Abstract
Background
The circadian system drives pervasive biological rhythms in plants. Circadian clocks integrate endogenous timing information with environmental signals, in order to match rhythmic outputs to the local day/night cycle. Multiple signaling pathways affect the circadian system, in ways that are likely to be adaptively significant. Our previous studies of natural genetic variation in Arabidopsis thaliana accessions implicated FLOWERING LOCUS C (FLC) as a circadian-clock regulator. The MADS-box transcription factor FLC is best known as a regulator of flowering time. Its activity is regulated by many regulatory genes in the "autonomous" and vernalization-dependent flowering pathways. We tested whether these same pathways affect the circadian system.
Results
Genes in the autonomous flowering pathway, including FLC, were found to regulate circadian period in Arabidopsis. The mechanisms involved are similar, but not identical, to the control of flowering time. By mutant analyses, we demonstrate a graded effect of FLC expression upon circadian period. Related MADS-box genes had less effect on clock function. We also reveal an unexpected vernalization-dependent alteration of periodicity.
Conclusion
This study has aided in the understanding of FLC's role in the clock, as it reveals that the network affecting circadian timing is partially overlapping with the floral-regulatory network. We also show a link between vernalization and circadian period. This finding may be of ecological relevance for developmental programing in other plant species.
Background
Most eukaryotes and some prokaryotes have evolved a circadian clock to adapt to the 24 h day/night cycle. These clocks drive biological rhythms in many aspects of metabolism, physiology, and behavior, all with a period close to 24 h [1]. Circadian rhythms affect fundamental processes of plant life, such as photosynthesis and cell elongation [2]. Day-length measurement (photoperiodism) also depends on the circadian clock, which thereby controls seasonal rhythms such as the timing of flowering [3,4]. Described molecular-genetic models from various circadian organisms has each included a gene circuit with negative-feedback elements, involving 24 h rhythms in the levels of positively- and negatively-acting transcriptional regulators [5]. In Arabidopsis, there is emerging evidence that a set of about 20 genes create one, or more, feedback circuits (the 'circadian oscillator') to generate the 24 h period [2,6], and this rhythmically regulates the level of around 6% of transcripts [7].
Circadian clocks, including those of Arabidopsis, are reset by light and temperature signals in a characteristic fashion that entrains the clock to the local time in its environment [6]. However, circadian period is buffered against long-term changes in temperature, such that the period remains close to 24 h when assayed at various constant temperatures, over a physiologically relevant range. Such 'temperature compensation' is another distinguishing feature of circadian clocks, including those in Arabidopsis [8,9]. Whereas the mechanisms of photic entrainment are being elucidated [2,6], those governing temperature entrainment and temperature compensation remain to be determined.
A circadian clock maintains accurate timing because it is buffered against many environmental changes, yet several environmental-signaling pathways must affect the circadian oscillator for entrainment to occur. Limiting the input connections to the circadian clock from the rest of the plant-signaling network provides a potential mechanism to balance the opposing requirements of homeostasis and entrainment. In the gene network that regulates flowering time, for example, the circadian clock is an integral part of the photoperiodic sensor, receiving input from light signaling [3,4]. Current models indicate that output from the photoperiod pathway converges with several other pathways that control flowering time, but the photoperiod sensor is thought to receive no input from those pathways [3,4]. Genetic variation among Arabidopsis accessions prompted us to reexamine this notion.
Substantial natural variation has been detected in clock-affecting genes, based upon assays of rhythmic leaf movement under constant light [9-11]. This assay allowed us to map Quantitative Trait Loci (QTL) that affect circadian period in recombinant populations derived from crosses between accessions Cape Verde Islands (Cvi) × Landsberg erecta (Ler) and Columbia (Col) × Ler [10]. In each population, we located a major QTL towards the top of chromosome 5, close to the map location of FLOWERING LOCUS C (FLC). The Ler allele of FLC is weakly functional due to a transposon insertion within an intron of FLC [12,13]. The populations that we used included Ler as one parent, therefore FLC function segregates in these recombinant populations [14]. The Ler allele of the QTL shortened the circadian period by 0.8 h, as did independent flc mutant alleles, leading us to conclude that the known allelic variation in FLC could account for the QTL [10].
FLC encodes a MADS-box transcription factor that had no known function in the circadian clock, but was well-characterized as a repressor of flowering. FLC expression is enhanced by FRIGIDA (FRI) and its paralogues, which are active in many late-flowering accessions [12,13]. FLC transcription is suppressed by genes of the autonomous floral-promotion pathways and by prolonged cold temperatures (vernalization, indicative of winter in nature; reviewed in [3,11,12]). As flc mutants harbor an altered circadian period, we hypothesized that other genetic and physiological regulators of FLC would have predictable effects on the circadian clock. We therefore tested whether the network of FLC regulators that was defined with respect to flowering time also functions in the control of circadian period. A substantial number of upstream regulators, a related gene, and a downstream target gene do have similar functions, but we also find clear distinctions between FLC-related pathways. The circadian period is also sensitive to vernalization, revealing a previously-unrecognized connection between the gene circuits involved in responses to daily and to seasonal rhythms.
