, 33), was amplified by PCR from λ DNA without its promoter sequence. When the construct is eventually inserted into the Acinetobacter genome, expression of these genes would be controlled by the upstream promoter sequence in the Acinetobacter chromosome. Additionally, the first of the two alternative start codons of the S gene was rendered inactive by nucleotide substitution, thus allowing for production of only an active form of holin, the product of the S gene, while its inhibitory variant, which is two residues longer (8, 33), would not be translated.
FIG. 3.
Artificially created lysis reporter strain of Acinetobacter strain ADP1 responding to hexadecane, an inducer of the alkM gene promoter. (A) Structure of the lysis cassette of bacteriophage λ, devoid of promoter and ligated to a Kmr gene. This construct was inserted into the chromosome of Acinetobacter strain ADP1 under the control of the alkM promoter. (B) Release of DNA from the obtained strain. Cells were grown to an OD600 0.4, and hexadecane was added at the indicated concentrations. After overnight incubation, cells were removed by centrifugation, and DNA released into the medium was isolated as described in Materials and Methods and separated by agarose gel electrophoresis. M, DNA molecular mass markers, with the upper band corresponding to a 10-kB DNA fragment.
The construct was tested for its ability to cause lysis of Acinetobacter cells when inserted into the chromosome under the control of a known inducible promoter. Specifically, it was placed into the coding region of the alkM gene, which is inducible by hexadecane (25). As Fig. 3B illustrates, hexadecane indeed induced cell lysis and the release of a large amount of DNA in the obtained strain, while having no such effect on the wild-type strain (data not shown).
In order to generate random insertions of the lysis cassette into the chromosome of Acinetobacter, it was flanked with 19-bp-long inverted repeats recognizable by Tn5 transposase and used to form transposomes (7) which were electroporated into Acinetobacter cells. The library of KAN-resistant transformants, consisting of ∼11,000 clones, was then subjected to treatment with a translational inhibitor, the antibiotic tetracycline, at its MIC (0.3 μg/ml). After 2 h of incubation, cells were removed by centrifugation and the released DNA was isolated from the medium.
This DNA was used to transform a fresh batch of Acinetobacter, and the obtained KAN-resistant transformants were again subjected to treatment with tetracycline. After four such selection cycles, the library began to display noticeable lysis upon treatment with tetracycline, and the amount of released DNA rose dramatically. At this point, the library was subcloned and a number of clones were tested for the release of DNA in the presence of tetracycline. Chromosomal DNA from clones displaying significant tetracycline-induced DNA release was directly sequenced to determine the location of the inserted lysis cassette. Two different groups of such clones were identified. In one group, the lysis cassette was inserted into the regulatory region of an Ilv operon, which displays strong homology to known Ilv operons encoding acetolactate synthase and ketol acyl reductoisomerase, enzymes involved in biosynthesis of branched-chain amino acids (31). In the other group, which demonstrated an even higher level of induction by tetracycline, the lysis cassette inserted into the regulatory region of one of the Acinetobacter rRNA operons, immediately downstream from the putative P2 promoter (5).
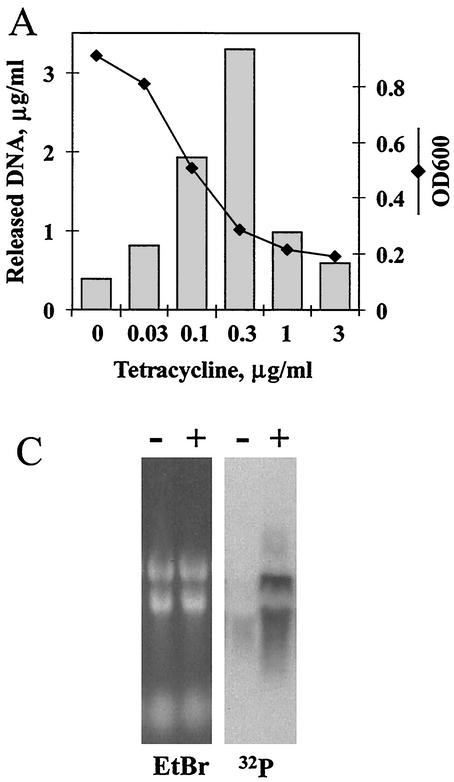
One of the clones of this latter group, designated LIT1 (lysis from inhibition of translation), was studied further. Figure 4A shows that the amount of DNA released by this clone depends on the concentration of tetracycline: it rises with an increase in antibiotic concentration and, after reaching a peak value at approximately the MIC of tetracycline, goes down at higher concentrations. This decline presumably occurs because at high concentrations tetracycline inhibits production of bacteriophage proteins causing cell lysis. It should be noted that during 2 h of incubation with tetracycline at its MIC, only a small fraction of LIT1 cells underwent lysis. Longer incubations (6 h and more) resulted in nearly complete lysis of LIT1 cells (data not shown).
