Abstract
To identify the independent spatial and temporal activities of the essential developmental gene the Otx2, the germline mutation of which is lethal at embryonic day 8.5, we floxed one allele and substituted the other with an inducible CreER recombinase gene. This makes ‘trans' self-knockout possible at any developmental stage. The transient action of tamoxifen pulses allows time-course mutation. We demonstrate efficient temporal knockout and demarcate spatio-temporal windows in which Otx2 controls the head, brain structures and body development.
Introduction
Although regulatory genes frequently control several developmental steps (Kmita & Duboule, 2003), most loss-of-function studies demonstrate only their earliest roles (Nagy et al, 2003). The successive functions of a given gene and the length of its activity have been inaccessible until recently. In mice, the CreER recombinase system is a promising tool for addressing these questions (Santagati et al, 2005). To disclose the many functions and the corresponding time-windows of an essential gene, Otx2, we have added a twist to CreER-based conditional knockout, to allow spatially and temporally controlled gene invalidation during short developmental periods. The Otx2 homeobox gene has key roles in early antero-posterior patterning (Kimura-Yoshida et al, 2005), neuroectoderm induction and maintenance (Rhinn et al, 1998), and rostral brain and cranio-facial development (Acampora et al, 1995; Matsuo et al, 1995; Ang et al, 1996). Conditional knockout studies using constitutive Cre mice have shown that Otx2 is required around mid-gestation for neuronal specification in the retina (Nishida et al, 2003) and in the ventral midbrain (Puelles et al, 2004; Vernay et al, 2005). To further explore the significance of its persistent expression in many brain structures (Frantz et al, 1994; Mallamaci et al, 1996), we performed time-course self-knockout between embryonic day (E)10.5 and E18.5, delineating the requirement of Otx2 during the later stages of development.
Results
Strategy
Temporally and spatially controlled ablation of the Otx2 gene can be achieved using alleles that are complementarily modified (Fig 1A): a pre-mutant flox allele carrying loxP sites flanking the essential Otx2 exon 2 and a recombinase-expressing CreER allele in which the tamoxifen-inducible CreERT2 encoding gene (Feil et al, 1997) replaces the Otx2 coding region (for the mouse lines, see supplementary Figs S1,S2 online). Breeding an Otx2flox/CreERT2 male with an Otx2flox/flox female yields 50% heterozygous Otx2flox/CreERT2 embryos that are prone to conditional invalidation and 50% Otx2flox/flox control embryos. A single pulse of tamoxifen triggers Otx2 invalidation in 6–48 h (Hayashi & McMahon, 2002) only in cells expressing Otx2 at that time. Importantly, the target gene is preserved in all other territories (Fig 1B), which enables one to discriminate between activities in independent domains. Strict spatial control necessitates that the recombinase be produced throughout the Otx2 expression domains. A comparison of the Otx2 and CreERT2 pattern in Otx2+/CreERT2 mice showed perfect superimposition at all stages (Fig 1C).
Figure 1.

The knock-in-based CreERT2 strategy. (A) The mating of an Otx2flox/CreERT2 male with a homozygous Otx2flox/flox female yields 50% Otx2flox/CreERT2 and 50% Otx2flox/flox embryos. Intraperitoneal tamoxifen injection (Tam i.p., blue) triggers gene deletion only in Otx2flox/CreERT2 embryos and solely in cells expressing Otx2. Purple and yellow boxes correspond to Otx2 and CreERT2 genes, respectively, and red triangles to loxP sites. (B) Potentialities of the method. During normal development (top), two spatially and temporally distinct Otx2 expression sites (purple circles 1 and 2) control the development of structures S1 and S2. Depending on when CreERT2 is activated (+tam, yellow stripes), the Otx2 gene is deleted in region 1 (middle) or 2 (bottom), perturbing development of structure S1 or S2, respectively. (C) In situ hybridization of Otx2+/CreERT2 embryos at embryonic day (E)6.5–E10.5 (whole mount) and E16.5 (sagittal sections) and adults (transversal sections) at the level of mesencephalon (top) and cerebellum (bottom) with CreERT2 and Otx2 probes (insets; for probes used, see supplementary Figs S1,S2 online). The arrow points to the isthmus. Scale bars, 100 μm for E6.5–E8.5, 500 μm for E10.5, E.16.5 and adult. cb, cerebellum; cp, choroid plexus; di, diencephalon; mes, mesencephalon; oe, olfactory epithelium; op, otic placode; ov, optic vesicle; sc, superior colliculi; tel, telencephalon.