Results
Dose-dependent effect of FLC on circadian period
FLC RNA abundance correlates with repression of flowering time and quantitatively mediates the vernalization response of flowering time [15]. We sought to determine whether FLC expression levels similarly regulated the circadian clock. To assay the plant's endogenous circadian period, rhythmic movement of the primary leaves of Arabidopsis seedlings were measured by video imaging under constant white light. Relatively large numbers of plants were tested in replicate experiments in order to increase the sensitivity of the assays, allowing us to detect small changes in circadian period (see experimental procedures). FLC RNA abundance was manipulated by two methods. Firstly, we tested a line expressing FLC from the CaMV 35S promoter (35S:FLC). This transgenic construction strongly delays flowering time. The 35S:FLC line had a circadian period lengthened by over 1 hour in multiple experiments (τ = 25.65 ± 0.31 [SEM] h vs. 24.44 ± 0.17 h, P < 0.0001) (Figure 1, Table 1). Secondly, circadian experiments were carried out on lines harboring the four possible homozygous combinations of FLC alleles with alleles of the FLC activator, FRIGIDA (FRI), all uniformly in a Col background. These comprised the functional allelesFRI-Sf2 and FLC-Col (FRI and FLC) and recessive alleles fri-Col and flc-3 (fri and flc) [16]. Lines harboring functional FLC had a lengthened circadian period compared to flc lines (Figure 1). Joint statistical analysis of 6 replicate experiments was used to reveal that FLC alone increased circadian period by an average of 0.6 hours, across both FRI genotypes (P = 0.004, Figure 1, Table 1). This is in agreement with the results of Swarup et al. (1999) [10], who showed that FLC lengthened circadian period in both FRI and fri backgrounds. Our findings are also consistent with the function of FLC in flowering time. fri;FLC plants flower slightly later than fri;flc mutants under non-inductive photoperiods, indicating FLC can function independently of FRI in the floral pathway [17]. Increasing the abundance of FLC transcript is therefore sufficient to increase the circadian period of the plant. The greater effect of 35S:FLC compared to endogenous FLC suggests that increasing FLC RNA levels increase circadian period in a graded manner, similar to FLC's dose-dependent delay of flowering [16,18]. It might be possible that a broader spatial domain of FLC expression in 35S:FLC plants contributed to this result, though such effects on the location of FLC expression have not previously been implicated in flowering-time control.
Figure 1.
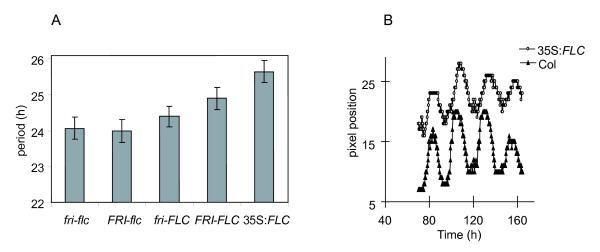
Effects of null flc and 35S: FLCon circadian period 1a) Circadian periods of FRI-FLC mutant combination and 35S:FLC plants assayed by rhythmic leaf movement in constant dim white light for approx. 1 week (n = 36–155). fri-flc τ = 24.07 ± 0.31 [SEM] h, FRI-flc τ = 24.00 ± 0.32 h, fri-FLC τ = 24.40 ± 0.29 h, FRI-FLC τ = 24.90 ± 0.31 h. 1b) Representative traces of rhythmic leaf movement of a 35S:FLC (white circles) and wild-type (filled triangles).
Table 1.
FLC effects on circadian period.
| genotype | fri;flc | FRI;flc | fri;FLC | FRI;FLC | Col | 35S:FLC |
| period(h) | 24.07 | 24.00 | 24.40 | 24.90 | 24.44 | 25.65 |
| s.e.m | 0.31 | 0.32 | 0.29 | 0.31 | 0.17 | 0.31 |
| n | 85 | 82 | 107 | 87 | 155 | 36 |
| P | _ | _ | *0.004 | *0.004 | _ | ** < 0.0001 |
Mean circadian periods of leaf movement in Arabidopsis mutant and wild-type seedlings, calculated using REML analysis.
* P from Wald test; indicates the statistical significance of FLC's effect alone, by comparison of FLC to flc irrespective of FRI/fri.
** P from the standard error of the difference; indicates the statistical significance of 35S:FLC effect, compared to Col wild-type.
s.e.m., standard error of the mean.
n, number of contributing leaf traces.
We suggest that functional FRI caused an additional increase in period in the presence of functional FLC (Figure 1; see also Figure 3). Although FRI;FLC had the longest mean period in each experiment, the interaction between FRI and FLC was not significant in the joint analysis (data not shown). This indicates that FRI can increase circadian period weakly and less consistently than FLC. In contrast, functional FRI strongly enhances FLC RNA levels and severely delays flowering [16,18], highlighting an obvious difference between circadian and flowering time control. Again, it is possible that FRI does not enhance FLC expression in the cells that regulate leaf movement as much as 35S:FLC.
Figure 3.
