Abstract
In multicellular organisms, changes in the DNA replication programme could act to integrate differentiation with cell division in various developmental and transcriptional contexts. Here, we have addressed the use of DNA replication origins during differentiation in the HoxB domain—a cluster of nine genes developmentally regulated in a collinear manner. In undifferentiated mouse P19 cells, we detected several DNA replication origins in the 100 kb HoxB locus, indicating a relaxed origin use when the locus is transcriptionally silent. By contrast, in retinoic-acid-induced differentiated cells, when HoxB transcription is activated, a general silencing of DNA replication origins occurs in the locus except one located downstream of Hoxb1, at the 3′ boundary of the HoxB domain. Silencing of the replication origins is associated with histone hyperacetylation, whereas the active Hoxb1 origin persists as a hypoacetylated island. These findings provide direct evidence for the differentiated use of origins in HoxB genes, and we suggest that this regulation might contribute to the regulated expression of HoxB genes during development.
Introduction
How particular sites in the genome are selected to initiate DNA replication in multicellular eukaryotes is poorly understood. Although sequence features such as A + T richness or the presence of CpG islands seem to be involved, they do not seem to be as stringent as those acting in prokaryotes or in unicellular Saccharomyces cerevisiae. Epigenetic modifications, such as alterations of chromatin structure, DNA methylation and changes in transcription factor activity, might select particular initiation sites from several potential origins of replication (for reviews, see DePamphilis, 1993; Aladjem & Fanning, 2004). Nevertheless, few metazoan origins have been mapped so far, and most of them have been studied in short regions that do not reach the average size of a replicon (100–120 kb). These results do not exclude other possible initiation sites and potential variations in the use of the origin. Therefore, the question of the existence of a single specified origin for a given chromosomal domain and the specification of replication origin in relation to different transcriptional programmes or cell fates remains unclear.
Here, we report a differentiation-regulated specification of DNA replication origins in the HoxB domain, a locus that is essential for embryonic development from Drosophila to humans, notably in patterning and determining anteroposterior identity (Krumlauf, 1994). The organization and regulation of the HoxB locus is remarkable, as it contains nine genes, spread over 100 kb, that are activated with temporal and spatial collinearity in embryos. Both the timing of activation of the genes and their anterior boundaries of expression follow the order of the genes in the locus (for reviews, see Duboule & Morata, 1994; Krumlauf, 1994). A connection between transcription and the replication programme has been proposed to contribute to the temporal and spatial collinearity of the HoxB gene domain during embryonic development (Duboule & Morata, 1994; Fisher & Méchali, 2003), but no DNA replication origin has yet been identified in this domain. The transcription of the HoxB domain can be induced by retinoic acid (RA; Simeone et al, 1990), and the mouse embryonic carcinoma P19 cell line recapitulates RA-regulated gene expression and neuroectodermal cell differentiation in vitro (McBurney, 1993), including the temporal collinearity of HoxB transcription (Jones-Villeneuve et al, 1982; Fisher & Méchali, 2003). Here, we show that several origins of replication can be used along the 120 kb domain in pluripotent undifferentiated cells. However, differentiation and activation of the locus for transcription leads to the silencing of replication origins in the locus and the establishment of a site-specific DNA replication origin at the 3′ boundary of the locus.
Results and Discussion
Multiple replication origins at the silent HoxB locus
We used the nascent strand relative abundance measurement as a robust assay for mapping replication origins (Giacca et al, 1994; Danis et al, 2004; Wang et al, 2004). DNA nascent strands were first prepared from asynchronous undifferentiated P19 cells, which do not express HoxB genes (Fisher & Méchali, 2003). In each preparation, we assessed the quality of the nascent strand purification by looking at the DNA replication origin previously defined in the c-myc gene promoter (McWhinney & Leffak, 1990; Vassilev & Johnson, 1990; Girard-Reydet et al, 2004). As shown in Fig 1A, a clear peak of tenfold nascent strand enrichment, which was indicative of a DNA replication origin, was detected upstream of the c-myc promoter.
Figure 1.
