Abstract
The emergence and increasing prevalence of multidrug-resistant bacterial pathogens emphasizes the need for new and innovative antimicrobial strategies. Lytic phages, which kill their host following amplification and release of progeny phage into the environment, may offer an alternative strategy for combating bacterial infections. In this study, however, we describe the use of a nonlytic phage to specifically target and deliver DNA encoding bactericidal proteins to bacteria. To test the concept of using phage as a lethal-agent delivery vehicle, we used the M13 phagemid system and the addiction toxins Gef and ChpBK. Phage delivery of lethal-agent phagemids reduced target bacterial numbers by several orders of magnitude in vitro and in a bacteremic mouse model of infection. Given the powerful genetic engineering tools available and the present knowledge in phage biology, this technology may have potential use in antimicrobial therapies and DNA vaccine development.
The emergence of antimicrobial resistance among a multitude of bacterial and fungal pathogens has become a critical problem in modern medicine. The economic impact of managing infections caused by antibiotic-resistant bacteria is substantial, and current costs are estimated to be more than US$4 billion annually (14). As resistance spreads among bacteria, there is grave concern that antimicrobial agents will become increasingly limited and, in some cases, nonexistent. Bacterial resistance is now a serious worldwide problem, and development of novel antibacterial therapies must become a high priority.
Lytic phage therapy has been proposed as a natural alternative approach to conventional antibiotics, and noncontrolled clinical studies have shown that phages can be effective in combating infections caused by a variety of pathogens in humans (2, 4, 19, 43, 44). Early animal studies by Smith and Huggins (39) demonstrated that a single intramuscular dose of an anti-K1 phage preparation (3 × 108 PFU) was more effective than multiple doses of antibiotics in protecting mice from an Escherichia coli O18:K1:H7 infection. Later studies showed that a single oral dose of E. coli-specific phage could successfully reduce, by many orders of magnitude, the number of bacteria in the alimentary tract of calves, piglets, and lambs infected with an enteropathogenic E. coli strain (40, 41). Recently, a phage with lytic activity against clinical isolates of Enterococcus faecium was shown to be effective in rescuing mice from a lethal vancomycin-resistant E. faecium infection (3).
In this report we describe an alternative approach involving the use of genetically engineered phage to deliver the instructions for cell death to bacteria. In contrast to lytic phage therapy, where bacteria are killed following amplification and release of progeny phage into the environment, this approach uses nonlytic phage to deliver DNA encoding bactericidal proteins (28). In bacteria, programmed cell death is mediated through “addiction modules” consisting of two components, a stable toxin and an unstable antidote that antagonizes the toxic effect; examples include the pemI-pemK genes of plasmid R100, the phd-doc genes of phage P1, and the ccdA-ccdB genes of plasmid F (6, 8, 10, 15). Addiction modules are thought to increase the stability of extrachromosomal elements by selectively killing plasmid-free cells, resulting in the proliferation of plasmid-harboring cells in the population (13). Analogous killer gene systems, such as mazE-mazF (also known as chpAI-chpAK), sof-gef, kicA-kicB, relB-relE, and chpBI-chpBK, have been identified on the E. coli K-12 chromosome (1, 9, 12, 23, 30). The function of these chromosomally encoded killing systems is unknown. However, it has been postulated that they may serve as a mechanism for altruistic cell death during nutritional stress (8, 10, 46). Biological containment systems based on the conditional expression of killing genes have been developed to control the release of organisms used as bioremediation agents and biopesticides. Suicide systems based on the controlled expression of gef, for example, have been developed for several bacterial species, including Pseudomonas putida, Pseudomonas aeruginosa, and E. coli (16, 18, 26, 35, 36, 42).
The availability of the well-characterized nonlytic M13 phage delivery system and the wealth of molecular tools available for E. coli made this an obvious host organism for testing the concept of using phage-delivered lethal agents as a means of controlling bacterial infections. The toxins of addiction modules represent an enormous resource of potentially useful antimicrobial agents. In view of the widespread development of multidrug resistance among bacterial pathogens, the delivery of such agents by phage could be used as the basis for alternative therapies and thus help solve what has become a major public health problem.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The following E. coli strains were used for the cloning, propagation, and infection experiments: strain XL1-Blue MRF′ [F′ proAB lacIqZΔM15 Tn10(Tetr)/Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac], strain ER2738 [F′ proAB lacIqΔ(lacZ)M15 zzf::Tn10(Tetr)/fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5], and ERAPlacI [ER2738 transformed with the vector construct APlacITpBHR]. The R408 helper phage (Promega Corporation, Madison, Wis.), which has a defective packaging signal, was used for packaging the phagemids. Bacterial cells were grown in Luria-Bertani (LB) broth, and for solid media, agar was added to a concentration of 1.5% (wt/vol). Appropriate antibiotics were added as required to select for the presence of plasmids at the following concentrations: ampicillin (50 μg/ml), kanamycin (50 μg/ml), and tetracycline (20 μg/ml). Isopropylthiogalactopyranoside (IPTG) was added to a final concentration of 1 mM unless otherwise stated. DNA manipulations were performed with standard methods (37).