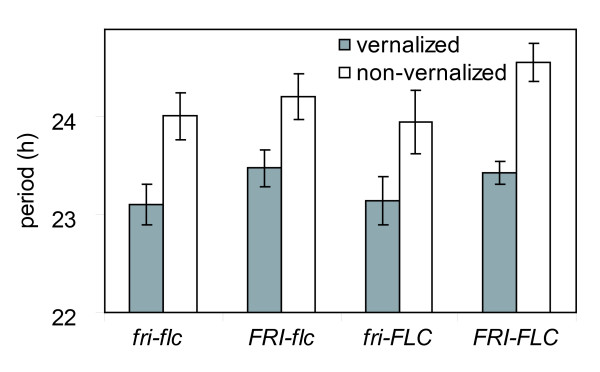
Effects of vernalization on the Arabidopsis clock Comparison of circadian periods of vernalized and non-vernalized Arabidopsis mutant seedlings harboring all possible combinations of FRI-FLC (n = 18–26). After vernalization treatment at 2°C for 8 weeks, plants were assayed for rhythmic leaf movement for approx. 1 week in constant dim white light. vern fri-flc τ = 23.10 ± 0.21 [SEM] h, non-vern fri-flc τ = 24.01 ± 0.24 h, vern FRI-flc τ = 23.48 ± 0.19 h, non-vern FRI-flc τ = 24.21 ± 0.23 h, vern fri-FLC τ = 23.14 ± 0.25 h, non-vern fri-FLC τ = 23.95 ± 0.32 h, vern FRI-FLC τ = 23.43 ± 0.12 h, non-vern FRI-FLC τ = 24.56 ± 0.19 h.
Effects of FLC regulators on circadian period
The above results lead us to the hypothesis that any factor that modulates FLC expression levels, including the genes of the autonomous pathway, would affect circadian period. In order to compare the network of circadian regulators to the pathways that regulates flowering time, we analyzed the circadian period of plants carrying mutations in several genes of the autonomous flowering-time pathway (Figure 2), all of which contribute to regulate FLC RNA abundance (reviewed in Henderson et al., 2003 [19]).
Figure 2.
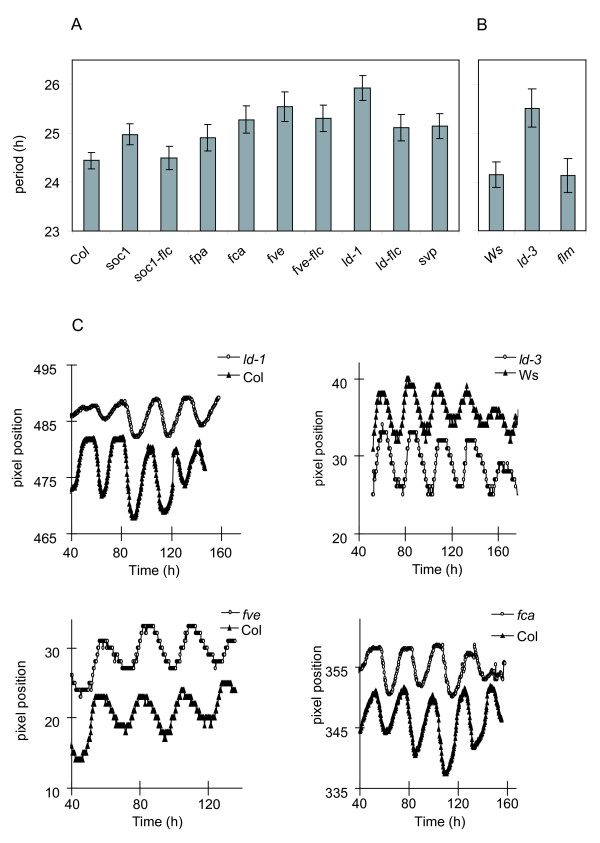
Circadian period of autonomous flowering time pathway mutants Circadian period estimates for mutants in the autonomous flowering-time pathway and their respective wild-types. Period was assayed by rhythmic leaf movement in constant white light for approx. 1 week (n = 25–155). 1a) genotypes in the Col-0 genetic background. 1b) Genotypes in the Ws-2 background. 1c) Representative leaf movement traces of Arabidopsis mutant (white circles) and wild-type seedlings (filled triangles), imaged in constant dim white light for approx. 1 week.
FCA
FCA is one of the genes that defines the autonomous flowering-time pathway. FCA encodes a nuclear-localized protein with RNA recognition motifs. It is involved in 3' RNA processing of FCA transcripts through physical interaction with the FY protein [20-22]. FCA promotes flowering by repression of FLC RNA levels [17,23]: plants with an fca lesion have high levels of FLC RNA and flower late. Assays on fca mutants revealed an increase in circadian period of nearly 1 hour compared to the Col-0 wild-type (τ = 25.28 ± 0.28 h vs. 24.44 ± 0.17 h, P = 0.002), in line with the hypothesis that elevated FLC levels increase circadian period (Figure 2 and 5, Table 2).
Figure 5.