FIG. 4.
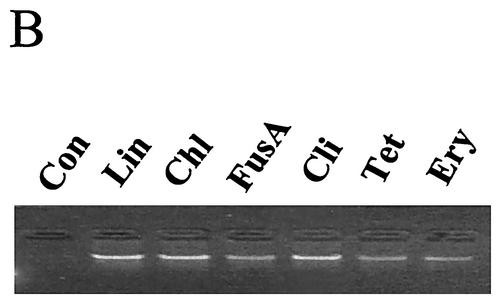
Properties of strain LIT1 of Acinetobacter strain ADP1 obtained by SDR. The lysis cassette in this strain is under transcriptional control of rRNA operon promoters. (A) Release of DNA from LIT1 cells induced by tetracycline. Cells at an OD600 of 0.2 were grown for 2 h with the indicated concentrations of tetracycline. Culture densities (OD600 [diamonds]) and the amount of released DNA (bars) are indicated. (B) Induction of DNA release from LIT1 cells by translational inhibitors. Antibiotics were added to exponentially growing LIT1 cells at an OD600 of 0.4 at their respective MICs determined for wild-type Acinetobacter. DNA was isolated from the medium after 2 h of incubation and separated by agarose gel electrophoresis. Con, control; Lin, linezolid; Chl, chloramphenicol; FusA, fusidic acid; Cli, clindamycin; Tet, tetracycline; Ery, erythromycin. (C) Northern blot analysis of the expression of the lysis cassette in strain LIT1 incubated for 20 min either without (−) or with (+) erythromycin at 3 μg/ml. The gel stained with ethidium bromide (EtBr) shows that the amounts of RNA in both lanes were equal. The Northern blot hybridization (32P) with radiolabeled probe corresponding to the Rz gene reveals erythromycin-induced overexpression of the lysis cassette.
DNA release in the LIT1 clone was induced not only by tetracycline but also by all tested inhibitors of translation, such as linezolid, chloramphenicol, fusidic acid, clindamycin, and erythromycin (Fig. 4B), but not by antibacterials affecting other processes, such as ethidium bromide or ciprofloxacin (data not shown). None of these antibiotics induced lysis and DNA release in wild-type Acinetobacter.
As expected, the induction of lysis in the LIT1 strain was caused by increased transcription of the lysis cassette (Fig. 4C). This result suggests that inhibition of protein synthesis induces a compensatory increase in the transcription of the rRNA operon. Detailed analysis showed that the insertion of the lysis cassette in LIT1 altered the −10 region of the P2 promoter and thus presumably inhibited constitutive expression from P2, leaving the lysis cassette under transcriptional control of the P1 promoter located ∼150 bp upstream of P2 and strongly responding to inhibition of translation. Indeed, primer extension analysis confirmed that the tetracycline-induced expression of the lysis cassette originated from the P1 promoter. Furthermore, the induction of expression of rRNA operons by tetracycline and erythromycin was confirmed by primer extension analysis of rRNA in wild-type cells (data not shown).
The molecular mechanism of this compensatory response of the P1 promoter to inhibition of translation is currently under investigation. Regardless of the mechanism, however, the LIT1 strain could be used as a lysis reporter strain for isolation of mutants of Acinetobacter hypersusceptible to translational inhibitors through the use of the SDR approach.
Selection of erythromycin-hypersusceptible mutants.