Plasmid constructions.
The levels of LacI repressor protein were controlled by endogenously expressed LacI (lacIq) and by a plasmid expressing the lacI gene. The lacI gene was amplified by PCR from E. coli DH5α with primers 5-CGAATTGGATCCGGAGGTGGAATGTGAAACCAGTAACG-3′ and 5′-TCGGCGGAATTCCTAATGAGTGAGCTAACT-3′ (restriction recognition sites in bold). The 1,140-bp BamHI- and EcoRI-digested PCR product was cloned into the corresponding sites of pBluescript II SK+, yielding lacIpSK. The plasmid APlacIpSK was generated by cloning an artificial promoter with consensus E. coli −35 and −10 hexamers (bold sequences), TTATGGTACCTGTTTCATCCCCTATTGACAATGAAACATCGGCTCGTATAATGTGTTTCATTGTGAGCATGAAACAGCGGCCGCGGTACCAACT, into the Klenow polymerase-treated SpeI site of lacIpSK. The TL17 transcriptional terminator derived from the E. coli α-operon (45) was cloned into the Klenow polymerase-treated EcoRI site of APlacIpSK. The resulting plasmid, APlacITpSK, was digested with XbaI and HindIII, treated with DNA polymerase I Klenow fragment, and cloned into the Klenow-treated EcoRI site of the broad-host-range vector pBBR122 (Mobitec), yielding plasmid APlacITpBHR.
The LacI-regulated promoter (UPRIP) containing a lacI operator site (italic sequence) flanked by consensus E. coli −35 and −10 hexamers (bold sequence), GGATCCTCAGAAAATTATTTTAAATTTCCAATTGACATTGTGAGCGGATAACAATATAATGTGTGGAGCTT, was cloned into the BamHI and HindIII sites of pBluescript II SK+, generating plasmid UPRIPpSK. Removal of the pBluescript lac promoter was accomplished by digesting UPRIPpSK with PvuII and SacI, treating the digested fragments with T4 polymerase, and religating the UPRIP promoter-containing fragment to the vector backbone to yield plasmid pUPRIP. The gene encoding Gef was amplified by PCR from E. coli XL1-Blue MRF′ with the upstream primer 5′-TACCGGAAGCTTGGAGGTGAGCAATGAAGCAGCATAAGGCGAT-3′ and downstream primer 5-ACAATTCTCGAGGAAGTGCCGGATCCGAA-3′. The 395-bp HindIII- and XhoI-digested PCR product was cloned into the corresponding sites of pUPRIP, generating UPRIPgefpSK. To prevent runaway transcription, the transcriptional terminator TL17 was cloned into the Klenow polymerase-treated XhoI site of UPRIPgefpSK, yielding pGef. The gene encoding ChpBK was amplified by PCR from E. coli XL1-Blue MRF′ with primers 5′-GCGTGTGGATCCGGAGGTGAAATATGGTAAAGAAAAGTG-3′ and 5′-ATTTTCGGATCCTTATTCCACCACCGCCT-3′. The 386-bp undigested PCR fragment was cloned into the Klenow polymerase-treated XhoI site of pUPRIP, generating plasmid pChpBK.
Preparation of phage stocks.