Efficient Otx2 deletion at all stages
We first asked whether a single tamoxifen injection could temporarily block the function of Otx2. To test this, we attempted to reproduce defects in its known early functions. We triggered mutagenesis at E7.5, expecting a head phenotype as found in hypomorphs or chimaeras (Matsuo et al, 1995; Rhinn et al, 1998). All tamoxifen-injected Otx2flox/CreERT2 embryos (23 out of 23) showed abnormal head development (Fig 2A), whereas Otx2flox/flox animals (20 out of 20) and solvent-injected embryos of both genotypes were normal. From E9.5 onwards, Otx2flox/CreERT2 embryos showed an absence or reduction of the forebrain, the eye and the inferior mandible, indicating a marked reduction in the dosage of Otx2 (Matsuo et al, 1995). Genotype-specific tamoxifen-induced deletion of Otx2 exon 2 was confirmed by PCR (Fig 2B; for oligos and probes, see supplementary Fig S1 online). The affected areas corresponded precisely to sites that were fate mapped by their CreERT2 expression at E7.5, as shown by the Z/AP reporter line (Lobe et al, 1999), which expresses alkaline phosphatase after Cre-mediated recombination (Fig 2C). Thus, a single early tamoxifen injection can induce typical Otx2 loss-of-function phenotypes. To study the late functions of Otx2, we then asked whether single injections at later stages yielded similar deletion efficiency. As tamoxifen exerts its effect in 48 h, we performed single injections at E10.5, E12.5, E14.5 and E16.5. Delayed PCR analysis confirmed efficient deletion of the floxed allele at all stages tested (Fig 2D). In addition, in situ analysis of Otx2 messenger RNA with exon 2 probe 30 h after injection showed a marked decrease in the signal, demonstrating the rapidity of tamoxifen action under all conditions (Fig 2E–L). Finally, ectopic onset of Math1 gene expression was found in the mesencephalon 30 h after injection (see supplementary Fig S3I,M online). As it is normally restricted to the more posterior regions (Fig 4J), this confirmed that Otx2 protein activity is quickly abolished. Thus, we could achieve Otx2 inactivation at its endogenous sites of expression at precise stages of development.
Figure 2.

Efficient Otx2 deletion after single tamoxifen injection. (A) Morphology of animals of indicated age and genotype after tamoxifen (Tam) or solvent (Mock) injection at embryonic day (E)7.5. Scale bars, 500 μm. (B) PCR detection of flox and deleted exon 2 (Δe2) alleles (for primers, see supplementary Fig S1 online) using DNA from the brain or head of embryos of indicated age and genotype. (C) Alkaline phosphatase activity in E9.5 Otx2+/CreERT2 ; Z/AP embryos that received tamoxifen or solvent at E7.5. Scale bar, 500 μm. (D) PCR analysis, as in (B), using DNA from the indicated part, and age of Otx2flox/CreERT2 embryos after tamoxifen (Tam) or solvent (mock) injection at the indicated stages. (E–L) In situ hybridization with Otx2 exon 2 probe and CreERT2 probe (insets; for probes, see supplementary Figs S1,S2 online) of sagittal sections of Otx2flox/CreERT2 embryos that received tamoxifen (Tam) or solvent (Mock) at indicated times and were collected 30 h later. Scale bars, 1 mm.
Figure 4.