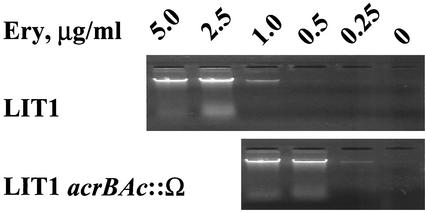
To verify that antibiotic-hypersusceptible mutants of strain LIT1 would indeed release more DNA into the medium at a low concentration of antibiotic, we artificially created such a mutant by disrupting the gene of a putative multidrug efflux pump, AcrAB. AcrAB of E. coli, composed of the integral membrane protein AcrB and periplasmic protein AcrA, is known to transport erythromycin out of bacterial cells, thus significantly reducing its effectiveness (21, 30). Homology search of the Acinetobacter genome identified an operon encoding highly homologous proteins (46 and 56% sequence identity, respectively), which we designated AcrAAc and AcrBAc. We disrupted the acrBAc gene by inserting an Ω cassette carrying a Spec-Sm resistance determinant (24). This led to an increased sensitivity of Acinetobacter to erythromycin: its MIC for this strain was 0.5 μg/ml, instead of 3 μg/ml for wild-type bacteria.
After transferring this gene disruption into the LIT1 strain by homologous recombination, the resulting strain, LIT1acrBAc::Ω, was tested for release of DNA at different concentrations of erythromycin. As expected, release of DNA in this strain was observed at approximately 5-times-lower concentrations of the antibiotic than in strain LIT1 (Fig. 5). This experiment suggested (i) that insertion of the Ω cassette into any other gene in LIT1 that results in an increased sensitivity to erythromycin (reduced MIC) would cause release of DNA from these cells at a low concentration of the drug and (ii) that such an insertion would be picked up and amplified by the SDR procedure.
FIG. 5.
Shift in the concentration dependence of erythromycin-induced DNA release in strain LIT1 upon disruption of the drug efflux pump gene acrBAc. The experiment was performed essentially as described in the legend to Fig. 3B.
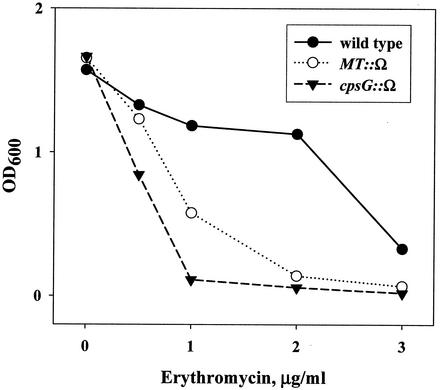
In order to create a library of random insertions of the Ω cassette in strain LIT1, the total chromosomal DNA isolated from the library of the Ω cassette in wild-type Acinetobacter was used to transform strain LIT1, yielding approximately 106 transformants resistant to SPT and STR. This library was then subjected to treatment with a low concentration of erythromycin (0.5 μg/ml). Released DNA was collected and used to transform fresh LIT1 cells. After completing four cycles of the selection procedure, DNA released into the medium was transformed into wild-type Acinetobacter sp. cells with selection for Spec-Sm resistance. This was done to eliminate any potential mutations that might cause enhanced lysis of strain LIT1 by affecting the regulatory mechanism controlling the expression of the lysis cassette rather than by increasing sensitivity to erythromycin. The MIC of erythromycin was determined for 92 randomly chosen clones and was found to be significantly reduced in 15 of them. Sequencing of the regions of the chromosome flanking the inserted Ω cassette in these clones identified three variants. In one group of clones, displaying a two- to threefold reduction of the erythromycin MIC, the Ω cassette inserted into a 23S rRNA gene and presumably made cells hypersensitive to erythromycin by reducing the number of ribosomes in the cell. In another group, displaying an approximately fourfold reduction of erythromycin MIC and 50% inhibitory concentration (IC50) values (Fig. 6), the insertion occurred in a gene encoding a putative membrane transporter that displays 12 transmembrane domains by TMpred analysis (available at the Expasy Molecular Biology server proteomic tools [http://us.expasy.org/tools]) and demonstrates homology to uncharacterized transporters in other bacterial species. Finally, in the third group of clones, displaying fivefold reductions of the erythromycin MIC and IC50 (Fig. 6), the Ω cassette inserted into a gene closely homologous to the E. coli gene cpsG, which encodes phosphomannomutase, an enzyme involved in the production of exopolysaccharide (18).
FIG. 6.
Increased erythromycin sensitivity of the two mutants isolated by SDR. Wild-type and mutant strains of Acinetobacter strain ADP1 were inoculated at an OD600 of 0.01 at different concentrations of erythromycin, and their ODs after 18 h of incubation were determined.