For each phage lysate, ERAPlacI cells harboring the toxic protein expression vector were grown overnight at 37°C in LB broth supplemented with tetracycline, ampicillin, and kanamycin. The cultures were diluted 1:100 and grown until an optical density at 600 nm (OD600) of 0.1 was reached. At the time of early exponential phase, the cells were infected with R408 helper phage at an input ratio of 10 phages per cell. The phage-infected cells were then incubated at 37°C with vigorous aeration for 6 h. After propagation, the cells were removed by centrifugation at 2,500 × g for 15 min at 4°C, and phage-containing supernatants were passed through a 0.2-μm-pore-size filter. The phages were precipitated overnight at 4°C with 5% polyethylene glycol 6000 and 0.5 M NaCl. After centrifugation at 16,000 × g for 30 min, the phage pellets were resuspended in sterile SM buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4). R408 helper phage were enumerated with the soft agar overlay technique. Phage containing the lethal-agent phagemid were measured by transduction of the pBluescript ampicillin resistance marker. Lysates were serially diluted with sterile SM buffer, mixed with 107 CFU of E. coli ER2738 or ERAPlacI, and overlaid on LB plates or plated on LB agar containing ampicillin and kanamycin, respectively. After overnight incubation at 37°C, the plates were examined for plaques or ampicillin-resistant colonies, respectively.
Phage infection and delivery of phagemids in vitro.
Bacteria were grown in LB broth with antibiotic selection at 37°C until an OD600 of 0.8 was reached. Bacterial cells were diluted in LB broth to a final density of 106 CFU/ml, and IPTG was added, if appropriate, to a final concentration of 1 mM. An aliquot of cells (105 CFU, 100 μl) was added with an equal volume of phage lysate (8 × 109 PFU/ml) and incubated for 30 min at 37°C without shaking. According to the kinetics of adsorption for M13 phage, 97.3% of cells should be bound by at least one phage within the 30-min incubation (17). Survival of cells in phage-infected cultures was determined in triplicate by plating serial dilutions of cultures onto LB agar supplemented with 1 mM IPTG. The surviving E. coli cells were enumerated and compared to the number of bacteria in a phage-free E. coli control culture. Phage-free cultures (containing only bacteria) and cell-free cultures (containing only phage) were used as controls in all experiments to demonstrate the absence of contamination.
Animal studies.
All experiments were approved by the Medical University of South Carolina Institutional Animal Care and Use Committee. Female ICR mice (20 to 25 g) were obtained from Harlan Sprague Dawley and Charles River Laboratories. Neutropenia was induced essentially by the method of Cryz et al. (7). Briefly, 6 days, 3 days, and 1 day before the experiment, 200 μl of cyclophosphamide (25 mg/ml; Sigma, St. Louis, Mo.) was administered to each mouse by intraperitoneal injection. In pilot experiments, this produced profound leukopenia for 3 to 4 days following the last dose and slowed the clearance of bacteria from the blood. On the day of the experiment, overnight cultures of E. coli ER2738 were diluted to an OD625 of 0.5, then held stationary at 37°C for 30 to 60 min, and adjusted to an OD625 of 1.0, which corresponded to approximately 5 × 108 CFU/ml. Mice were then injected sequentially within 5 min with 200 μl of E. coli ER2738 at 5 × 108 CFU/ml, 200 μl of phage lysate adjusted to 1.2 × 1010 phagemid-containing particles/ml, and 100 μl of 250 mM IPTG. All injections were intraperitoneal, with bacteria and phage lysate administered in the left abdomen and IPTG in the right abdomen. Peripheral blood from the tip of the tail was collected at 1, 3, and 5 h following injections and immediately plated, either neat or diluted in ice-cold LB, onto LB agar plates containing tetracycline (20 μg/ml). Unpaired t tests were performed with GraphPad Prism version 3.0a for Macintosh (GraphPad Software, San Diego, Calif.).
RESULTS AND DISCUSSION
Construction of inducible gef and chpBK expression plasmids.
Conditionally lethal genes associated with bacterial plasmids or the E. coli chromosome are neutralized by their cognate antidote protein or an antisense RNA that inhibits the translation of the toxin-encoding mRNA. Translation of chromosomally encoded gef is normally coupled to that of an overlapping reading frame, orf69, which in turn is negatively regulated by the trans-acting antisense RNA sof (31). Annealing between sof and orf69 mRNAs leads to the formation of a stem-loop structure that sequesters the ribosome-binding site of gef, thereby preventing gef from being translated. For the purpose of inducing bacterial cell death, plasmid pGef does not contain orf69 and is therefore not regulated by the antisense RNA sof. In contrast to Gef, ChpBK is a member of the proteic killer gene system and is neutralized by the chromosomally encoded chpBI gene. In wild-type cells, ChpBK will have to overcome the presence of the neutralizing antidote protein ChpBI in order to function as a lethal agent.