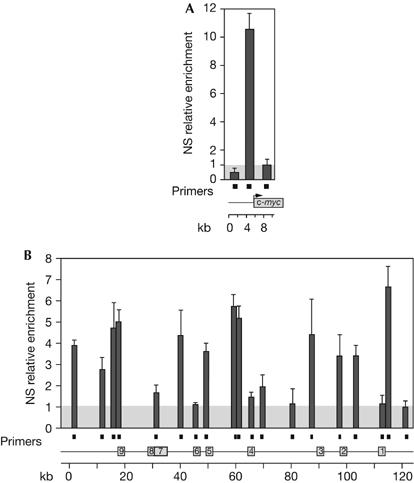
DNA replication initiates at several sites in the HoxB domain in undifferentiated P19 cells. (A) Real-time quantitative PCR was performed on P19 nascent strands, using primer pairs (filled squares) located along the c-myc locus. Enrichment was calculated as fold enrichment over the background level (grey box; see the supplementary information online). The results shown are the average of three independent nascent strand preparations that were quantified twice. Error bars indicate the standard error of the mean. Genes are indicated by grey boxes. (B) Same as (A), but for the HoxB locus. kb, kilobase; NS, nascent strands.
Quantification of nascent strand abundance was then carried out using primer pairs designed along the 100 kb of the HoxB locus. Fig 1B shows that, in undifferentiated P19 cells, several sites showed a high frequency of DNA replication initiation. Data obtained from three independent nascent strand preparations, each showing a clear enrichment for the c-myc promoter as an internal control, demonstrated that at least five DNA replication origins (close to Hoxb1, Hoxb3, Hoxb4, between b5 and b6, and close to b9) were four- to sixfold higher than the background level (Fig 1B). Other positions along the locus showed intermediate levels of enrichment that were significantly above the background level, and such sites were close to Hoxb2.
These results show that, in undifferentiated P19 cells, in which the HoxB genes are not transcribed, several positions along the locus can be used as DNA replication origins. Several preferred sites were detected in the hundred kilobases of the HoxB domain, located in both intergenic and promoter regions. We conclude that the domain shows relaxed origin selection when transcriptionally silent. Large initiation zones have been described, such as the dihydrofolate reductase locus, in which replication starts at several positions along 55 kb (Dijkwel et al, 2002), and the β-globin locus, which contains highly enriched regions spread over 50 kb (Aladjem et al, 2002). The observed pattern might reflect flexibility in the choice of origin between cell cycles in the cell population.
HoxB induction silences the use of the origin in the locus
P19 cells were induced to differentiate and express HoxB genes by RA treatment. The transcriptional level of Oct4, a marker of the undifferentiated state that is negatively regulated by RA (Schoorlemmer et al, 1994), is high in P19 cells but decreases to undetectable levels on addition of RA (Fig 2A). The induction of HoxB gene transcription was confirmed at the quantitative level by real-time PCR (Fig 2A). In addition, Hoxb1 messenger RNA in situ hybridization shows that all the cells respond to RA (Fig 2B). We also showed that Hoxb1 shows a specific, transitory pattern of induction (Fig 2A): Hoxb1 mRNA is abundant 24 h after RA addition but is no longer detectable after 2 days, as previously described (Simeone et al, 1990).
Figure 2.
