Abstract
The coordination of chromatin assembly with DNA replication, which is essential for genomic stability, requires the combined activation of histone deposition with the firing of replication origins. We report here the direct interaction of chromatin assembly factor 1 (CAF1), a key factor involved in histone deposition, with the replication kinase Cdc7-Dbf4. We isolated a complex containing both the largest subunit of CAF1 (p150) and the Cdc7-Dbf4 kinase specifically in S phase and thus prove the existence of this interaction in vivo. We then show that the Cdc7-Dbf4 kinase efficiently phosphorylates p150. This event induces a change in p150 oligomerization state, which promotes binding to proliferating cell nuclear antigen (PCNA). Conversely, CAF1 recruitment is reduced in a PCNA/DNA loading assay using Cdc7-depleted extracts. Our data define p150 as a new target for this kinase with implications for the coordination between DNA replication and CAF1 functions.
Keywords: CAF1, Cdc7-Dbf4, chromatin assembly, replication, PCNA
Introduction
Eukaryotic genomes are organized into chromatin (Kornberg, 1977), the dynamics of which have proved to be of importance in genome function (Vaquero et al, 2003). Histone deposition onto DNA is the first step in the process of chromatin assembly to build up the core particle of chromatin, which consists of 146 bp of DNA wrapped around a histone octamer (H2A–H2B)2(H3–H4)2. Biochemical approaches have identified crucial factors and histone chaperones involved in this process (Mello & Almouzni, 2001; Loyola & Almouzni, 2004). Among these, the conserved chromatin assembly factor 1 (CAF1) composed of three histone-binding proteins, p150, p60 and p48 (Kaufman et al, 1995; Verreault et al, 1996) is the only characterized histone chaperone that promotes nucleosome assembly coupled to DNA synthesis in the context of DNA replication (Smith & Stillman, 1989) and nucleotide excision repair (Gaillard et al, 1996; Green & Almouzni, 2003). In both cases, its targeting to sites of DNA synthesis involves a physical interaction between its largest subunit, p150, and the homotrimeric sliding clamp, proliferating cell nuclear antigen (PCNA), a DNA polymerase accessory factor (Shibahara & Stillman, 1999; Moggs et al, 2000). Thus, the interaction between p150 and PCNA represents an attractive control point (Mello & Almouzni, 2001) that could be exploited for the coordination of CAF1 recruitment with the activation of each replication origin or with an individual repair site. How such regulation can take place remains an open issue. Interestingly, both p150 and p60 are phosphoproteins (Smith & Stillman, 1991) and CAF1 function in vitro can be regulated by cell-cycle kinases/phosphatases (Martini et al, 1998; Keller & Krude, 2000). Moreover, p150 can dimerize in vitro, another property crucial for its function (Quivy et al, 2001). Here, we investigate how such modifications might participate in regulating CAF1 function. We show that Dumb bell former 4 (Dbf4), a regulatory subunit of the cell division cycle 7 (Cdc7) kinase, which is essential for initiation of DNA replication (Bell & Dutta, 2002), interacts directly with and phosphorylates p150 in vitro. This phosphorylation stabilizes p150 as a monomer and favours its interaction with PCNA, which can thus promote CAF1 recruitment at single-strand DNA breaks (intermediates found at replication and repair sites). We discuss the implications of our findings for both CAF1 and Cdc7-Dbf4 functions.
Results
Dimeric and monomeric state of p150
We have previously shown by sedimentation in a glycerol gradient that recombinant p150 can dimerize in vitro and we have defined a domain of 36 amino acids crucial for this dimerization (Quivy et al, 2001; Fig 1A). We now set up an in vitro crosslink assay on purified recombinant p150 and mutant p150Δ (deleted in the region crucial for dimerization) to establish that purified p150 can be found in a monomer–dimer equilibrium (supplementary Fig S1 online). To verify whether p150 could be detected as a dimer in vivo, we generated nuclear extracts from human 293 cells transfected with wild-type (wt) green fluorescent protein (GFP)–p150 or mutant GFP–p150Δ constructs and carried out immunoprecipitation with an anti-GFP antibody. Using p150 antibody, we detected by western blot both endogenous and exogenous p150. Whereas wt GFP–p150 was able to co-immunoprecipitate endogenous p150, the mutant GFP–p150ΔCAF1 could not (Fig 1B). Thus, wt p150 can oscillate between a dimeric and a monomeric state in vivo. Given that p150 is a phosphoprotein in vivo (Smith & Stillman, 1991), we wondered whether such an equilibrium could be regulated by kinase activity.