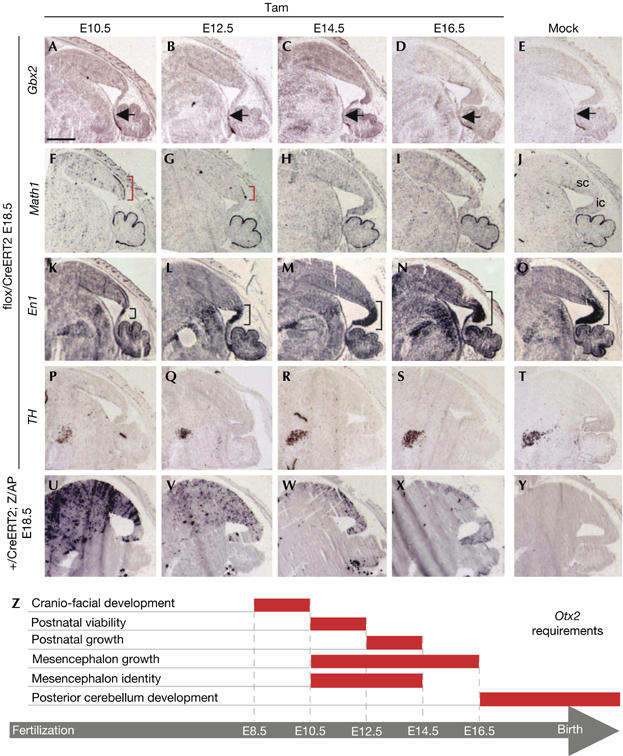
Midbrain specification under Otx2 temporal control. (A–T) In situ hybridization of sagittal sections of embryonic day (E)18.5 brains from Otx2flox/CreERT2 embryos that received tamoxifen (Tam) at indicated times or solvent (Mock) with Gbx2 (A–E), Math1 (F–J), En1 (K–O) and TH (P–T). Rostral limit of Gbx2 expression is marked by black arrows. Ectopic Math1 expression is marked by red brackets. En1 labelling extension is indicated by black brackets. cb, cerebellum; ic, inferior colliculus; sc, superior colliculus. (U–Y) Alkaline phosphatase activity in sagittal sections of E18.5 Otx2+/CreERT2 ; Z/AP animals that received tamoxifen at time corresponding to the panels above. Scale bar, 1 mm. (Z) Schematic representation of the temporal and spatial delineation (red bars) of the multiple requirements of Otx2 during the later stages of embryonic development.
Specific time windows for late activities of Otx2
To study Otx2 functions during the later stages of development, we used the same injection intervals as above. When injected at E10.5, only 5% of Otx2flox/CreERT2 animals reached postnatal day 10 (P10), although their proportion was normal at E18.5 (Fig 3A). This shows that around E10.5, Otx2 controls a developmental step that is necessary for postnatal survival. By contrast, viability was normal and animals were present at the expected ratio (Acampora et al, 1995) when injection was performed at E12.5 or later. Thus, Otx2 is crucial for postnatal survival around E10.5 and its requirement ends at E12.5. Similarly, we could mark out the temporal limit of the requirement of Otx2 for cranial development. Contrary to earlier inactivation, tamoxifen injection at E10.5 and later resulted in morphologically normal embryos (Fig 3B). This implies that Otx2 acts before E10.5 to determine cranial morphogenesis. We also found that Otx2flox/CreERT2 animals injected at E12.5, but not E14.5 and E16.5 mutants, were consistently smaller than their Otx2flox/flox littermates at 10 weeks (Fig 3C). These animals were born with a normal weight, but then accumulated up to 25% growth retardation (Fig 3D). This indicates the contribution of the Otx2 gene between E12.5 and E14.5 to postnatal growth.
Figure 3.