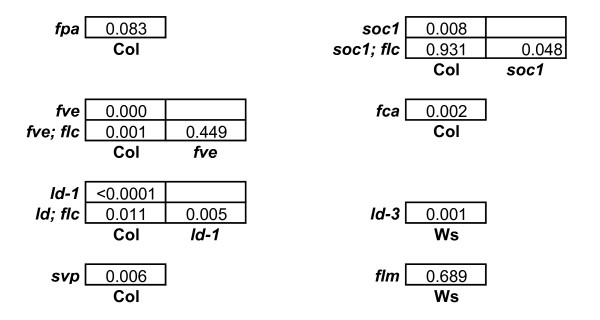
Statistical significance testing for circadian period effects of floral pathway mutant lines. Measures of significance for comparisons among circadian periods from Table 2, calculated from the standard errors of the differences.
Table 2.
Circadian period of floral pathway mutant lines.
| genotype | Col | fpa | soc1 | soc1;flc | fve | fve;flc | fca |
| period(h) | 24.44 | 24.91 | 24.98 | 24.49 | 25.54 | 25.30 | 25.28 |
| s.e.m | 0.17 | 0.27 | 0.22 | 0.24 | 0.30 | 0.27 | 0.28 |
| n | 155 | 46 | 69 | 70 | 36 | 36 | 39 |
| genotype | Col | ld-1 | ld;flc | Ws | ld-3 | ||
| period(h) | 24.44 | 25.93 | 25.12 | 24.15 | 25.51 | ||
| s.e.m | 0.17 | 0.25 | 0.27 | 0.26 | 0.39 | ||
| n | 155 | 53 | 43 | 49 | 19 | ||
| genotype | Ws | flm | Col | svp | |||
| period(h) | 24.15 | 24.13 | 24.44 | 25.15 | |||
| s.e.m | 0.26 | 0.35 | 0.17 | 0.25 | |||
| n | 49 | 25 | 155 | 53 | |||
Mean circadian periods of Arabidopsis mutant and wild-type seedlings, tested as in Figure 1.
LUMINIDEPENDENS (LD)
LD represses FLC expression and encodes a predicted nuclear protein with a glutamine rich Carboxy terminus, suggesting a role as a transcriptional regulator [16,24,25]. Of the autonomous pathway genes tested, LD had the most striking effect on circadian period. A period increase of approximately 1.5 hours in ld mutants was observed relative to the wild type, in two genetic backgrounds (τ = 25.93 ± 0.25 h vs. 24.44 ± 0.17 h for ld-1 in a Col background, P= <0.0001 and τ = 25.51 ± 0.39 h vs. 24.15 ± 0.26 h for ld-3 in a Ws-2 background, P= 0.0012) (Figure 2 and 5, Table 2). In order to test whether the circadian effect of LD required FLC, we assayed the period of an ld; flc double mutant [17,23]. The ld; flc double mutant lines decreased circadian period by 0.8 h relative to the ld single mutant (τ = 25.12 ± 0.27 h vs. 25.93 ± 0.25 h, P = 0.005) (Figure 2). The ld; flc double mutant had a longer circadian period than wild-type plants (τ = 25.12 ± 0.27 h vs. 24.44 ± 0.17 h, P = 0.01) (Figure 2 and 5, Table 2), which in turn had a longer period than the flc mutants (Figure 1). Thus the ld;flc double mutant has an intermediate phenotype, with a significantly longer period than the flc single mutant but shorter than the ld single mutant. We conclude that the effect of LD on period is at most partly dependent on FLC, andLD function must also influence the circadian clock independently of FLC. This contrasts with the FLC-dependence of the effects of LD on flowering [17].
FVE
FVE encodes a component of a histone-acetylation complex, which functions to strongly suppresses FLC RNA abundance, to thus function in the flowering-time pathway [23,26,27]. Additionally, FVE affects morphological traits such as leaf shape and inflorescence patterns [28]. The fve mutant increased circadian period by over 1 hour (τ = 25.54 ± 0.30 h vs. 24.44 ± 0.17 h, P = 0.0002). To test whether this was due to an increase in FLC activity in the mutant, we assayed the fve; flc double mutant. We found a statistically insignificant decrease in circadian period (0.24 h) in the double mutant compared to the fve single mutant (τ = 25.30 ± 0.27 h vs. 25.54 ± 0.30 h, P= 0.45), so the period of the fve; flc double mutant retained a significantly lengthened period (Figure 2 and 5, Table 2). These results indicate to us that FVE regulates circadian timing in a manner that is largely independent of FLC, again in contrast to its effect on flowering [17].
FPA
FPA is predicted to encode a protein that contains RNA recognition motifs and also represses FLC RNA and function in the flowering-time pathway [29]. The fpa mutant showed only a small increase in period (0.47 h) compared to the wild type, and this modest effect was not statistically significant (τ = 24.91 ± 0.27 h vs 24.44 ± 0.17 h, P = 0.08) (Figure 2 and 5, Table 2). Therefore, FPA seems to have less effect on the circadian clock of Arabidopsis than the other mutants tested. Though FPA has been functionally linked with FVE in the flowering time network, it differs from FVE in that FPA is less sensitive to FLC gene dosage [23], FPA interacts with genes of the photoperiodic-response pathway [30], and has been implicated in the gibberellin response [31]. These differences may reflect a range of molecular functions that limits the effect of FPA on circadian timing.