To ensure adequate but controlled expression of the lethal genes in E. coli, gef and chpBK were cloned into a high-copy-number plasmid and placed under the control of a LacI/IPTG-regulated promoter (PUPRIP). The UPRIP promoter differs from conventional lac and tac promoters in the position of the lacI operator, which was placed between the consensus −35 and −10 E. coli promoter elements. In addition, an adenine- and thymine-rich sequence derived from the rRNA rrnB promoter, which has been shown to increase promoter activity (33), was placed upstream of the −35 hexamer. The Shine-Dalgarno sequences for gef (5′-GGAGA-3′) and chpBK (5′-GGTGA-3′) were replaced during construction of the PUPRIP fusion to the lethal-agent genes with the consensus Shine-Dalgarno sequence (5′-GGAGG-3′).
gef and chpBK encode a cell-killing function.
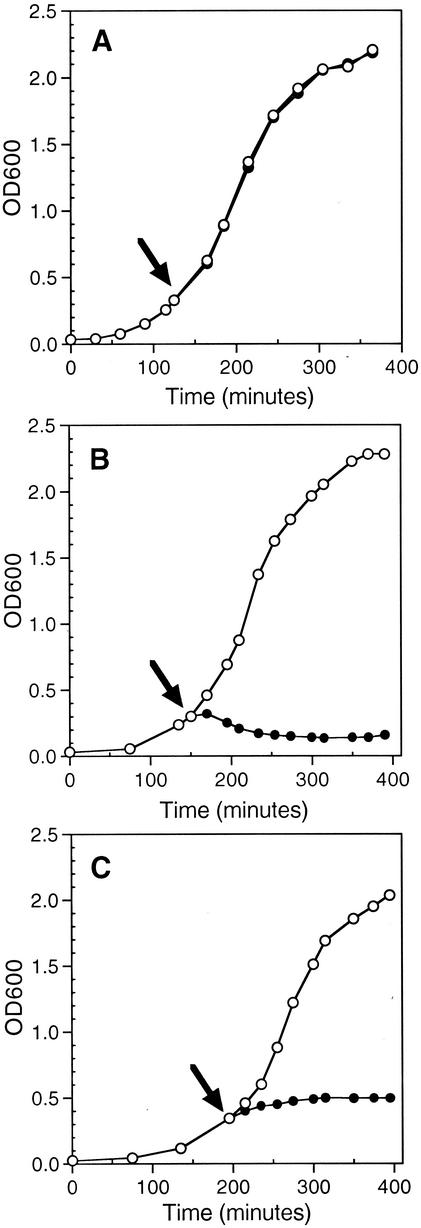
The expression plasmids carrying the IPTG-inducible lethal agents were transformed into E. coli ERAPlacI cells containing the gene for endogenously expressed LacI protein (lacIq) and a plasmid expressing the lacI gene (APlacITpBHR). Upon induction of pGef with IPTG, the optical density of the culture stopped increasing almost immediately and shortly thereafter decreased (Fig. 1B). Induction of the cultures resulted in the appearance of a characteristic ghost cell morphological phenotype (11, 30). The ghost cells were observed by phase-contrast microscopy as translucent cells with polar bodies of condensed material (data not shown). Induction of Gef protein synthesis has been shown to result in collapse of cell membrane potential, arrest of respiration, and release of cytoplasmic contents (30, 32, 34). The dramatic change in cell morphology following induction of Gef is thought to be the consequence of irreversible damage and permeabilization of the host cell membrane.
FIG. 1.
Kinetics of gef and chpBK induction. Cells of E. coli ERAPlacI harboring (A) pUPRIP, (B) pGef, and (C) pChpBK were grown overnight at 37°C and then diluted 1:100 into fresh LB medium containing appropriate antibiotics. At early log phase (OD600 of 0.3 to 0.4), the culture was divided equally (indicated by arrow) and incubated in the presence (•) or absence (○) of 1 mM IPTG. The growth of the culture was monitored spectrophotometrically at a wavelength of 600 nm. Each graph is representative of at least two experiments.
Upon induction of pChpBK with IPTG, the optical density of the culture increased at a slower rate until it reached a plateau after 120 min (Fig. 1C). Although the chpB locus is thought to be a structural and functional homolog to the pem locus of plasmid R100, the cellular target of ChpBK is not yet known (23). Microscopic evaluation of the cells following induction of ChpBK did not reveal any gross morphological change (data not shown). It should be noted that after overnight growth of both strains harboring pGef and pChpBK in the presence of IPTG, the cultures resumed growth (data not shown), suggesting that a small proportion of the cell population escaped cell death.