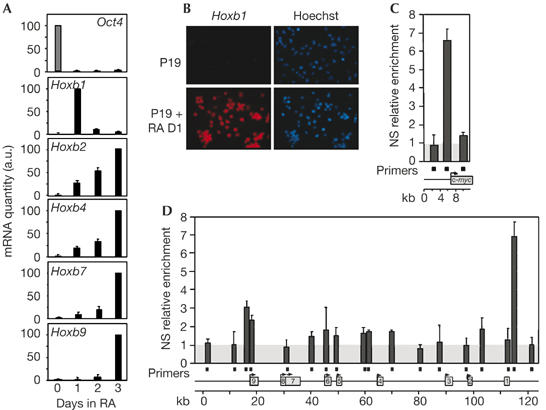
Retinoic-acid-triggered differentiation silences replication origins in the HoxB locus. (A) Total RNA was collected from retinoic-acid-treated P19 cells on the indicated days. Complementary DNA was generated by reverse transcription and quantified by real-time PCR to analyse transcription of Hoxb1, Hoxb2, Hoxb4, Hoxb7, Hoxb9 and Oct4, a marker of undifferentiated cells. For each gene, quantities were calculated by setting the value obtained for the higher point to 100%. Averages and standard errors from two independent experiments are shown. (B) Untreated P19 cells (P19) and P19 cells treated with retinoic acid for 1 day (P19+RA D1) were hybridized with an antisense Hoxb1 probe and coloured by Hoechst. No signal was detected with a sense Hoxb1 probe. (C) Real-time quantitative PCR was performed on nascent strands purified from P19 cells treated with retinoic acid for 3 days, using primer pairs (filled squares) located along the c-myc locus. Enrichment was calculated as fold enrichment over the background level (grey box; see the supplementary information online). The results are the average of three independent nascent strand preparations that were quantified twice. Error bars indicate the standard error of the mean. Genes are indicated by grey boxes. (D) Same as (C), but for the HoxB domain. a.u., arbitrary units; kb, kilobase; NS: nascent strands; RA, retinoic acid.
The use of a replication origin in the HoxB locus was determined after RA-induced differentiation. Fig 2C confirms that the c-myc origin of replication was still detected under these conditions (Fig 2C). Three independent experiments were carried out in differentiated cells, and the results showed a clear restriction in origin use across the domain, with the Hoxb1 origin becoming the predominant DNA replication initiation site specified in the domain (compare Fig 2D with Fig 1B). We found that the relative enrichment level around Hoxb1 was comparable with that of the c-myc origin, whereas the other potential origins that had been active in undifferentiated P19 cells were now downregulated. Weak activity of replication initiation might remain at the 5′ border of the locus, upstream from the Hoxb9 gene. We conclude that a restriction in origin use occurs when cell differentiation and transcription of the HoxB domain are induced, resulting in the silencing of DNA replication origins inside the HoxB domain, to the benefit of a replication origin at the boundary of the domain.
Hoxb1 origin does not depend on Hoxb1 transcription
We analysed this origin more precisely, with a resolution of 1 kb, using other primer pairs (Fig 3). Interestingly, the origin overlaps with an enhancer that mediates the early expression pattern of Hoxb1 and contains an RA response element (RARE; Marshall et al, 1994), a sequence bound by RA receptors after RA treatment. As Hoxb1 is transcriptionally active after 1 day of RA treatment but is repressed after 3 days, we examined whether the origin was linked to the transcriptional state of the gene. The use of this Hoxb1 origin was therefore analysed in untreated P19 cells (P19), P19 cells after 1 day of RA treatment (RA D1) and P19 cells after 3 days of treatment (RA D3; Fig 3). We found that the DNA replication origin was present in all three cases. We conclude that the use of the Hoxb1 origin is not linked to the transcription of the gene, but could be specified in relation to the co-regulated expression of the whole HoxB domain.
Figure 3.
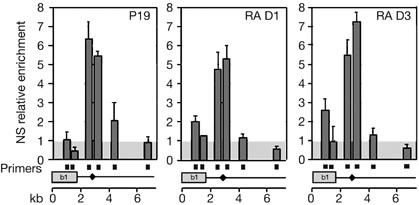
Hoxb1 origin of replication is unaffected by transcription of the gene. Nascent strands (NS) were isolated from P19 cells (P19) and P19 cells treated with retinoic acid (RA) for 1 (RA D1) or 3 (RA D3) days. Real-time quantitative PCR was performed using the indicated primer pairs (filled squares), and the level of enrichment was calculated as fold enrichment over the background level (grey box; see the supplementary information online). Results are the average of three independent NS preparations that were quantified twice. Error bars indicate the standard error of the mean. Filled diamond indicates the position of the RA response element. kb, kilobase.