Figure 1.
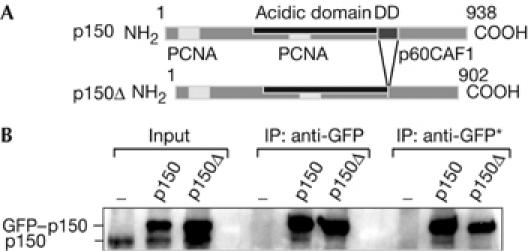
A dimeric state of wild-type p150 in vivo. (A) Scheme of wild-type p150 (p150) and mutant p150 (p150Δ). PCNA, p60 binding sites and the region crucial for dimerization (DD) are indicated. (B) An anti-GFP antibody immunoprecipitated GFP fusion proteins from 293 cell nuclear extracts transiently expressing GFP–p150 or GFP–p150ΔCAF1. Western blot (anti-p150) shows endogenous p150 and exogenous GFP–p150 fusion proteins in the input and immunoprecipitated material. Negative control (−) corresponds to 293 cells transfected with an empty vector. IP anti-GFP and IP anti-GFP* correspond to 0.15 and 0.5 M NaCl washes, respectively. GFP, green fluorescent protein; IP, immunoprecipitation; PCNA, proliferating cell nuclear antigen.
Cdc7-Dbf4 phosphorylates p150
As kinases that can modify p150 have not been identified so far, we used a yeast two-hybrid screen (see supplementary information online), which facilitates the detection of transient interactions in vivo, to search for a kinase interacting with p150. Along with already known p150 interacting partners such as p150 itself, PCNA and histones, we isolated one clone corresponding to Xenopus Dbf4 (xDbf4), a regulatory subunit of Xenopus Cdc7 kinase (xCdc7), which is involved in the initiation of replication (Furukohri et al, 2003). In a yeast two-hybrid assay, activation of reporter genes occurred only with xDbf4 but not with our negative controls (ras and LexA), and not with the smallest subunit of CAF1 (xp48CAF1; supplementary Fig S2A online). We confirmed this interaction with glutathione S-transferase (GST)–xDbf4 (and not GST alone), which pulled down xp150 when coexpressed in bacteria (Fig 2A). Similarly, the human p150 also interacted with xDbf4 (not shown), which indicated the conservation of this interaction from Xenopus to human. We then examined in vitro phosphorylation by the Cdc7-Dbf4 kinase of its known substrate GST–Mcm2 in comparison with p150 (supplementary Fig S2B,C online). The level of p150 phosphorylation by Cdc7-Dbf4 was comparable with that of GST–Mcm2 (supplementary Fig S2C online) and occurred in a dose-dependent manner (Fig 2B). Preliminary attempts using various truncated forms of p150 failed to identify phosphorylation sites on p150, but might indicate that these sites are widely distributed over the protein. This is not surprising given that p150 contains more than 100 serine/threonine residues that could be potentially phosphorylated by Cdc7-Dbf4. Finally, we verified that the p150 subunit was phosphorylated by Cdc7-Dbf4 not only as an individual subunit but also in the whole CAF1 complex, which also contains the p60 and p48 subunits. In this context, only the p150 subunit, but not the p60 and p48 subunits, was phosphorylated, which proves the specificity of the Cdc7-Dbf4 kinase for the p150 subunit (Fig 2C). Taken together, these data demonstrate that p150 is a significant substrate for Cdc7-Dbf4.
Figure 2.