The work on characterization of these mutations and identification of other hypersusceptibility mutations is ongoing. The results presented here demonstrate the relative ease with which such mutations can be selected using the SDR approach.
DISCUSSION
The SDR strategy described in this work is conceptually different from the majority of other selection schemes. Instead of selecting surviving cells, it selects marker insertions into the chromosome that cause a specific phenotype. By its nature, the applicability of SDR is limited to bacterial species with a high rate of transformation and homologous recombination, such as the Acinetobacter strain used in this work. Potentially, it can also be applied to Streptococcus pneumoniae and several other highly transformable organisms. In spite of this limitation, this technique can be useful in a variety of tasks in bacterial genetics.
For example, as illustrated here, the SDR strategy allows for identification of promoters responding to particular stimuli. Using a library of random insertions of the lysis cassette, we selected strains that lyse in the presence of translational inhibitors. This selection identified two promoters that are activated upon inhibition of translation: those of the Ilv operon and of the rRNA operon. The mechanisms of this activation are currently being investigated. Activation of transcription of the rRNA operon by translational inhibitors such as chloramphenicol and SPT has previously been reported (2, 28).
More generally, the SDR strategy can be employed for identification of a variety of conditional lethal mutations. It is directly applicable to situations in which mutants lyse and release DNA under certain conditions. However, as we demonstrate here, SDR can be used even under conditions at which lysis does not normally occur. This can be achieved by following the approach described here: first, selection of a lysis reporter strain responding to a particular condition (chemical agent, temperature, medium composition, etc.) and then selection of mutants hypersensitive to this condition. Alternatively, DNA used for transformation can be isolated directly from dead bacteria if methods are developed to separate them from live bacteria either on the basis of altered physicochemical properties (e.g., buoyant density, staining with fluorescent dyes, etc.) or biosynthetic properties (e.g., failure to produce green fluorescent protein, or presence of a surface antigen). In this work we demonstrated that the SDR strategy works, and we anticipate that other researchers may find it useful in a variety of applications in bacterial genetics.
Here we used the SDR strategy with a practical purpose in mind: to identify antibiotic hypersusceptibility mutations, which can lead to the development of medically useful antibiotic potentiators as well as reveal previously obscure aspects of antibiotic action. Importantly, some of the identified mutations could have been predicted from the existing literature. For example, we have found that knock out of a gene encoding murein transglycosylase Slt70 leads to ampicillin hypersusceptibility. Previously it has been demonstrated that an inhibitor of Slt70, bulgecin, increases sensitivity of E. coli to another β-lactam antibiotic, cefoxitin (16). Similarly, disruption of the prc gene, which in our experiments also led to ampicillin hypersusceptibility, was found to increase susceptibility of E. coli to a number of antibiotics (29). The majority of ampicillin hypersusceptibility mutations discovered by SDR selection to date affect genes involved in the production of exopolysaccharides. Earlier, it was reported that a mutant of a close relative of Acinetobacter, S. marcescens, with a defect in O-antigen production, also demonstrates ampicillin hypersusceptibility (23). It is important to note, however, that these parallels with previously published results have become apparent only in hindsight, and the exact nature of the mutations selected by SDR could hardly be predicted. The advantage of the SDR strategy is that it allows for identification of mutants with the strongest levels of hypersusceptibility, thus directly elucidating the most promising targets for antibiotic potentiators.
In addition to antibiotics, the SDR strategy can be applied for selection of mutants hypersusceptible to other antimicrobial agents, such as disinfectants, as well as mutants hypersusceptible to human serum. The latter mutants may potentially identify targets for a new class of antimicrobial drugs that synergize with antimicrobial defense mechanisms in the serum, such as the complement system. The work on SDR selection and characterization of a wide range of Acinetobacter mutations leading to hypersusceptibility to antimicrobial agents is ongoing.
Acknowledgments
We thank Ellen Neidle for providing us with a plasmid used in this work and helpful advice and Penelope N. Markham for critical reading of the manuscript. We are especially grateful to Michael Fonstein and research staff of Integrated Genomics, Inc. for providing us with access to the ERGO database and Acinetobacter genome information.