The restriction of a single origin located at the border of the HoxB domain is in agreement with modification in origin use, which is observed during development in the Xenopus ribosomal DNA domain, Drosophila DNA polymerase-α gene and Sciara coprophila II/9A locus (Hyrien et al, 1995; Sasaki et al, 1999; Lunyak et al, 2002). Recently, a global observation of the IgH domain identified a mixed pattern of activation of origins during B-cell development (Norio et al, 2005). Our results show that the activation of a transcriptional programme in a co-regulated cluster of developmental genes results in the silencing of the DNA replication origin not only in the coding regions of the HoxB domain but also in the intergenic regions. We conclude that the regulation of DNA replication origin selection that we describe here for the HoxB domain cannot be explained simply by the interference between transcriptional and replication machineries, as described for other domains (Saha et al, 2004).
Origin restriction correlates with histone acetylation
We analysed nucleosome density in the HoxB locus by chromatin immunoprecipitation using an antibody against unmodified histone H3. RA treatment induces a global decrease (two- to threefold) in the amount of unmodified H3 in the central part of the HoxB domain (Hoxb7–Hoxb3 genes; Fig 4A), which could reflect decondensation and looping of the HoxB domain (Chambeyron & Bickmore, 2004). We further analysed histone acetylation throughout the HoxB domain. In undifferentiated cells, a strong acetylation signal for H3 (K9 and K14) was detected at the c-myc promoter, as expected (Gombert et al, 2003), whereas the level of acetylation was low throughout the HoxB domain. By contrast, a strong increase in histone H3 acetylation was detected after RA induction, consistent with the transcriptional activation of the domain. Interestingly, the acetylation was not restricted to the promoter regions (Fig 4B). Nevertheless, the Hoxb1 region containing the replication origin remained hypoacetylated. A similar profile was detected for acetylated histone H4 (K5, K8, K12, K16). RA treatment induces an increase in H4 acetylation throughout the HoxB domain, except for the Hoxb1 region (Fig 4C). We also noted that the Hoxb1 origin remains hypoacetylated even when the gene is transcribed, after 1 day of RA treatment (data not shown). We conclude that, in this large cluster of coordinately activated genes, restriction in origin use is associated with acetylation of histones.
Figure 4.
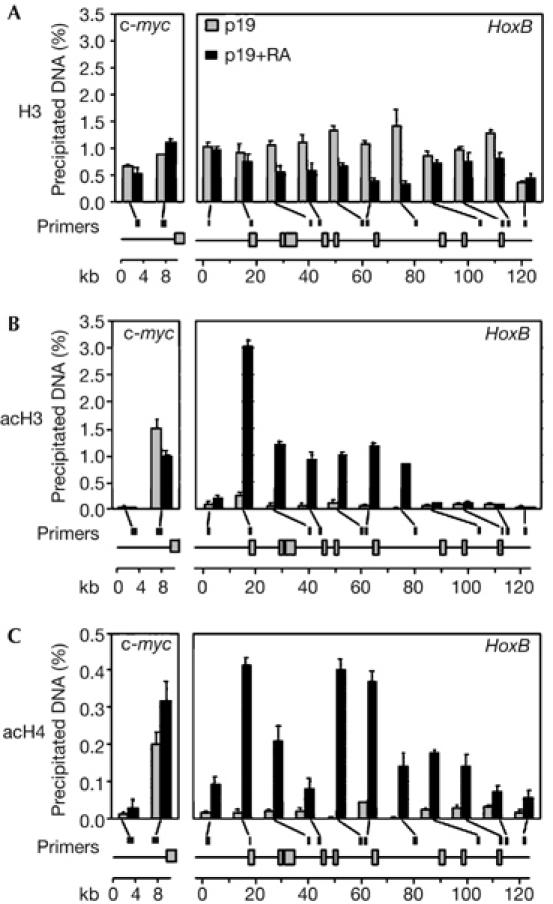
Retinoic acid treatment induces H3 and H4 acetylation at the HoxB locus. Levels of (A) H3, (B) acetylated H3 (acH3) and (C) acetylated H4 (acH4) were analysed by chromatin immunoprecipitation and real-time PCR quantification using the indicated primer pairs (filled squares) before (P19) and after (P19+RA) 3 days of RA-induced differentiation. As a reference, the acetylation level from the c-myc promoter is shown in the left subpanel. The results are the average of two independent immunoprecipitation preparations that were quantified twice. Error bars indicate the standard error of the mean. Genes are indicated by grey boxes. kb, kilobase; RA, retinoic acid.