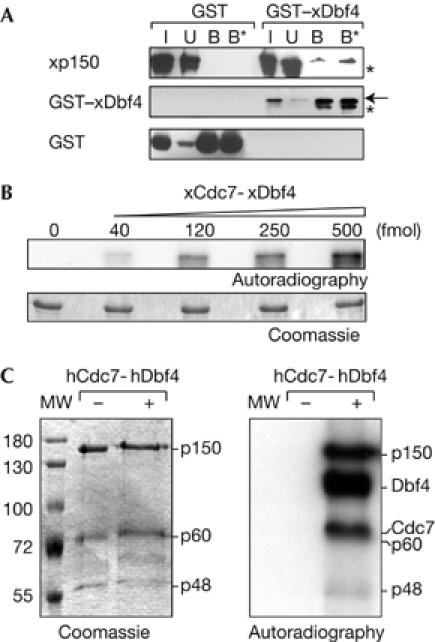
p150 interacts with and is phosphorylated by Cdc7-Dbf4 kinase in vitro. (A) Extract from bacteria coexpressing His6–xp150 with GST–xDbf4 or GST (used as control) was mixed with glutathione resin and pulled-down proteins were analysed by western blot. xp150, GST–xDbf4 and GST in the input (I, 10%), unbound (U) and bound fractions (B and B*, 0.5 and 1 M NaCl washes, respectively) are shown. The arrow indicates the position of full-length GST–xDbf4 and asterisks indicate degradation products. (B) Purified recombinant p150 was phosphorylated with ATPγ32P and increasing amounts of xCdc7-xDbf4 (in fmol) and resolved by SDS–PAGE. Coomassie-stained gel and its autoradiography are shown. (C) The whole CAF1 complex containing the three p150, p60 and p48 subunits was incubated with ATPγ32P without (−) or with (+) hCdc7-hDbf4 and resolved by SDS–PAGE. Coomassie-stained gel and its autoradiography are shown. Molecular weight (MW) markers and migration position of the CAF1 subunits, phosphorylated Dbf4 and Cdc7 are indicated. CAF1, chromatin assembly factor 1; Cdc7, cell division cycle 7 (hCdc7, human Cdc7; xCdc7, Xenopus Cdc7); Dbf4, Dumb bell former 4 (hDbf4, human Dbf4; xDbf4, Xenopus Dbf4); GST, glutathione S-transferase; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
To explore further the connection between Cdc7-Dbf4 and CAF1 in vivo, we determined whether they were present in a common complex within cells. Given that Cdc7-Dbf4 is activated at the onset of S phase, we searched for this complex during that phase. We generated a human 293Flp-In cell line that stably expresses an epitope-tagged p150 (Flag tag). We prepared nuclear extracts from asynchronous cells and from cells synchronized in G1, S and G2/M phases by a thymidine block, a release from this block and a nocodazole block, respectively (Fig 3B) and performed Flag–p150 immunoprecipitation with anti-Flag antibodies. Using western blot analysis with specific antibodies (supplementary Fig S3 online), we detected a p150/Cdc7-Dbf4 complex in the immunoprecipitated material from S-phase cells (Fig 3C). Remarkably, in the corresponding fraction, we also found PCNA (Fig 3C). We then verified that the Cdc7-Dbf4 kinase associated with p150 was active, because, in contrast to a control Flag immunoprecipitation, the Flag–p150 material phosphorylated recombinant Mcm2 (Fig 3D). The association in vivo of p150 with (i) an active Cdc7-Dbf4 kinase and (ii) PCNA in S-phase cells is suggestive of control of CAF1 and PCNA interaction by this kinase.
Figure 3.
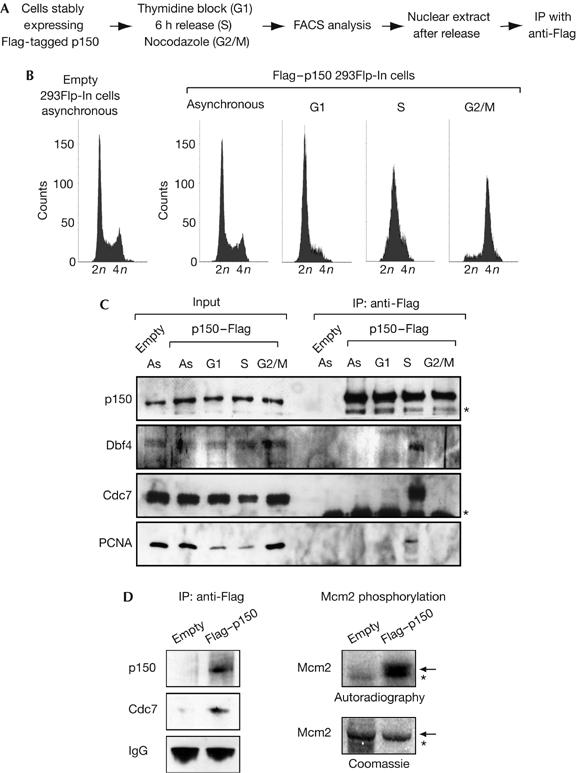
p150 interacts with an active Cdc7-Dbf4 kinase in vivo. (A) Experimental scheme. (B) Flow cytometry profiles of asynchronous 293Flp-In cells stably expressing an empty construct (left), and 293Flp-In cells stably expressing Flag–p150 (right) in either asynchronous or G1, S and G2/M phases (thymidine block, 6 h release and nocodazole block, respectively). (C) p150, Dbf4 and Cdc7 form a complex during S phase. p150 complexes were immunoprecipitated with an anti-Flag antibody from nuclear extracts generated from asynchronous 293Flp-In cells stably expressing an empty construct or 293Flp-In cells stably expressing Flag–p150 (right) in either asynchronous or G1, S and G2/M phases as in (B). Western blot shows p150, Dbf4, Cdc7 and PCNA in the extract (Input, 5%) and in the immunoprecipitated material. Asterisks indicate nonspecific or degradation products (p150 blot) and IgG chains (Cdc7 blot). (D) The Cdc7-Dbf4 co-immunoprecipitated with p150 is active. Left: western blot shows the Cdc7–p150 complex immunoprecipitated as above. IgG heavy chains are used as controls. Right: autoradiography and Coomassie staining of recombinant hMcm2 (arrow) phosphorylated with ATPγ32P by the Cdc7–p150 complex. Flag immunoprecipitate from cells stably expressing an empty construct is used as a negative control. Asterisk indicates a contaminant. Cdc7, cell division cycle 7; Dbf4, Dumb bell former 4; PCNA, proliferating cell nuclear antigen.