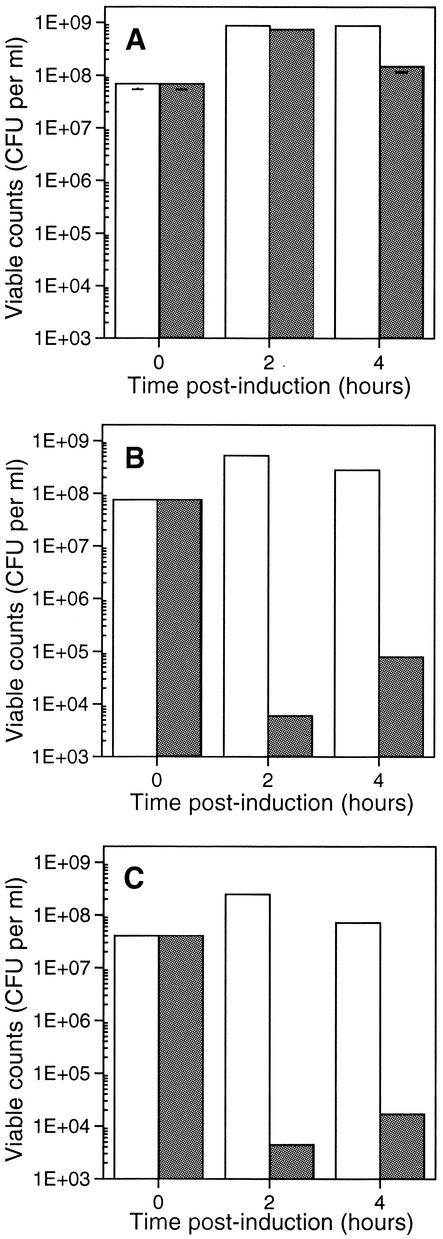
To investigate whether Gef and ChpBK were exerting a bacteriostatic or bactericidal effect in E. coli, samples were removed from the cultures immediately prior to and following IPTG induction and allowed to recover overnight under repressed conditions (LB plates containing kanamycin and ampicillin but lacking IPTG). The viable counts of the Gef and ChpBK cultures dropped by four orders of magnitude within 2 h (Fig. 2B and 2C). A reduction in CFU was not observed for the control cultures containing the vector plasmid alone (Fig. 2A). The results of the present study are consistent with the findings of Poulsen et al. (30), who showed that Gef expression leads to growth arrest and ultimately cell death. In agreement with the results of Masuda et al. (23), we found that ChpBK affects cell growth; however, we further extended these results to show that ChpBK also affects cell viability. Together, these data suggest that Gef and ChpBK exert a bactericidal effect on E. coli.
FIG. 2.
Gef and ChpBK are bactericidal to E. coli. Cells of E. coli ERAPlacI harboring (A) pUPRIP, (B) pGef, and (C) pChpBK were grown overnight at 37°C and then diluted 1:100 into fresh LB medium containing appropriate antibiotics. At early log phase (OD600 of 0.3 to 0.4), the culture was divided equally and incubated in the presence (▪) or absence (□) of 1 mM IPTG. At the indicated times (0, 2, and 4 h postinduction), samples from a dilution series of the culture were plated under repressed conditions on LB plates containing the appropriate antibiotics. Viable counts are expressed as mean CFU, counted on noninductive plates. Each graph is representative of at least two experiments.
Phage delivery of lethal agents Gef and ChpBK results in loss of viability.
One of the challenges of molecular therapy is how to deliver the therapeutic agent to the offending bacterium. Phage delivery systems provide the opportunity to target specific bacterial cells at a high frequency. To test the concept of using phage as a lethal-agent delivery system, we took advantage of the M13 phagemid system. The genes encoding Gef and ChpBK were cloned into a vector carrying the f1 intergenic region, and male E. coli cells carrying this phagemid were infected with helper phage R408. Using this helper phage allowed the preferential packaging of phagemid DNA over helper phage DNA, resulting in lysates with a high percentage of particles containing only the lethal-agent phagemid (95%; data not shown).
