Since Charles Darwin published The Origin of Species in 1859, his theory of evolution has been a central axiom for the biological sciences. Scientists have devoted considerable effort to unravelling the evolutionary mechanisms of biological, and other, systems. Their work—in particular the discovery of DNA as the carrier of hereditary information—has led to the now nearly universally accepted idea that evolution causes the increasing complexity of most biological systems. However, although the basic mechanisms of evolution—mutation and selection—are clear, accumulating data suggest that the means to increase complexity have also become more complex. This begs the question how such mechanisms have evolved over time.
Thanks to rapid progress in the biological sciences—in particular in the field of genomics—and the information sciences including linguistics, we have amassed an enormous amount of data on genomes and biological evolutionary mechanisms. These are providing an unprecedented opportunity to investigate the possible transformation of evolutionary strategies of evolving systems, such as genomes and languages. In fact, the information contained in genomes has long been compared with languages, and many linguistic methodologies are now used to analyse genomes (Searls, 2002). If we compare the evolution of genomes and language, we serendipitously find that both systems have undergone similar strategic shifts to attain increasing complexity, which suggests that this is an intrinsic property of many evolving systems, not just biological ones.
During the primary stage in the evolution of both genomes and languages, increasing complexity was achieved mainly by increasing the number of basic information-carrying elements: nouns and verbs in language, and genes in biology. The Chinese language is a good example to use in this context, because of its history of more than 6,000 years and its continuing evolution. The early Chinese written language, called Oracle, consisted of a few thousand characters, which gradually increased to include more than 47,000 individual characters, as the ancient Chinese continually invented characters to refine their ability to describe their environment (Table 1; Ji, 1989). For example, more than 70 characters described horses on the basis of their colour, age and gender, and more than 80 characters described their behaviour (Chen, 1936). According to a statistical analysis of 800 million ancient characters, one would need to know about 22,000 characters to attain 99.99% coverage of ancient Chinese (Zhang, 2004). The same phenomenon can be observed in biological evolution. Simple and early organisms, such as prokaryotes, manage to survive and proliferate with a few thousand genes, whereas the number of genes in higher organisms is about a magnitude higher—some plants, such as maize, have more than 50,000 genes (Messing et al, 2004).
Table 1.
Evolution of Chinese characters (a complete list of sources is listed in Ji (1989))
| Era | Quantity of characters |
| Shang Dynasty (Oracle bone inscriptions) (around 17th century BC to 11th century BC) | 5,000 |
| Eastern Han Dynasty (around 25 AD to 220 AD) | 9,353 |
| Jin Dynasty (around 265 AD to 420 AD) | 12,824 |
| Liang Dynasty (around 502 AD to 557 AD) | 16,917 |
| Tang Dynasty (around 618 AD to 907 AD) | 22,561 |
| Northern Song Dynasty (around 960 AD to 1127 AD) | 31,319 |
| Ming Dynasty (around 1368 AD to 1644 AD) | 33,179 |
| Qing Dynasty (around 1644 AD to 1912 AD) | 47,035 |
In the second stage of their evolution, both systems began to rearrange existing elements into new combinations to increase complexity further. Interestingly, this leads to a decrease in the quantity of elements. For example, modern Chinese uses considerably fewer characters than did the ancient language: only 4,600 characters are now needed to attain 99.99% coverage (Zhang, 1997). However, modern Chinese is definitely more powerful than its ancient predecessor, because it is able to describe a much more complex world. The most frequently used 3,500 characters—covering about 99.87% of modern Chinese—can be combined to form more than 70,000 words (Zhang, 1997), which include the meanings of most ancient characters. Genomes have undergone a similar evolutionary shift. Mammals—such as Homo sapiens and the mouse—have about 25,000 genes (International Human Genome Sequencing Consortium, 2004; Guénet, 2005), which is much fewer than some plants: for example, rice has 43,000 genes (Paterson et al, 2005) and maize has 59,000 (Messing et al, 2004). The greater complexity of mammals is explained partly by the recombination of existing genes, through mechanisms such as alternative splicing (Johnson et al, 2003) and tandem chimerism (Parra et al, 2006). In fact, mammals depend much more on gene recombination to achieve higher complexity than do plants (Messing, 2001).