Biological consequences of Otx2 invalidation at embryonic day (E)10.5, E12.5, E14.5 or E16.5. (A) Genotype distribution, at E18.5 and P10, of F1 animals (Fig 1A) that received tamoxifen (Tam) at the indicated time or solvent (Mock). The number of individuals analysed is indicated at the top of the bars. (B) Morphology of E14.5 and P0 Otx2flox/CreERT2 mice after tamoxifen injection at E10.5. Scale bars, 2 mm. (C) Size comparison of 10-week littermates of the indicated genotypes that received tamoxifen at E12.5. (D) Weight distribution of animals of the indicated genotypes at P10, 3 weeks and 10 weeks after birth versus time of tamoxifen (Tam) or solvent (Mock) injection. Histograms represent mean value and error bars are standard deviation. The number of animals in each series is shown at the bottom. *P<0.001 after Student's t-test. (E–L) Anatomical and histological comparison of midbrain/hindbrain development at P10. Dorsal view of the brain (E–H) and Nissl-stained sagittal sections (I–L) of animals that received tamoxifen at E12.5 (E,I), E14.5 (F,J), E16.5 (G,K) or solvent (H,L). Arrows point to the inferior colliculi. The arrowhead points to the granular-like staining of the mesencephalon. Asterisk shows size and foliation defects of posterior cerebellum. ic, inferior colliculus; sc, superior colliculus. (M–P) Alkaline phosphatase activity in sagittal sections of brains of P10 Otx2+/CreERT2 ; Z/AP animals that received tamoxifen at time corresponding to the panels above. Scale bars, 2 mm. (Q–T) Histological organization of ectopic cerebellar-like structure (Ecto) and endogenous cerebellum (Endo) of an adult Otx2flox/CreERT2 animal that received a single injection of tamoxifen (Tam) at E12.5. Haematoxylin–eosin staining (Q), calbindin immunofluorescence (white arrowheads) (R), GAD67 in situ hybridization (S) and calretinin immunofluorescence (T) on sagittal sections. Scale bar, 500 μm.
Examination of the brain of P10 animals injected at E12.5 and E14.5 showed mesencephalon abnormalities, with early injections causing the strongest phenotypes. A large medial portion of inferior colliculi was missing in animals injected at E12.5 (Fig 3E). In animals injected at E14.5, inferior colliculi showed improved development but still did not join medially; however, they seemed to be normal in animals injected at E16.5 (Fig 3F–H).
Sagittal sections showed sequential roles of Otx2 in midbrain/hindbrain development. In animals injected at E12.5, a posterior medial part of the dorsal midbrain adopted a cerebellar histology, with formation of granular cell layers (Fig 3I). In adults, this ectopic structure showed true cerebellar organization with Purkinje cells, interneurons and internal granular cells, as shown by calbindin, GAD67 and calretinin expression (Fig 3Q–T). In animals injected at E16.5, the mesencephalon and anterior cerebellum seemed to be normal, but a marked reduction of posterior cerebellum development and foliation was observed (Fig 3K). This chronology of Otx2-controlled steps corresponds to dynamic changes of the Otx2 expression areas. Analysis of the progeny of Otx2-expressing cells with the Z/AP mouse line shows that, as development proceeds, a decreasing number of cells express Otx2 in the mesencephalon, whereas an increasing number of posterior cerebellum founders start to express Otx2 (Figs 3M–P, 4U–Y).
The different functions and their temporal limits identified in this time-course experiment are summarized in Fig 4Z. We analysed the midbrain changes in greater detail, whereas other studies might focus on the postnatal survival, growth defect and cerebellum phenotypes.
Temporal control of mesencephalon patterning
To study temporal activities of Otx2 in the mesencephalon of all mutants, their brains were analysed at E18.5. This avoided the perinatal death of embryos injected at E10.5 and allowed complete tamoxifen action in embryos injected at E16.5.