Cdc7-Dbf4 maintains p150 as a monomer
We next decided to test the potential impact of phosphorylation by Cdc7-Dbf4 on the p150 oligomerization state using an in vitro crosslinking assay (supplementary Fig S1 online). To discriminate between an effect due to phosphorylation by any other kinase (charge effect), we tested in parallel cyclin-dependent kinase 2 (Cdk2)-Cyclin A. This was previously shown to phosphorylate p60 in vitro (Keller & Krude, 2000), which interacts with and phosphorylates p150 in vitro (supplementary Fig S4 online). We phosphorylated purified p150 in vitro using hCdk2-hCyclin A or xCdc7-xDbf4 and added increasing concentrations of crosslinking reagent (Fig 4A). Both unphosphorylated and Cdk2-Cyclin A-phosphorylated p150 provided a typical dimer–monomer pattern (Fig 4B, black and white arrowheads). In contrast, the specific phosphorylation of p150 by Cdc7-Dbf4 gave rise to a single band migrating as a monomer (Fig 4B, white arrowhead). To confirm further that the phosphorylation is crucial for stabilizing p150 as a monomer, we generated a xCdc7 kinase-dead mutant. Sequence alignment of several kinases including Cdc7 identified a conserved D residue in the kinase subdomain VII (Hanks & Hunter, 1995). This residue, conserved from yeast to human at position 165 in xCdc7, was replaced by an N to generate the xCdc7 kinase-dead mutant (D165N), which was unable to phosphorylate p150, Dbf4 and itself (supplementary Figs S4C,5 online). In contrast to the wt Cdc7-Dbf4, the kinase-dead Cdc7(D165N)-Dbf4 failed to stabilize p150 as a monomer, and provided the typical dimer–monomer pattern that was identical to the control or a Cdk2-Cyclin A phosphorylation (Fig 4C). Thus, we conclude that although both Cdc7-Dbf4 and Cdk2-Cyclin A kinases phosphorylate p150, only phosphorylation by Cdc7-Dbf4 stabilizes p150 as a monomer.
Figure 4.