This correlation might be surprising because histone acetylation was found to be associated with origin specification in some cases, which was suggested to facilitate the access of replication initiation proteins to DNA (Aggarwal & Calvi, 2004; Danis et al, 2004). Nevertheless, different DNA replication origins of the chicken β-globin locus have been shown to have opposite epigenetic modifications (Prioleau et al, 2003). The regulation of origin use in the Hoxb domain might therefore not simply be linked to an open or closed state of the chromatin structure. Different mechanisms of regulation could have evolved to select the position of a given DNA replication origin in relation to specific developmental states or to the specific organization of the chromatin domain for the regulation of its transcription programme.
Replication origins and regulation of the HoxB domain
In conclusion, we have shown that DNA replication origins in the HoxB locus are developmentally regulated and that induction of the locus is associated with specification of an origin at its boundary. The HoxB domain is unusual both in its high gene density (nine genes over 100 kb) and in the collinear regulation of its transcription. The selection of a DNA replication origin just at the edge of this cluster of nine transcriptionally co-regulated genes is particularly intriguing. Interestingly, the origin downstream from Hoxb1, which is used in both undifferentiated and differentiating P19 cells, colocalizes with an RARE. The presence of an origin of replication at this site could be part of a pre-existing organization of the chromatin domain for its developmental regulation and might contribute to the regulation of transcription over this domain (Fig 5). This possibility is strengthened by the observation that passage through mitosis in the presence of RA is sufficient to induce Hoxb1, whereas transcriptional induction of Hoxb2 to Hoxb9 requires the S phase in a collinear manner (Fisher & Méchali, 2003). We propose that the location of the replication origin at the 3′ boundary of the domain could act as a structural element in the domain, preparing the chromatin to respond to RA and to activate Hoxb1. The silencing of the other replication origins in the domain during differentiation might therefore be important for both preventing replication origin assembly from interfering with transcription of the nine genes in the cluster and also allowing the replication fork to move in a single direction along the HoxB locus, a direction that follows the collinearity of their expression.
Figure 5.
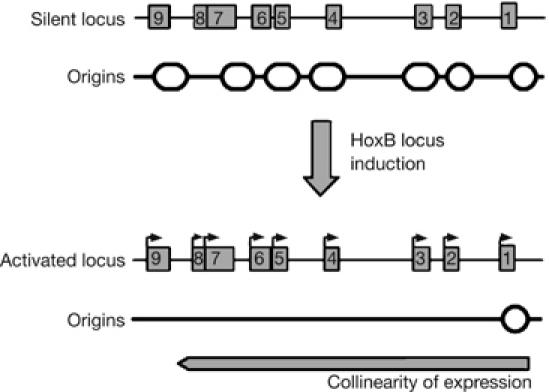
Silencing of DNA replication origins in the HoxB domain during transcriptional induction. DNA replication origins can be present at several positions along the cluster of the nine HoxB genes at a stage when the domain is not transcribed. Induction of differentiation by retinoic acid leads to the progressive transcription of the nine genes, in a collinear manner. Transcriptional activation correlates with a global silencing of DNA replication origins inside the cluster. An important DNA replication origin remains at the 3′ border of Hoxb1, the first gene transcriptionally activated in the locus, suggesting that the HoxB cluster forms a single co-regulated chromatin unit.
Methods
Purification of nascent DNA strands. Nascent strands were purified from P19 or P19+RA cells as described in the supplementary information online.
Crosslinking and chromatin immunoprecipitation. Formaldehyde crosslinking was performed as described in Gombert et al (2003), with modifications (see the supplementary information online). Chromatin immunoprecipitation was performed as described in Orlando et al (1997) with anti-H3 from Abcam (Cambridge, UK; #ab1791) and anti-H3(acetyl K9,K14) and anti-H4(acetyl K5,K8,K12,K16) from Upstate (Dundee, UK; #06-599 and #06-866, respectively).