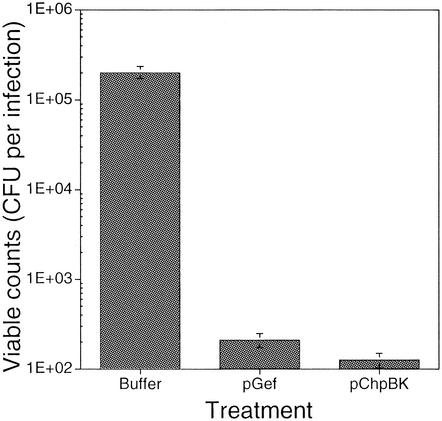
CFU assays were performed to evaluate the effects of the phage-delivered lethal agents on the viability of E. coli ER2738 (Fig. 3). Cells expressing the F pilus were incubated, in the presence of IPTG, with different phage lysates (actual multiplicity of infection of 3.6 [17]) for 30 min. The fraction of surviving cells was determined following dilution and plating of the infected culture onto nonselective (LB) and selective (LB-ampicillin) plates containing 1 mM IPTG. Following overnight incubation at 37°C, phage delivery of the lethal agents had resulted in a significant reduction in viable-cell counts (948- and 1,579-fold reduction, respectively). This loss in cell viability was thought to be a consequence of Gef- and ChpBK-mediated cell death because filamentous phages have been shown to be nonlytic DNA viruses.
FIG. 3.
In vitro phage delivery of lethal agents to E. coli. A CFU assay was performed to evaluate the effects of the phage-delivered lethal agents on the killing of E. coli ER2738. Target cells were grown to mid-exponential phase (OD600 of 0.8) in LB broth containing tetracycline and diluted to approximately 106 CFU/ml in LB broth containing 1 mM IPTG. An aliquot of cells (105 CFU, 100 μl) was incubated at 37°C with an equal volume of phage lysate (8 × 109 PFU/ml). Control experiments were carried out in the absence of phage lysate. Treatments were as follows: (first bar) cells plus buffer, (second bar) cells plus Gef phagemid lysate, and (third bar) cells plus ChpBK phagemid lysate. Treatments reflect viable cell counts following 30 min of incubation at 37°C. Viable counts were determined following dilution and plating of the infection on LB plates containing 1 mM IPTG. The figure is representative of at least two experiments with each infection performed in triplicate. All values are means ± standard deviation.
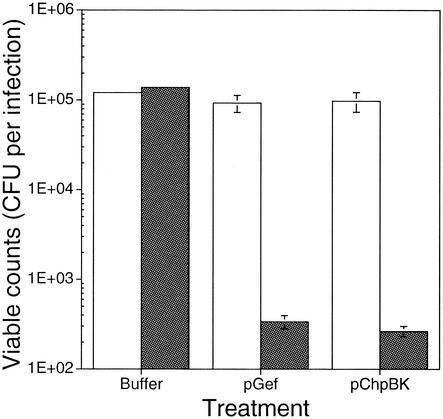
Although the data supported the concept that the lethal agents mediated the loss in viability, the above experiment did not rule out the possibility that the effect was due to nonspecific lysis. In order to confirm that this was not the case, an additional killing assay was performed in which phage lysates (pGef and pChpBK) were incubated with E. coli ERAPlacI in the presence and absence of inducer. After an incubation time of 30 min, infected cultures were plated on selective plates (LB containing kanamycin and ampicillin) under inducing or repressed conditions. As expected, the ERAPlacI cells were protected from the lethal action of pGef and pChpBK due to the presence of excess LacI, which effectively repressed the UPRIP promoter (Fig. 4). Importantly, phage delivery of the lethal-agent lysates in the absence of inducer did not affect the viability of ERAPlacI. However, in the presence of IPTG, the number of viable cells dropped 275- and 370-fold following exposure to pGef and pChpBK lysates, respectively.
FIG. 4.
Cell death following phage delivery of pGef and pChpBK is IPTG dependent. A CFU assay was performed to evaluate the effects of the phage-delivered lethal agents in the presence and absence of IPTG. E. coli ERAPlacI cells were grown to mid-exponential phase (OD600 of 0.8) in LB broth containing appropriate antibiotics (kanamycin and tetracycline) and diluted to approximately 106 CFU/ml in LB with (▪) or without (□) 1 mM IPTG. An aliquot of cells (100 μl, 105 CFU/ml) was incubated at 37°C with an equal volume of phage lysate (8 × 109 PFU/ml). Control experiments were carried out in the absence of phage lysate. Treatments were as follows: (first bar) cells plus buffer, (second bar) cells plus Gef phagemid lysate, and (third bar) cells plus ChpBK phagemid lysate. Treatments reflect viable-cell counts following 30 min of incubation at 37°C. Viable counts were determined following dilution and plating of the infection on LB plates with (▪) or without (□) 1 mM IPTG. The figure is representative of at least two experiments with each infection performed in triplicate. The values are means ± standard deviation.