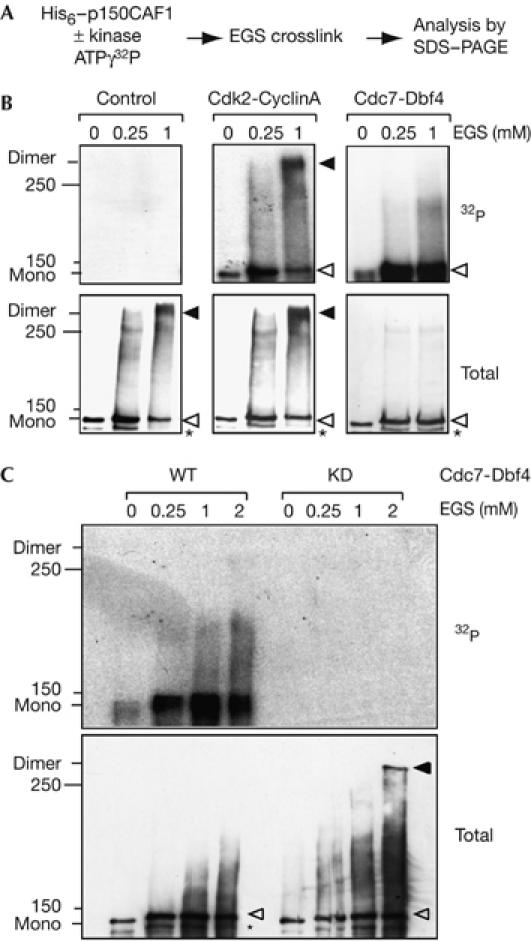
Specific phosphorylation of p150 by Cdc7-Dbf4 stabilizes p150 as a monomer. (A) Scheme: p150 was phosphorylated with ATPγ32P using either hCdk2-hCyclin A or xCdc7-xDbf4, crosslinked with EGS and subjected to SDS–PAGE to resolve monomeric and dimeric forms of p150. (B) Monomer and dimer of p150 indicated by white and black arrowheads, respectively, are shown by autoradiography (top, 32P) and western blot (bottom, total). As a control, kinase was omitted. Asterisk denotes degradation products, and the position of 150 and 250 kDa molecular weight (MW) marker is indicated. (C) Analysis as in (B) using either wild-type Cdc7-Dbf4 (WT) or the kinase-dead Cdc7(D165N) mutant (KD). Cdc7, cell division cycle 7 (xCdc7, Xenopus Cdc7); Dbf4, Dumb bell former 4 (xDbf4, Xenopus Dbf4); EGS, ethylene glycol bis[succinimidyl succinate]; hCdk2, human cyclin-dependent kinase 2; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
Cdc7-Dbf4 promotes interaction of p150 with PCNA
As the association of Cdc7-Dbf4 kinase with p150 occurs in S-phase cells, we examined the impact of Cdc7-Dbf4 activity on the interaction of p150 with PCNA, its key partner during S phase (Fig 3C). First, we asked whether a Cdc7-Dbf4- and/or Cdk2-Cyclin A-dependent phosphorylation could promote interaction of purified p150 with GST–PCNA in vitro (Fig 5A). Under these in vitro conditions, phosphorylation of p150 by Cdc7-Dbf4 readily stimulated interaction with GST–PCNA but not with GST alone (Fig 5A, right, compare lanes 6 and 9). By contrast, phosphorylation by Cdk2-Cyclin A failed to promote such interaction (Fig 5A, lane 5). Second, to strengthen these observations, we used S100 extracts, which contain native PCNA and are competent for nucleosome formation when complemented with p150 (Kaufman et al, 1995). We found that after Cdc7 immunodepletion (Fig 5B, right), binding of PCNA to p150 was severely reduced compared with a mock depletion (three- to fourfold, not shown; Fig 5B, left, compare lanes 5 and 6). Importantly, binding of PCNA to p150 was restored after complementation of the depleted extract with wt xCdc7-xDbf4 (Fig 5B, lane 7) but not with the kinase-dead xCdc7(D165)-Dbf4 (Fig 5B, lane 8). This is in agreement with the absence of interaction between p150 and PCNA in the presence of Cdc7-Dbf4 but under conditions that prevent phosphorylation events (supplementary Fig S6 online). We thus conclude that the sole interaction of Cdc7 with p150 is not sufficient, and that active phosphorylation events involving Cdc7 are required for p150–PCNA interaction. Given that after phosphorylation by Cdc7, p150 is stabilized as a monomeric form, we tested whether interaction of p150 with PCNA is increased when p150 is present as a monomer. We used either wt p150, which can oscillate between a monomeric and dimeric state (supplementary Figs S1, S4 online), or the constitutive monomeric p150Δ mutant (supplementary Fig S1 online) to pull down PCNA from an S100 extract. In contrast to the wt p150, the interaction of monomeric p150Δ mutant with PCNA was resistant to 500 mM NaCl (Fig 5C, compare lanes 7 and 12). This indicates that the interaction between PCNA and p150 is more stable when p150 is in a monomeric conformation. Taken together, our data show that Cdc7-Dbf4 phosphorylation stabilizes p150 in a monomeric state and facilitates its interaction with PCNA. Given that the interaction between p150 and PCNA mediates recruitment of CAF1 at sites of DNA synthesis, we decided to monitor the role of Cdc7-Dbf4 in this process. To address this issue, we used a PCNA/DNA loading assay in which DNA immobilized on paramagnetic beads is used as a template for PCNA loading and CAF1 recruitment (Moggs et al, 2000; Mello et al, 2004). Given that nicks provide reaction intermediates, which efficiently trigger PCNA loading, we used nicked DNA molecules generated by