Effects of other MADS-box genes on circadian period
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1)
SOC1 (also referred to as AGL20) is a MADS-box gene that activates flowering, in response to signals from the autonomous and photoperiodic flowering-time pathways [32,33]. Unlike the genes tested above, SOC1 is described as a target of FLC-mediated transcriptional repression in the flowering-time pathway [17]. The soc1 mutant has a small increase in circadian period relative to wild type (τ = 24.98 h ± 0.22 h vs. 24.44 ± 0.17 h, P = 0.008), indicating that SOC1 normally shortens circadian period (Figure 2 and 5, Table 2). If FLC represses SOC1 expression, this result is consistent with the idea that flc mutants shorten circadian period at least in part via increased expression of SOC1. We assayed the soc1; flc double mutant in order to test this notion, which would predict a long period in the soc1; flc double mutant, similar to the soc1 single mutant. The period of the double mutant was however intermediate between the single mutants, being slightly reduced compared to the soc1 single mutant (τ = 24.49 ± 0.24 h vs. 24.98 ± 0.22 h, P= 0.048). The resulting period of soc1; flc was identical to that of Col (Figure 2 and 5, Table 2), but longer than that of the flc null mutant (Figure 1). This result suggests that FLC may increase circadian period in part by repressing SOC1, but its effect also occurs by other means, consistent with the function of FLC in the floral-repression pathway [17].
FLM and SVP
FLC is a member of a six-gene sub-family of the MADS class, based on sequence similarity. FLOWERING LOCUS M (FLM) (also known as MAF1/AGL27), is the member of this family with the highest level of identity with FLC [34]. Over-expression studies have shown FLM to have a similar function in the repression of flowering as FLC. However, unlike FLC, FLM expression is not increased by FRI, and FLM does not contribute to the vernalization requirement [34,35]. Leaf-movement assays on the flm mutant revealed that it had no circadian defect (P = 0.69) (Figure 2 and 5, Table 2).FLM functions with another MADS box gene, SVP (SHORT VEGETATIVE PHASE) in repressing flowering time [35,36]. When we tested for circadian period, the svp mutant showed a significant lengthening in period of 0.7 h compared to wild type (τ = 25.15 ± 0.25 h vs. 24.44 ± 0.17 h, P = 0.006) (Figure 2 and 5, Table 2), in contrast to the significantly shortened circadian period of the flc mutant. These results indicate that FLC may have a unique function in shortening the circadian period of Arabidopsis, which is not shared by other MADS box transcription factors that function in the flowering-time pathways.
Seasonal regulation of the circadian clock by vernalization
FLC expression represses flowering in biennial and winter-annual plants, causing overwintering in the vegetative state. Vernalization relieves the floral repression in part by stably suppressing FLC expression, so flowering can proceed in the Spring [37]. flc mutants retain some vernalization response [17], which may be mediated by other MADS-box genes [38]. Based on the prediction that any effect on FLC-expression levels, whether genetic or physiological, should alter circadian period, we sought to test the effect of vernalization on circadian period and the effect of flc mutations on the clock after vernalization. We assayed circadian period in seedlings harboring the FRI and FLC alleles, as described above, comparing seedlings that had been vernalized to controls grown without vernalization. Flowering-time analysis of these plants, following the leaf movement assays, confirmed the effectiveness of the vernalization treatments in accelerating flowering (data not shown). We found in duplicate experiments that vernalization consistently decreased circadian period (P < 0.001) but that none of the single-gene or gene interaction effects was significant (Figure 3, Tables 3 and 4). Therefore, vernalization consistently shortened circadian period, regardless of FLC. There is a possibility that development during the vernalization period may have caused the change in circadian period. However, prior to these experiments, we identified growth conditions for the vernalized and non-vernalized plants such that in these experiments, seedlings of both groups were phenotypically indistinguishable from one another. Furthermore, as the same primary leaf pair was assayed in both treatment groups, a directly comparable developmental trait comparison was made (see Materials and Methods).
Table 3.
Circadian period of lines pre- and post-vernalization.
| Vernalized plants | ||||
| genotype | fri;flc | FRI;flc | fri;FLC | FRI;FLC |
| period(h) | 23.10 | 23.48 | 23.14 | 23.43 |
| s.e. | 0.21 | 0.19 | 0.25 | 0.12 |
| n | 20 | 24 | 20 | 26 |
| Non-vernalized plants | ||||
| genotype | fri;flc | FRI;flc | fri;FLC | FRI;FLC |
| period(h) | 24.01 | 24.21 | 23.95 | 24.56 |
| s.e. | 0.24 | 0.23 | 0.32 | 0.19 |
| n | 22 | 18 | 23 | 24 |
Mean circadian period of Arabidopsis mutant seedlings, tested as in Figure 1, with or without vernalization at 2°C for 8 weeks.
Table 4.
Statistical significance testing for period effects of vernalization.
| Gene factor × vernalization | *P |
| vernalization(minus FRI, FLC factors) | <0.001 |
| FRI | 0.842 |
| FLC | 0.692 |
| FRI × FLC | 0.461 |
* P from Wald test; indicates the statistical significance of vernalization treatment effects alone and vernalization × gene effects on circadian period.