Oligonucleotide primers and real-time quantitative PCR. Nascent DNA strands and immunoprecipitated DNA were analysed by quantitative real-time PCR using primers encompassing the c-myc or HoxB loci. See the supplementary information online for the sequences of the PCR primers, real-time PCR quantification and normalization procedures.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank J.M. Lemaître, M. Lutzmann and P. Pasero for a critical reading of the manuscript and helpful discussions, F. Maillet and N. LeFloc'h for help in HoxB induction characterization and nascent strand preparation and P. De Santa Barabra for help in RNA in situ hybridization experiments. This work was supported by the CNRS, the Ligue contre le Cancer, l'Association pour la Recherche contre le Cancer (ARC) and the Agence Nationale de la Recherche.
References
- Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Fanning E (2004) The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep 5: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem MI, Rodewald LW, Lin CM, Bowman S, Cimbora DM, Brody LL, Epner EM, Groudine M, Wahl GM (2002) Replication initiation patterns in the β-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol Cell Biol 22: 442–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18: 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Méchali M (2004) Specification of a DNA replication origin by a transcription complex. Nat Cell Biol 6: 721–730 [DOI] [PubMed] [Google Scholar]
- DePamphilis ML (1993) Origins of DNA replication in metazoan chromosomes. J Biol Chem 268: 1–4 [PubMed] [Google Scholar]
- Dijkwel PA, Wang S, Hamlin JL (2002) Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol Cell Biol 22: 3053–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D, Morata G (1994) Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet 10: 358–364 [DOI] [PubMed] [Google Scholar]
- Fisher D, Méchali M (2003) Vertebrate HoxB gene expression requires DNA replication. EMBO J 22: 3737–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacca M et al. (1994) Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci USA 91: 7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard-Reydet C, Gregoire D, Vassetzky Y, Mechali M (2004) DNA replication initiates at domains overlapping with nuclear matrix attachment regions in the Xenopus and mouse c-myc promoter. Gene 332: 129–138 [DOI] [PubMed] [Google Scholar]
- Gombert WM, Farris SD, Rubio ED, Morey-Rosler KM, Schubach WH, Krumm A (2003) The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol Cell Biol 23: 9338–9348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Méchali M (1995) Transition in specification of embryonic metazoan DNA replication origins. Science 270: 994–997 [DOI] [PubMed] [Google Scholar]
- Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnins VI (1982) Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol 94: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R (1994) Hox genes in vertebrate development. Cell 78: 191–201 [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Ezrokhi M, Smith HS, Gerbi SA (2002) Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol Cell Biol 22: 8426–8437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H, Studer M, Popperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R (1994) A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 370: 567–571 [DOI] [PubMed] [Google Scholar]
- McBurney MW (1993) P19 embryonal carcinoma cells. Int J Dev Biol 37: 135–140 [PubMed] [Google Scholar]
- McWhinney C, Leffak M (1990) Autonomous replication of a DNA fragment containing the chromosomal replication origin of the human c-myc gene. Nucleic Acids Res 18: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL (2005) Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20: 575–587 [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11: 205–214 [DOI] [PubMed] [Google Scholar]
- Prioleau MN, Gendron MC, Hyrien O (2003) Replication of the chicken β-globin locus: early-firing origins at the 5′ HS4 insulator and the ρ- and β(A)-globin genes show opposite epigenetic modifications. Mol Cell Biol 23: 3536–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Shan Y, Mesner LD, Hamlin JL (2004) The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev 18: 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Sawado T, Yamaguchi M, Shinomiya T (1999) Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol Cell Biol 19: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J, van Puijenbroek A, van Den Eijnden M, Jonk L, Pals C, Kruijer W (1994) Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol Cell Biol 14: 1122–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F (1990) Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 346: 763–766 [DOI] [PubMed] [Google Scholar]
- Vassilev L, Johnson EM (1990) An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol Cell Biol 10: 4899–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lin CM, Brooks S, Cimbora D, Groudine M, Aladjem MI (2004) The human β-globin replication initiation region consists of two modular independent replicators. Mol Cell Biol 24: 3373–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