limited DNase-1 digestion (Moggs et al, 2000). We performed our assay with mock- or Cdc7-depleted S100 extracts complemented with wt Cdc7-Dbf4 or kinase-dead Cdc7(D165N)-Dbf4, as in Fig 5B. Remarkably, after 90 min of reaction, similar amounts of PCNA were loaded onto DNA for each condition. In contrast to the mock-depleted extract, only traces of p150 were detected in the Cdc7-depleted extract (Fig 5D, lane 2). Importantly, recruitment of p150 was restored after complementation with purified recombinant wt xCdc7-xDbf4 (Fig 5D, lane 3) but not with kinase-dead xCdc7(D165N)-xDbf4 (Fig 5D, lane 4). We thus conclude that a phosphorylation event mediated by Cdc7-Dbf4 is crucial for the recruitment of p150 onto DNA-bound PCNA. Given that p150 recruitment to PCNA is required to couple DNA synthesis and nucleosome formation (Shibahara & Stillman, 1999; Moggs et al, 2000), we decided to test whether the presence of Cdc7-Dbf4 had an impact on nucleosome formation mediated by CAF1, using a supercoiling assay (Gaillard et al, 1996). Importantly, a single nick on a circular molecule is sufficient to initiate nucleosome assembly in a supercoiling assay (Gaillard et al, 1997). Thus, a single PCNA–p150 loading event is required for the propagation of a wake of nucleosome assembly. As the Cdc7 depletion is not complete (Fig 5B), a limited amount of p150 recruited onto DNA could be sufficient to propagate histone deposition and generate supercoiled molecules. Nevertheless, when using a limiting amount of CAF1, a modest but reproducible decrease in supercoiling is observed if Cdc7-Dbf4 is depleted from the extract (supplementary Fig S7 online). We conclude from these data that the initial step in the control of CAF1 function—that is, p150–CAF1 recruitment onto DNA-bound PCNA, is controlled by Cdc7-Dbf4.
Figure 5.
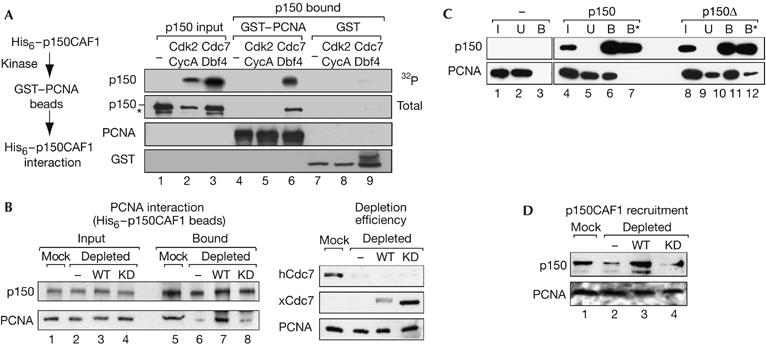
Cdc7-Dbf4 specifically promotes p150–PCNA interaction. (A) Left: experimental scheme. Right: GST–PCNA or GST beads were used to pull down p150, non-phosphorylated (−) or phosphorylated with ATPγ32P by hCdk2-hCyclin A or xCdc7-xDbf4. p150 in the input (5%) and bound (25%) fractions is shown by autoradiography (32P) and western blot (total). Amounts of bead-linked GST–PCNA and GST were equivalent in each condition. Asterisk denotes degradation products. (B) Cdc7 activity favours p150–PCNA interaction in the S100 extract. Left: p150 pull-down assay. p150 beads were incubated with S100 extracts that were mock depleted (Mock), Cdc7 depleted (−) or Cdc7 depleted and complemented with wild-type (wt) xCdc7-xDbf4 (WT) or with the kinase-dead Cdc7(D165N) mutant (KD). Western blot shows pulled-down PCNA and immobilized p150 in the input (20%) and bound fractions. Right: control of Cdc7 depletion and complementation of the S100 extract. Equivalent amounts of S100 extracts (6 μg) of mock, depleted (−) or depleted and complemented with wt xCdc7-xDbf4 or the kinase-dead xCdc7(D165N) (KD) were subjected to SDS–PAGE. Efficiency of depletion (hCdc7) and complementation with wt xCdc7 (WT) or kinase-dead xCdc7(D165N) (KD) is verified by western blot. PCNA is used as a loading control. (C) Pull-down of PCNA from the S100 extract by wt p150 (p150) or monomeric mutant p150Δ beads (p150Δ). Western blot analysis of immobilized p150 and pulled-down PCNA in the input (I, 16%), unbound (U) and bound (B and B*) fractions is shown. B and B* represent resistant material after 150 and 500 mM NaCl washes, respectively. (D) Cdc7 activity promotes p150 recruitment onto DNA in a PCNA/DNA loading assay. DNA beads and purified p150 were incubated with S100 extracts that were mock depleted (−), Cdc7 depleted or Cdc7 depleted and complemented with purified wt xCdc7-xDbf4 (WT) or kinase-dead xCdc7(D165N)-xDbf4 (KD). After incubation for 90 min, DNA-bound PCNA and p150 were detected by western blot. Cdc7, cell division cycle 7 (hCdc7, human Cdc7; xCdc7, Xenopus Cdc7); Dbf4, Dumb bell former 4 (xDbf4, Xenopus Dbf4); GST, glutathione S-transferase; hCdk2, human cyclin-dependent kinase 2; PCNA, proliferating cell nuclear antigen; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