Discussion
We report here that the genes of the autonomous floral-promotion pathway, and FLC itself, modulate the period of a circadian clock in Arabidopsis. The effects on period are modest, but our data measurements are accurate, as detected in plots of relative amplitude error (RAE) versus period length for individual cotyledon traces of a genotype in an experiment (see Additional file 1). Additionally, there is no noticeable increase in arrhythmic mutant plants relative to wild-type (data not shown). Data for all genotypes tested in this study were collected from multiple experiments and in each case the period effects relative to the wild type were consistent. The only exception to this was the fpa mutant, which had first a long period and then a short period, relative to wild-type, in two independent experiments. As expected from this result, statistical analysis of these data was unable to assign a function for FPA in the circadian clock. It should also be mentioned that in each of the multiple FRI;FLC mutant combination studies (Figure 1A, Table 1), fri-flc double mutant period was always reduced compared to FRI-FLC period. Direct comparison with other genotypes between replicate experiments was not reasonable without the statistical analysis we performed using REML.
The mechanisms by which the floral-promotion genes affect circadian period are similar, but not identical, to their control of flowering time. Regulators other than FLC must be involved because, for example, LD and FVE affect the clock significantly in the flc mutant background. We also reveal a vernalization-dependent shortening of the circadian period. Recent studies in chestnut seedlings have shown that during the chilling period, circadian expression of genes homologous to Arabidopsis core-clock genes are suppressed, with cyclic expression of these genes resumes post-chilling [39]. Our studies however identify alterations in the Arabidopsis clock after the cold-exposure period. FLC would be an obvious candidate gene to mediate this response, however, our studies did not fully support this possibility. Circadian analysis of mutants in other vernalization-responsive MADS-box genes, such as MAF2 [34,40] may shed light on the how the clock is altered by vernalization. It is unclear which components of the circadian-clock mechanism are the targets that mediate these period changes. Rhythmic, transcriptional-translational feedback loops are important in circadian timing and the genes tested here are regulators of gene expression, though FLC is not thought to be rhythmically regulated [41]. It is possible that the expression level of one of the clock genes is FLC-dependent, for example. Given the modest effects we observed upon circadian period, the FLC-dependent change in expression level might be very slight.
From an ecological perspective, the stable effects of vernalization allow plants to distinguish between Spring and Autumn, even though both seasons have an equal day length. Our work suggests that an additional mechanism may contribute to this distinction, namely that the circadian clocks of plants run "faster" in Spring than in Autumn.
Environmental changes can thus have "after-effects" on the circadian clock, in Arabidopsis, as in other organisms. These are typically observed as an alteration in the circadian period immediately after exposure to exotic light-dark or warm-cold cycles [11,42]. The period-shortening effect of vernalization is expected to be much longer-lasting. The physiological consequences of the period shortening will depend on which rhythms are affected, and how the change in period under constant conditions affects the rhythms under day-night cycles. Short-period mutations can reduce the critical photoperiod in Arabidopsis, leading to earlier flowering under shorter photoperiods [43]. If the shortened circadian periods due to FLC repression affected rhythmic CO expression, this would reinforce the acceleration of flowering in Spring days compared to Autumn days, by induction through the photoperiod pathway, in addition to the removal of autonomous-pathway flowering repression. Such cross-talk between the autonomous pathway and the circadian system emphasizes the networked structure of plant-signaling circuits. These include the circuits adapted to mediate plastic responses to rhythmic, daily, and seasonal environmental signals.
Conclusion
We demonstrated that several genes in the Arabidopsis autonomous-flowering pathway are also involved in regulation of the circadian clock. We identify FLC as a dose-dependant regulator of circadian period and identify autonomous-pathway genes regulating the clock in both an FLC dependant and independent manner. As FLC expression levels are reduced by vernalization, we tested the hypothesis that circadian period was altered in vernalized FLC wild-type plants. Though we could not firmly establish FLC as the mediator of vernalization's effect on circadian period, we showed conclusively that vernalization alters circadian period in Arabidopsis. Figure 4 is an illustrated schematic of how vernalization and the genes we tested may be regulating circadian period in Arabidopsis.
Figure 4.
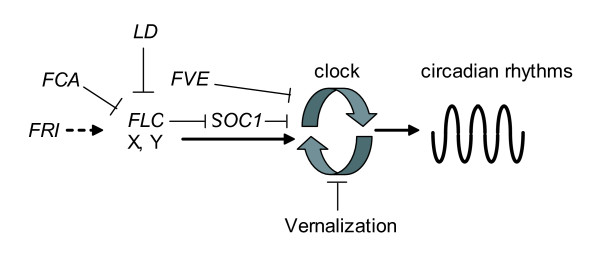
Schematic of FLC's role in the Arabidopsis circadian clock. A schematic of FLC's role in regulating circadian period. FLC function increases circadian period (arrow). This function is marginally enhanced by FRI (dashed arrow) and perhaps mediated in part by repressing (blunt arrow) SOC1, which decreases period (blunt arrow). Flowering-time genes of the autonomous pathway such as LD and FCA maintain shorter circadian periods in part by repressing FLC but with significant contributions via other components (X and Y, drawn with FLC for clarity only). Vernalization also regulates circadian period by an unidentified mechanism. Not all genes of the autonomous flowering pathway influence circadian timing (e.g. FPA, not shown).