In comparison to strain ER2738 (Fig. 3), a larger proportion of ERAPlacI cells survived exposure to the lethal agents pGef and pChpBK. This finding is not surprising and most likely reflects the additional levels of LacI repressor produced from the plasmid APlacITpBHR present within ERAPlacI but absent from ER2738; consequently, while the promoter system is more efficiently repressed, it is more difficult to attain full induction (38). These results support the hypothesis that phage can efficiently deliver phagemids expressing bactericidal proteins and that intracellular induction of these agents results in cell death.
Although the lethal-agent delivery system resulted in a dramatic reduction in cell viability, a fraction of the population survived exposure to the therapeutic. Fluctuation tests have estimated the mutation rate for Gef containment systems to occur at a frequency of about 10−5 to 10−6 per cell per generation when the regulatory elements of the biological containment system were plasmid borne (5). However, the rate of mutation could be reduced further by at least two orders of magnitude when the control and killing elements were incorporated into the host chromosome or multiple copies of the killing cassette were introduced into the cell (16, 27, 34). Therefore, one way to reduce the rate of mutation would be to deliver multiple killing systems, either by duplication of one system or by combining several different systems in the phagemid. In addition, since alteration or loss of the phage receptor (21) may also lead to resistance, it may be prudent to employ phage cocktails containing several different phages or a single genetically engineered phage that recognizes different cell surface receptors (22).
Reduction of bacterial load following phage delivery of gef and chpBK in mice.
Preliminary studies determined that the F pilus-expressing E. coli strain ER2738 was nonpathogenic in mice. In mice pretreated with cyclophosphamide to produce a neutropenic, immunocompromised state (7), a 2 × 109 CFU dose of ER2738 caused death in only 1 of 18 animals (data not shown). However, it was possible to produce transient bacteremia provided that the cyclophosphamide-treated mice were challenged with a single intraperitoneal dose of 108 CFU. In order to compare the efficacy of the phage-delivered lethal agents in vivo, we used this nonlethal model of infection and collected peripheral blood samples at various time points following bacterial challenge and administration of the phage lysates. The mean bacterial titers in the blood 1, 3, and 5 h after bacterial and phage inoculation are illustrated in Fig. 5A.
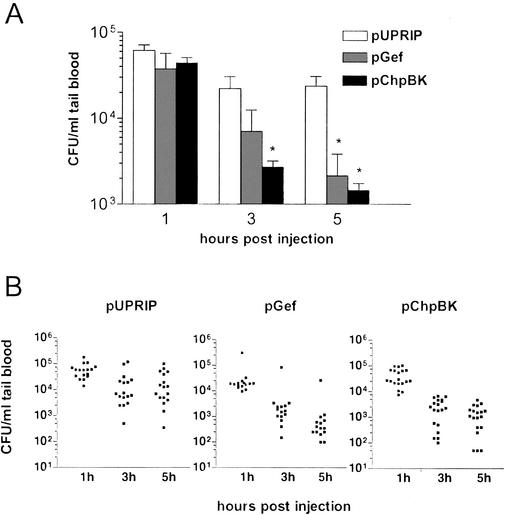
FIG. 5.
Phage-delivered lethal agents significantly reduce the level of viable bacteria in the blood of mice. A single dose of E. coli ER2738 (200 μl, 5 × 108 CFU/ml), IPTG (100 μl, 250 mM), and phage preparation (200 μl, 1.2 × 1010 phagemid-containing particles/ml) was administered by intraperitoneal injection to cyclophosphamide-treated mice. Mice were treated with a single dose of phage preparation containing phagemid pGef, pChpBK, or pUPRIP (actual multiplicity of infection of 3.6). The control vector pUPRIP is identical to the phagemids pGef and pChpBK except that it lacks a gene encoding a lethal agent. At the indicated time points, blood samples were taken and bacterial counts were determined by plating onto LB plates containing tetracycline (20 μg/ml). Mice with tail blood containing less than 20 CFU/ml (lowest level of detection) at 1 h were eliminated from the analysis. The viable bacterial counts in the blood were plotted as the mean plus standard deviation for (A) each treatment group and (B) each animal within each treatment group. Statistical analysis with the unpaired t test indicated significantly fewer viable bacteria in the blood at the times indicated compared to the control pUPRIP group for the experimental pGef and pChpBK groups (*, P ≤ 0.05, two-tailed, adjusted for multiple comparisons).