Discussion
In this study, we show the existence of a novel link between CAF1 and Cdc7-Dbf4, a key enzyme involved in the initiation of DNA replication. Our data enable us to propose a model in which p150 modification by Cdc7-Dbf4 controls CAF1 interaction with PCNA, which occurs precisely during S phase when Cdc7-Dbf4 activity is at a maximum. Such a control would participate in the tight coordination between DNA replication and CAF1 function to ensure the accurate perpetuation of genetic and epigenetic information and to avoid an excess of unused histone pools (Groth et al, 2005; Gunjan et al, 2005).
We hypothesize that p150 oscillates between monomeric and dimeric forms. After Cdc7-Dbf4 phosphorylation of p150, the transition towards a monomeric state would favour an interaction with PCNA and thus ensure targeting of CAF1 to sites of DNA synthesis. We have focused our attention here on the initial event—PCNA binding—but it is clear that other conformational transitions are required. First, the necessity of a switch back to a dimeric state for p150 is supported by the fact that the constitutive monomeric p150Δ mutant cannot support histone deposition (Quivy et al, 2001) whereas it binds to PCNA (this work). Second, a protein phosphatase 1 activity, which could favour the transition from a monomeric to a dimeric state of p150, by antagonizing Cdc7-Dbf4 phosphorylation, has been reported to be necessary for CAF1 activity (Keller & Krude, 2000). Thus, one should not consider the monomeric or the dimeric forms of p150 as the ‘active' or ‘inactive' form of p150, but rather consider both conformations to be a requirement for CAF1 activity. Future work should aim at deciphering how the sequence of events that follow the targeting of CAF1 onto DNA can be orchestrated by the successive action of kinases/phosphatase, protein–protein interactions and/or other signals.
Importantly, we should also consider that the impact of Cdc7 in the recruitment of p150 to PCNA at the replication site could occur at a level that is distinct from nucleosome assembly. In mammals, p150 is involved in the deposition of a replicative pool of HP1α during replication of pericentric heterochromatin (Quivy et al, 2004), and a connection between HP1 and Cdc7-Dbf4 has already been reported in Schizosaccharomyces pombe (Bailis et al, 2003). We thus propose that the effect of Cdc7 on p150 might relate to heterochromatin functions. We believe that this work could open up avenues concerning a new role for the Cdc7-Dbf4 kinase in connection to p150 function, with implications for temporal and spatial coordination of nucleosome assembly, DNA synthesis and/or heterochromatin organization in the context of DNA damage or DNA replication.
Methods
Cells. The 293Flp-In cell line stably expressing an epitope-tagged p150–Flag was generated with the Flp-In system (Invitrogen, Carlsbad, CA, USA; supplementary information online). Cells were synchronized in G1 phase by a single thymidine block (5 mM, 20 h). S-phase cells were collected 6 h after release from this block in the presence of 30 μM deoxycytidine. Cells were synchronized in G2/M phase by adding nocodazole (100 ng/ml, 20 h). FACScan and CellQuest software allowed cell-cycle analysis (BD Biosciences, San Jose, CA, USA). p150 and mutant p150Δ complementary DNAs were cloned into the peGFPc2 vector (Clontech Laboratories Inc., Palo Alto, CA, USA) to generate GFP–p150 and GFP–p150Δ expression plasmids. For transfection, we used the Effectene reagent (Qiagen GmbH, Hilden, Germany).
Antibodies, recombinant proteins, pull-down, kinase assays and immunological procedures. See the supplementary information online.