As early midbrain Otx2 and hindbrain Gbx2 expression controls the positioning of the isthmus (Martinez-Barbera et al, 2001), we checked its location in our mutants. At E18.5, this seemed to be normal in all cases, as shown by Gbx2 expression (Fig 4A–E). Detection of isthmus markers Gbx2 and En1 30 h and 48 h after E10.5 injection confirmed that the midbrain–hindbrain border was unchanged (supplementary Fig S3A–H online). In embryos injected at E10.5 and E12.5, cerebellum markers Math1 (Fig 4F–J) and Pax6 (not shown) labelled a dorsal area anterior to the isthmus. Hence, this ectopic cerebellum resulted from a local change in mesencephalon identity and not from a rostral shift of the hindbrain. Ectopic Math1 expression was already evident 30 h after injection (supplementary Fig S3I,M online). In embryos injected at E12.5, rostral extension of the labelled area was reduced compared with the E10.5 mutant, whereas its caudal limit was identical in both cases. Immediately posterior was the caudal midbrain, which expresses En1 and gives rise to inferior colliculi (Wurst et al, 1994). Interestingly, this domain never expressed Math1. However, it was also affected by the loss of Otx2 activity: its size was markedly reduced in embryos injected at E10.5 and progressively recovered to normal in embryos injected at E12.5, E14.5 and E16.5 (Fig 4K–O), perfectly correlating with the inferior colliculi phenotype in adults (Fig 3E–H). To explain this defect, we studied cell proliferation and apoptosis in embryos injected at E10.5. Two days after tamoxifen injection, KI-67 antibody showed a strong decrease in proliferation throughout the dorsal mesencephalon (supplementary Fig S3O,P online), whereas cleaved caspase 3 detection showed no change in the rate of apoptosis (not shown). This differently affected the parts of the midbrain, because, at this stage, superior colliculi have nearly reached their definitive size, whereas inferior colliculi are still to develop. Consistently, we observed thinner superior colliculi and reduced inferior colliculi (Fig 4K).
In the ventral midbrain, Vernay et al (2005) have shown that Otx2 ablation before E12.5 generally results in a decreased number of dopaminergic neurons. We analysed this population in our mutants using tyrosine hydroxylase (Fig 4P–T) and Lmx1b (not shown) markers. The dopaminergic population seemed to be reduced in embryos injected at E10.5 and E12.5 but was normal elsewhere. In agreement with Vernay et al (2005), we found no gross difference in cell proliferation (supplementary Fig S3O,P online) and apoptosis in the ventral midbrain. This indicates that Otx2 is required up to E14.5 to allow full differentiation of this neuronal population. Fate mapping of midbrain Otx2-expressing cells (Fig 4U–Y) showed that E14.5 is the time at which Otx2-expressing precursors no longer contribute to the ventral midbrain.
Together, these results discriminate between different functions of Otx2 in the mesencephalon: it is necessary up to E14.5 to prevent a dorsal area fated to form superior colliculi from adopting a cerebellar fate and to allow differentiation of ventral dopaminergic neurons; it is necessary up to E16.5 to ensure normal growth of the caudal midbrain (Fig 4Z).
Discussion
Here, we report the use of a knock-in CreERT2 allele over a floxed allele combined to transient CreERT2 activation by single tamoxifen pulses to capture temporal gene activity. The strategy has several advantages. First, it accurately delivers Cre enzyme to the sites of expression of the gene of interest. Second, it can be used during as broad a developmental period as the gene of interest is expressed. Third, contrary to ubiquitously expressed CreER genes, which, once activated, lead to thorough deletion of the floxed gene, the knock-in CreERT2 gene only disrupts the floxed gene in cells in which it is expressed at the time of tamoxifen injection. This spares other areas in which later gene expression will occur.
We focused on Otx2 phenotypes between E10.5 and E18.5. Efficient conditional knockout was proved by the reproduction of loss-of-function phenotypes and by the consistency of defects after injection at each stage. Previously, it was shown that Otx2 is required for cranio-facial development (Matsuo et al, 1995). Conditional knockout studies have shown the need for Otx2 around E10.5–E12.5 for correct patterning of the mesencephalon along the antero-posterior and dorso-ventral axes (Puelles et al, 2004; Vernay et al, 2005). The study by Vernay et al (2005) showed that beyond E12.5, although both patterns are established, Otx2 is still necessary to maintain mesencephalon identity and allow development of the posterior cerebellum. Using our allelic combination, we could, in a single time-course experiment, reproduce these findings and uncover new specific requirements of Otx2 for inferior colliculi development, survival at birth and postnatal body growth. Furthermore, we marked out temporal limits beyond which Otx2 is no longer necessary for developmental activities. Our data show that the requirement of Otx2 for cranio-facial development ends at E10.5. Similarly, we demarcate temporal windows for Otx2 control of survival after birth (E10.5–E12.5), body growth (E12.5–E14.5), superior colliculi identity (E10.5–E14.5), ventral midbrain neuronal differentiation (E10.5–E14.5), caudal mesencephalon development (E10.5–E16.5) and posterior cerebellum development (E16.5 and onwards). Cross-checking this information with Otx2 expression pattern should help in identifying the developing structures that are affected by the mutation.