This work was supported by National Institutes of Health grant AI 49214.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1988. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 2.Barker, M. M., and R. I. Gourse. 2001. Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J. Bacteriol. 183:6315-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burtnick, M. N., and D. E. Woods. 1999. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob. Agents Chemother. 43:2648-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, H. Y., J. H. Lee, W. L. Deng, T. F. Fu, and H. L. Peng. 1996. Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb. Pathog. 20:255-261. [DOI] [PubMed] [Google Scholar]
- 5.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummelsmith, J., and C. Whitfield. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 19:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 18:97-100. [DOI] [PubMed] [Google Scholar]
- 8.Graschopf, A., and U. Blasi. 1999. Molecular function of the dual-start motif in the lambda S holin. Mol. Microbiol. 33:569-582. [DOI] [PubMed] [Google Scholar]
- 9.Hara, H., Y. Yamamoto, A. Higashitani, H. Suzuki, and Y. Nishimura. 1991. Cloning, mapping and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J. Bacteriol. 173:4799-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hare, R. S., S. S. Walker, T. E. Dorman, J. R. Greene, L. M. Guzman, T. J. Kenney, M. C. Sulavik, K. Baradaran, C. Houseweart, H. Yu, Z. Foldes, A. Motzer, M. Walbridge, G. H. Shimer, Jr., and K. J. Shaw. 2001. Genetic footprinting in bacteria. J. Bacteriol. 183:1694-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 13.Holtje, J. V. 1996. Lytic transglycosylases. EXS 75:425-429. [DOI] [PubMed] [Google Scholar]
- 14.Judson, N., and J. J. Mekalanos. 2000. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 8:521-526. [DOI] [PubMed] [Google Scholar]
- 15.Kloos, D. U., M. Stratz, A. Guttler, R. J. Steffar, and K. N. Timmis. 1994. Inducible cell lysis system for the study of natural transformation and environmental fate of DNA released by cell death. J. Bacteriol. 176:7352-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraft, A. R., J. Prabhu, A. Ursinus, and J. V. Holtje. 1999. Interference with murein turnover has no effect on growth but reduces beta-lactamase induction in Escherichia coli. J. Bacteriol. 181:7192-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 18.Marolda, C. L., and M. A. Valvano. 1993. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1). J. Bacteriol. 175:148-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurry, L. M., and S. B. Levy. 1987. Tn5 insertion in the polynucleotide phosphorylase (pnp) gene in Escherichia coli increases susceptibility to antibiotics. J. Bacteriol. 169:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 21.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:215-218. [PubMed] [Google Scholar]
- 22.Palmen, R., P. Buijsman, and K. J. Hellingwerf. 1994. Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch. Microbiol. 162:344-351. [Google Scholar]
- 23.Palomar, J., M. Puig, R. Montilla, J. G. Loren, and M. Vinas. 1995. Lipopolysaccharide recovery restores susceptibility levels towards beta-lactams in Serratia marcescens. Microbios 82:21-26. [PubMed] [Google Scholar]
- 24.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 25.Ratajczak, A., W. Geissdorfer, and W. Hillen. 1998. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J. Bacteriol. 180:5822-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubires, X., F. Saigi, N. Pique, N. Climent, S. Merino, S. Alberti, J. M. Tomas, and M. Regue. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, vol. 1, p. 7.21-7.50. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schlessinger, D., M. Ono, N. Nikolaev, and L. Silengo. 1974. Accumulation of 30S preribosomal ribonucleic acid in an Escherichia coli mutant treated with chloramphenicol. Biochemistry 13:4268-4271. [DOI] [PubMed] [Google Scholar]
- 29.Seoane, A., A. Sabbaj, L. M. McMurry, and S. B. Levy. 1992. Multiple antibiotic susceptibility associated with inactivation of the prc gene. J. Bacteriol. 174:7844-7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umbarger, H. E. 1996. Biosynthesis of the branched-chain amino acids, p. 442-457. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 32.Vaara, M. 1993. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 37:2255-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Micriobiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 34.Wong, R. S., L. M. McMurry, and S. B. Levy. 2000. “Intergenic” blr gene in Escherichia coli encodes a 41-residue membrane protein affecting intrinsic susceptibility to certain inhibitors of peptidoglycan synthesis. Mol. Microbiol. 37:364-370. [DOI] [PubMed] [Google Scholar]