Crosslinking assays. We crosslinked purified His6–p150 and His6–p150Δ (100 ng) for 30 min at 23°C with 0.25, 1 or 2 mM EGS (ethylene glycol bis[succinimidyl succinate]; Pierce Biotechnology, Rockford, IL, USA) and terminated the reaction with 50 mM Tris–HCl (pH 7.5). We then added sample loading buffer and analysed reaction products by SDS–polyacrylamide gel electrophoresis.
PCNA/DNA loading assay. For PCNA/DNA loading assays, we used DNase-1-nicked DNA (300 ng of DNA) linked to paramagnetic beads (Dynal, Oslo, Norway; Mello et al, 2004) and incubated with S100 extracts (120 μg) and purified His6–p150 (75 ng) at 37°C. After washes with 150 mM NaCl, 20 mM Hepes–KOH (pH 7.6), 5 mM MgCl2, 0.1% N-lauroyl sarcosine and 0.05% NP-40, DNA-bound proteins were analysed by western blot.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information and Figures
Acknowledgments
We thank J. Diffley for Dbf4 antibodies, B. Stillman for S100 extracts, T. Hunter and W. Jiang for Cdc7-Dbf4 baculoviruses, A. Verreault for recombinant baculovirus CAF1 complex, J. Walter for xCdc7 antibody, J. Chow, D. Ray-Gallet and S. Polo for comments and P. Grandi for the two-hybrid screen. This work was supported by La Ligue Nationale Contre le Cancer (Equipe labellisée and S.K.), Délégation Générale pour l'Armement (DGA; A.G.), the Commissariat à l'Energie Atomique (CEA; LRC no. 26), Contract RTN (HPRN-CT-2002-00238), PIC Paramètres Epigénétiques, The Epigenome Network of Excellence (NoE) from the European Union (EU; LSHG-CT-2004-503433) and Action Concertée Incitative-Dynamique et Réactivité des Assemblages Biologiques (ACI-DRAB; no. 04393).
References
- Bailis JM, Bernard P, Antonelli R, Allshire RC, Forsburg SL (2003) Hsk1–Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat Cell Biol 5: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Furukohri A, Sato N, Masai H, Arai K, Sugino A, Waga S (2003) Identification and characterization of a Xenopus homolog of Dbf4, a regulatory subunit of the Cdc7 protein kinase required for the initiation of DNA replication. J Biochem (Tokyo) 134: 447–457 [DOI] [PubMed] [Google Scholar]
- Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G (1996) Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86: 887–896 [DOI] [PubMed] [Google Scholar]
- Gaillard P-H, Moggs JG, Roche DMJ, Quivy J-P, Becker PB, Wood RD, Almouzni G (1997) Initiation and bidirectional propagation of chromatin assembly from a target site for nucleotide excision repair. EMBO J 16: 6281–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CM, Almouzni G (2003) Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J 22: 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G (2005) Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell 17: 301–311 [DOI] [PubMed] [Google Scholar]
- Gunjan A, Paik J, Verreault A (2005) Regulation of histone synthesis and nucleosome assembly. Biochimie 87: 625–635 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596 [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B (1995) The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Keller C, Krude T (2000) Requirement of Cyclin/Cdk2 and protein phosphatase 1 activity for chromatin assembly factor 1-dependent chromatin assembly during DNA synthesis. J Biol Chem 275: 35512–35521 [DOI] [PubMed] [Google Scholar]
- Kornberg RD (1977) Structure of chromatin. Annu Rev Biochem 46: 931–954 [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G (2004) Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 1677: 3–11 [DOI] [PubMed] [Google Scholar]
- Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol 143: 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello JA, Almouzni G (2001) The ins and outs of nucleosome assembly. Curr Opin Genet Dev 11: 136–141 [DOI] [PubMed] [Google Scholar]
- Mello JA, Moggs JG, Almouzni G (2004) Analysis of DNA repair and chromatin assembly in vitro using immobilized damaged DNA substrates. Methods Mol Biol 281: 271–282 [DOI] [PubMed] [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G (2000) A CAF-1–PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol 20: 1206–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Grandi P, Almouzni G (2001) Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J 20: 2015–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G (2004) A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J 23: 3516–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, Stillman B (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585 [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58: 15–25 [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B (1991) Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. J Biol Chem 266: 12041–12047 [PubMed] [Google Scholar]
- Vaquero A, Loyola A, Reinberg D (2003) The constantly changing face of chromatin. Sci Aging Knowledge Environ 2003: RE4. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87: 95–104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information and Figures