In the chick, FGF8 signalling from the isthmic organizer induces a cerebellar fate, and this is counteracted in the mesencephalon by Otx2 expression (Sato et al, 2001). A recent study suggests that in zebrafish, the primary function of FGF8 is to keep Otx proteins, which are potent repressors of cerebellar fate, out of the rostral hindbrain (Foucher et al, 2006). The transformation of superior colliculi into cerebellum, which we found after E10.5 Otx2 ablation, supports similar mechanisms in the mouse. In addition, the temporal reduction of transformed area shows anterior to posterior progressive stabilization of superior colliculi identity. Strikingly, the most caudal region of the mesencephalon, which gives rise to inferior colliculi, does not undergo cerebellum transformation, although it requires Otx2 activity up to E16.5 to develop to its normal size. This shows different Otx2 activities in subdivisions of the caudal mesencephalon. The existence of such subsegments has recently been documented in the chick embryo (Hidalgo-Sanchez et al, 2005). Finally, both inferior colliculi and posterior cerebellum defects point to a potential role for Otx2 in the control of proliferation. This is in line with the Otx2 overexpression found in human medulloblastomas, which are aggressive tumours arising from cerebellar granular cell precursors (Di et al, 2005).
Methods
Mouse production and tamoxifen administration. Animals were maintained in the 129/Sv background (for details, see the supplementary information online). Day of vaginal plug was taken as E0.5. Tamoxifen (Sigma-Aldrich, St Louis, MO, USA), diluted at 10 mg/ml in sunflower oil, was administrated at 1 mg per 20 g of body weight. The protocols were approved by the French CGG and CREA committees.
Analyses of embryos and brains. For whole-mount analyses, samples were dissected in PBS and fixed in 4% paraformaldehyde (PFA) for in situ analyses, or in 0.2% glutaraldehyde for AP staining. For analyses of the sections, samples were dissected in PBS, fixed in 4% PFA, dehydrated and embedded in paraffin for Nissl staining, or protected in 30% sucrose and frozen in cryomount for haematoxylin–eosin staining and immunofluorescence, or directly frozen on solid CO2 for alkaline phosphatase and in situ studies. Sections (5–12 μm) were processed and Nissl and haematoxylin–eosin staining was carried out according to standard protocol. Alkaline phosphatase staining and whole-mount in situ hybridization were carried out as described by Wilkinson (1992) and Lobe et al (1999). Probes for in situ hybridization were as follows: 0.15 kb Otx2 exon 2, Cre coding sequence (see supplementary Figs S1,S2 online); Otx2, Math1, Gbx2, En1, GAD67 and TH probes were gifts from S. Vincent, B. Vernay, G. Martin, A. Joyner and M. Wassef. Immunofluorescence was performed as described by Vernay et al (2005). Primary antibodies used were 1:5,000 rabbit anti-calbindin D-28k (Swant, Bellinzona, Switzerland) and 1:5,000 rabbit anti-calretinin (Chemicon, CA, USA).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Data
Acknowledgments
We thank S. Mazan and D. Acampora for Otx2 clones, D. Metzger for the pCreERT2 plasmid, A. Nagy for the Z/AP mouse, B. Vernay, G. Martin and A. Joyner for probes, S. Vincent for probes and discussions, M. Wassef for probes, antibody and discussions, L. Quignodon for antibody, students M. Régent and M. Feyeux, the Pôle de Biologie Expérimentale de la Souris for mice and C. Scutt for critical reading. N.F. received fellowships from the French Ministry of Research, Retina France and the Association pour la Recherche sur le Cancer. This work was supported by grants from the Centre National de la Recherche Scientifique, Retina France and the Ligue Nationale contre le Cancer.