The third stage of evolution witnesses the arrival of ‘virtual', or modifying, elements, which have an important role as regulatory components. In language, adverbs, auxiliary words, prepositions and conjunctions are all virtual elements, whereas nouns and verbs are the main carriers of information. In fact, the five most frequently used characters in modern Chinese contain two ‘empty' elements (Fig 1). Although virtual words were quite rare in Oracle, they are much more frequent in modern Chinese. Similarly, non-coding RNA has a virtual role in genomes, whereas protein-coding genes are the main carriers of information. In mammals, it is RNA, not protein, that mainly controls gene activity (Mattick, 2004), which also helps to explain the unexpectedly low number of protein-coding genes in mammalian genomes.
Figure 1.
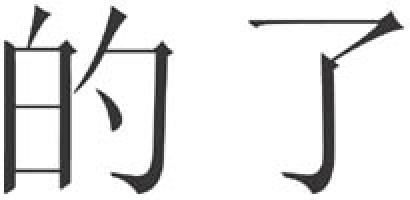
Examples of ‘virtual' elements in modern Chinese. The symbol on the left means ‘of' and the symbol on the right conveys a form of past tense (National Council of Language and Writing, 1989).
In summary, the evolution of both genomes and language has led to increased complexity, but has also developed novel strategies to enhance this process further. Thus, evolution has undergone several paradigm shifts: from increasing the number of basic elements to shuffling existing building blocks into new combinations, and finally to creating virtual regulatory elements. These strategic shifts improve the overall complexity and performance of the systems at a relatively low cost: the ability to combine elements reduces the number of building blocks needed, and virtual elements simplify regulation. Using RNA rather than proteins as regulators circumvents protein translation, and the average length of such RNA signals—22 nucleotides—is almost two orders of magnitude shorter than that required to encode an average protein (Mattick, 2004). It is therefore reasonable to propose that this “evolution of evolutionary strategies” is an intrinsic property of evolving systems. It will be a great challenge to provide further proof—for example, by establishing a model to simulate such higher-level evolution—especially in silico.
Acknowledgments
This study was supported by the National Basic Research Program of China and the National Natural Science Foundation of China.
References
- Chen TJ (1936) Kangxi Dictionary. Shanghai, China: World Press [Google Scholar]
- Guénet JL (2005) The mouse genome. Genome Res 15: 1729–1740 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Ji XL (1989) Encyclopedia of China: Language and Character. Beijing, China: Encyclopedia of China Publishing House [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Mattick JS (2004) RNA regulation: a new genetics? Nat Rev Genet 5: 316–323 [DOI] [PubMed] [Google Scholar]
- Messing J (2001) Do plants have more genes than humans? Trends Plant Sci 6: 195–196 [DOI] [PubMed] [Google Scholar]
- Messing J et al. (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci USA 101: 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Council of Language and Writing (1989) Frequency Statistics of Frequently Used Modern Chinese Characters. Beijing, China: Yuwen Press [Google Scholar]
- Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigó R (2006) Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res 16: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Freeling M, Sasaki T (2005) Grains of knowledge: genomics of model cereals. Genome Res 15: 1643–1650 [DOI] [PubMed] [Google Scholar]
- Searls DB (2002) The language of genes. Nature 420: 211–217 [DOI] [PubMed] [Google Scholar]
- Zhang K (1997) Statistical analysis on basic characters constituting Chinese words. Yuyan Jiaoxue Yu Yanjiu 1997: 43–51 [In Chinese] [Google Scholar]
- Zhang ZC (2004) Large-scale character-usage statistical analysis on Chinese ancient characters. In Proceedings of Third Symposium on Databases of Chinese Writing and History, pp 107–119. Beijing, China: National Committee for Programming Publishing House and National League of Ancient Book Publishing House [Google Scholar]