Methods
Plant materials
The following mutant lines in the Columbia-0 background have been described previously:
FRI-Sf2;FLC-Col, FRI-Sf2;flc-3, fri-Col;FLC-Col, fri-Col;flc-3, 35S:FLC [16]; soc1-2, soc1-2;flc-3 (previously agl20, agl20;flc-3) [44];fve-4, fve-4;flc-3 [45];ld-1, ld-1;flc-3, fpa-7 [17]. Mutants in the Ws-2 background have been described: flm [46], ld-3 [24]. We are grateful for seed of fca-9 [47] from Caroline Dean (Norwich), of svp-41 [36] from Peter Huijser (Cologne), and of ld-1 from the Nottingham Arabidopsis Stock Centre.
Growth and imaging conditions
Seedlings were grown and followed by assays of rhythmic leaf movements by video imaging, as described by Dowson-Day and Millar, 1999; Millar et al. 1995 [48,49]. Briefly, surface-sterilized seeds were plated on 1.5% Murashige-Skoog agar medium plates [50] that included 3% sucrose, and were stratified for 3 days at 4°C. Seedlings were germinated under 30–40 μmolm-2sec-1 continuous cool white fluorescent light for 7 days, at 21–22°C, followed by entrainment to three 12 h-light, 12 h-dark cycles. Growth of the first pair of primary leaves was recorded under 30–40 μmolm-2sec-1 continuous cool white fluorescent light, at 21–22°C for 7 days. Seedlings were arranged randomly with respect to genotype within each experiment, to avoid positional bias in the imaging arrays. For vernalization, stratified seed were germinated for 4 days, as described above (at which point cotyledons were expanding), then incubated at 2°C for 8 weeks under low-intensity white light (0.5–1.0 μmol m-2sec-1). After the vernalization treatment, seedlings were transferred to continuous light for 4 days, then entrained and imaged as above. In order to confirm the effectiveness of vernalization, imaged seedlings were transferred from agar to soil immediately after the leaf-movement assay. Flowering time of these lines was measured as the number of rosette leaves when the floral bolt was 1 cm high. These studies confirmed the expected effects of FLC and FRI upon vernalization-responsiveness (data not shown).
Data analysis
Leaf movement data were analyzed by Fast Fourier Transform non-linear least squares program FFT-NLLS [51], essentially as described in Dowson-Day and Millar, 1999 [48]. The circadian period of each genotype was estimated as the variance-weighted mean of the most significant period within the circadian range (15–35 h) for each leaf. In order to make the most efficient use of data gathered in separate experiments, the data from all the experiments were analyzed jointly using REML [52] in the statistical package GENSTAT 5 [53]. REML is a generalization of analysis of variance and is appropriate for the analysis of unbalanced data. Genotypes to be compared directly were included in the same experiments. In the analyses, experiment, camera within experiment, plant within camera, and cotyledon within plant were taken as random factors, with mutant line as a fixed factor. The analysis was weighted to allow for the inherent variabilities of estimation of period from the different traces. The period estimate for each leaf recording was weighted for analysis by the reciprocal of the error associated with the period, as estimated by FFT-NLLS. Significance of FRI-FLC interactions and vernalization effects on period were assessed using the Wald test with variances derived from REML. The significance of the differences between the mean period of pairs of genotypes was assessed using the standard error of each difference, derived from REML. Figures 1, 2,3 report the conventional SE of each genotype mean.
Authors' contributions
NS, AJM, and SJD conceived the experiments and wrote the paper, with critical experimental and intellectual revisions from SDM and RMA. SDM and RMA generated reagents. NS carried out the experiments. JRL conducted statistical analysis.
Supplementary Material
Robustness of rhythmicity of all lines examined MS EXCEL file containing 3 worksheets. These include RAE plots for all genotypes examined (sorted with respect to Figure 1, 2, 3 in the main article body).
Acknowledgments
Acknowledgements
Work in R.M.A's laboratory was supported by the College of Agricultural and Life Sciences and the Graduate School of the University of Wisconsin, and by National Science Foundation grants 0133663 and 0209786. N.S. was supported by a CASE studentship from Horticulture Research International and the Biotechnology and Biological Sciences Research Council. S.J.D. was a Department of Energy fellow of the Life Sciences Research Foundation. The imaging facility at Warwick was funded by grants from the Biotechnology and Biological Sciences Research Council, Royal Society and Gatsby Charitable Foundation to A.J.M.
Contributor Information
Neeraj Salathia, Email: nsalathia@cgr.harvard.edu.
Seth J Davis, Email: davis@mpiz-koeln.mpg.de.
James R Lynn, Email: james.lynn@warwick.ac.uk.
Scott D Michaels, Email: michaels@indiana.edu.
Richard M Amasino, Email: amasino@biochem.wisc.edu.
Andrew J Millar, Email: andrew.millar@ed.ac.uk.