References
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P (1995) Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121: 3279–3290 [DOI] [PubMed] [Google Scholar]
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J (1996) A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development 122: 243–252 [DOI] [PubMed] [Google Scholar]
- Di C et al. (2005) Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res 65: 919–924 [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237: 752–757 [DOI] [PubMed] [Google Scholar]
- Foucher I, Mione M, Simeone A, Acampora D, Bally-Cuif L, Houart C (2006) Differentiation of cerebellar cell identities in absence of Fgf signalling in zebrafish Otx morphants. Development 133: 1891–1900 [DOI] [PubMed] [Google Scholar]
- Frantz GD, Weimann JM, Levin ME, McConnell SK (1994) Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci 14: 5725–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP (2002) Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 244: 305–318 [DOI] [PubMed] [Google Scholar]
- Hidalgo-Sanchez M, Martinez-de-la-Torre M, Alvarado-Mallart RM, Puelles L (2005) A distinct preisthmic histogenetic domain is defined by overlap of Otx2 and Pax2 gene expression in the avian caudal midbrain. J Comp Neurol 483: 17–29 [DOI] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Nakano H, Okamura D, Nakao K, Yonemura S, Belo JA, Aizawa S, Matsui Y, Matsuo I (2005) Canonical Wnt signaling and its antagonist regulate anterior–posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev Cell 9: 639–650 [DOI] [PubMed] [Google Scholar]
- Kmita M, Duboule D (2003) Organizing axes in time and space; 25 years of colinear tinkering. Science 301: 331–333 [DOI] [PubMed] [Google Scholar]
- Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A (1999) Z/AP, a double reporter for cre-mediated recombination. Dev Biol 208: 281–292 [DOI] [PubMed] [Google Scholar]
- Mallamaci A, Di Blas E, Briata P, Boncinelli E, Corte G (1996) OTX2 homeoprotein in the developing central nervous system and migratory cells of the olfactory area. Mech Dev 58: 165–178 [DOI] [PubMed] [Google Scholar]
- Martinez-Barbera JP, Signore M, Boyl PP, Puelles E, Acampora D, Gogoi R, Schubert F, Lumsden A, Simeone A (2001) Regionalisation of anterior neuroectoderm and its competence in responding to forebrain and midbrain inducing activities depend on mutual antagonism between OTX2 and GBX2. Development 128: 4789–4800 [DOI] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S (1995) Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev 9: 2646–2658 [DOI] [PubMed] [Google Scholar]
- Nagy A, Perrimon N, Sandmeyer S, Plasterk R (2003) Tailoring the genome: the power of genetic approaches. Nat Genet 33: 276–284 [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T (2003) Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci 6: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, Czerny T, Brodski C, Ang SL, Wurst W, Simeone A (2004) Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development 131: 2037–2048 [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dierich A, Shawlot W, Behringer RR, Le Meur M, Ang SL (1998) Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development 125: 845–856 [DOI] [PubMed] [Google Scholar]
- Santagati F, Minoux M, Ren SY, Rijli FM (2005) Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development 132: 4927–4936 [DOI] [PubMed] [Google Scholar]
- Sato T, Araki I, Nakamura H (2001) Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development 128: 2461–2469 [DOI] [PubMed] [Google Scholar]
- Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, Ang SL (2005) Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci 25: 4856–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D (1992) Whole mount in situ hybridisation of vertebrate embryos. In In Situ Hybridisation: A Practical Approach, Wilkinson DG (ed) pp 75–83. Oxford, UK: IRL [Google Scholar]
- Wurst W, Auerbach AB, Joyner AL (1994) Multiple developmental defects in Engrailed-1 mutant mice: an early mid–hindbrain deletion and patterning defects in forelimbs and sternum. Development 120: 2065–2075 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data