References
- Jay C, Dunlap JJL, Patricia J. DeCoursey: Chronobiology: Biological Timekeeping. Sinauer Associates; 2003. [Google Scholar]
- McClung CR. Circadian Rhythms in Plants. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:139–162. doi: 10.1146/annurev.arplant.52.1.139. [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell. 2004;16:S18–31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Coupland G. Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol. 2003;6:13–19. doi: 10.1016/S1369-5266(02)00011-0. [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nature Reviews Genetics. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Millar AJ. The circadian clock. A plant's best friend in a spinning world. Plant Physiol. 2003;132:732–738. doi: 10.1104/pp.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170:387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ. Natural allelic variation identifies new genesin the Arabidopsis circadian system. Plant Journal. 1999;20:67–77. doi: 10.1046/j.1365-313X.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;132:1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:10102–10107. doi: 10.1073/pnas.1531467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, vandenBosch N, Pot J, Kuiper MTR. Development of an AFLP based linkagemap of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant Journal. 1998;14:259–271. doi: 10.1046/j.1365-313X.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. Vernalization and epigenetics: how plants remember winter. Curr Opin Plant Biol. 2004;7:4–10. doi: 10.1016/j.pbi.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Shindo C, Dean C. The need for winter in the switch to flowering. Annu Rev Genet. 2003;37:371–392. doi: 10.1146/annurev.genet.37.110801.142640. [DOI] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/S0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/S0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- Quesada V, Macknight R, Dean C, Simpson GG. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. Embo J. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda SL, Amasino RM. Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Molecular and General Genetics. 1996;251:69. doi: 10.1007/BF02174346. [DOI] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM. Isolation of _LUMINIDEPENDENS_: A gene involved in the control of flowering time in Arabidopsis. Plant Cell. 1994;6:75–83. doi: 10.1105/tpc.6.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M, Koornneef M, Putterill J. Flowering on time: genes that regulate the floral transition. Workshop on the molecular basis of flowering time control. EMBO Rep. 2001;2:1078–1082. doi: 10.1093/embo-reports/kve254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Van der Veen JH. A genetic and physiological analysis of late flowering mutants in _Arabidopsis thaliana_. Molecular and General Genetics. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Jarillo JA, Cruz-Alvarez M, Rold n M, Salinas J. _Arabidopsis_ late-flowering _fve_ mutants are affected in both vegetative and reproductive development. Plant Journal. 1995;7:543–551. doi: 10.1046/j.1365-313X.1995.7040543.x. [DOI] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse DT, Sheldon CC, Bagnall DJ, Peacock WJ, Dennis ES. FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 2002;29:183–191. doi: 10.1046/j.0960-7412.2001.01210.x. [DOI] [PubMed] [Google Scholar]
- Meier C, Bouquin T, Nielsen ME, Raventos D, Mattsson O, Rocher A, Schomburg F, Amasino RM, Mundy J. Gibberellin response mutants identified by luciferase imaging. Plant J. 2001;25:509–519. doi: 10.1046/j.1365-313x.2001.00980.x. [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci K, Michaels SD, Amasino RM. Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol Biol. 2003;52:915–922. doi: 10.1023/A:1025426920923. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 2000;21:351–360. doi: 10.1046/j.1365-313x.2000.00682.x. [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant Journal. 2003;33:867–874. doi: 10.1046/j.1365-313X.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Ramos A, Perez-Solis E, Ibanez C, Casado R, Collada C, Gomez L, Aragoncillo C, Allona I. Winter disruption of the circadian clock in chestnut. Proc Natl Acad Sci U S A. 2005;102:7037–7042. doi: 10.1073/pnas.0408549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Roden LC, Song HR, Jackson S, Morris K, Carre IA. Floral responses to photoperiod are correlated with the timing of rhythmic expression relative to dawn and dusk in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13313–13318. doi: 10.1073/pnas.192365599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003;35:613–623. doi: 10.1046/j.1365-313X.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Lee MH, Moon J, Lee I, Kim J. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet. 2004;36:167–171. doi: 10.1038/ng1298. [DOI] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant Journal. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- Page T, Macknight R, Yang CH, Dean C. Genetic interactions of the Arabidopsis flowering time gene FCA, with genes regulating floral initiation. Plant Journal. 1999;17:231–239. doi: 10.1046/j.1365-313X.1999.00364.x. [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant Journal. 1999;17:63–71. doi: 10.1046/j.1365-313X.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA. The Regulation of Circadian Period by Phototransduction Pathways in Arabidopsis. Science. 1995;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue. Physiologia Plantarum. 1962;15:493–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. Journal of Biological Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–554. doi: 10.2307/2334389. [DOI] [Google Scholar]
- Payne RW, Lane P, Digby P, Harding S, Leech P, Morgan G, Todd A, Thompson R, Tunnicliffe W, Welham S, White R. Genstat 5 Release 3 Reference Manual. Oxford: Oxford University Press; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Robustness of rhythmicity of all lines examined MS EXCEL file containing 3 worksheets. These include RAE plots for all genotypes examined (sorted with respect to Figure 1, 2, 3 in the main article body).