The experiment demonstrated that phage delivery of the lethal-agent phagemids pGef and pChpBK resulted in a significant reduction in circulating bacteria compared to the time-matched control, pUPRIP (unpaired t test, P ≤ 0.05). Phage titers in the blood were also determined at 3 h postinjection and ranged from 1 × 107 to 5 × 108 phagemid-containing particles per ml, showing that both bacteria and phage migrated readily from the injection site. At 5 h postinjection, the mice receiving the pGef and pChpBK lysates showed a 98% and 94% reduction in blood bacterial titers, respectively, compared to the control pUPRIP group. Future analysis with a lethal mouse model of E. coli infection will be necessary to show directly that the lethal-agent delivery system reduces the bacterial load in vivo. These studies will also be useful for examining the kinetics and limitations of lethal-agent delivery systems in a more clinically relevant model.
A number of problems may restrict the use of phage in the prevention and/or treatment of bacterial infections of animals and humans. One of these hurdles is the rapid clearance of phage by the spleen, liver, and other filtering organs of the reticuloendothelial system. Phage M13 is cleared slowly from the circulation (plasma half-life of approximately 4 h in 129/ICR white mice), with the liver and spleen being mainly responsible for M13 uptake (25). In addition, we found that M13 helper phage are naturally long circulating in ICR mice, and no significant improvement in half-life was obtained by serial passage methods designed to isolate mutants that resist sequestration (24).
While the specificity of phages for bacteria could be considered a drawback, it also offers the advantage of preserving the patient's normal microflora and reducing the chance of opportunistic infections. A relatively narrow host range, however, would require the infecting bacterial strain to be identified or necessitate the use of phages with broad activity against most of the pathogenic strains within a given species. It is also unclear whether diseases caused by intracellular pathogens could be successfully treated by phages, since bacteria located within human cells are unlikely to be accessible to phage. Additional problems include the development of bacterial resistance to phages and phage-neutralizing antibodies, both of which could hamper the effectiveness of subsequent treatments by the same phage delivery system.
In conclusion, the results of this study reinforce the view that phage delivery systems have immense potential in the management of bacterial infections in a medical and veterinary setting. In addition to using phage in the prophylaxis and treatment of bacterial infections in humans, lethal-agent delivery systems also have immense potential at the preharvest stage in the biocontrol of E. coli O157:H7 in animals and fresh foods (20) and could play a role in preventing transmission of fish pathogens (29). The ability to clone and manipulate almost any given piece of DNA, together with our present knowledge of phage genetics, may make it possible to adapt this technology for a multitude of bacterial pathogens.
Acknowledgments
This work was supported by Hexal Gentech Forschungs GmbH.
DNA sequencing data were obtained by the Biotechnology Resource Laboratory of the Medical University of South Carolina.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alisky, J., K. Iczkowski, A. Rapoport, and N. Troitsky. 1998. Bacteriophages show promise as antimicrobial agents. J. Infect. 36:5-15. [DOI] [PubMed] [Google Scholar]
- 3.Biswas, B., S. Adhya, P. Washart, B. Paul, A. N. Trostel, B. Powell, R. Carlton, and C. R. Merril. 2002. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 70:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlton, R. M. 1999. Phage therapy: past history and future prospects. Arch. Immunol. Ther. Exp. 47:267-274. [PubMed] [Google Scholar]
- 5.Contreras, A., S. Molin, and J. L. Ramos. 1991. Conditional-suicide containment system for bacteria which mineralize aromatics. Appl. Environ. Microbiol. 57:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couturier, M., E. M. Bahassi, and L. Van Melderen. 1998. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 7.Cryz, S. J., E. Furer, and R. Germanier. 1983. Simple model for the study of Pseudomonas aeruginosa infections in leukopenic mice. Infect. Immun. 39:1067-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 9.Feng, J., K. Yamanaka, H. Niki, T. Ogura, and S. Hiraga. 1994. New killing system controlled by two genes located immediately upstream of the mukB gene in Escherichia coli. Mol. Gen. Genet. 243:136-147. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes, K., A. P. Gultyaev, T. Franch, K. Pedersen, and N. D. Mikkelsen. 1997. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31:1-31. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes, K., L. K. Poulsen, T. Thisted, A. K. Nielsen, J. Martinussen, and P. H. Andreasen. 1990. The hok killer gene family in Gram-negative bacteria. New Biol. 2:946-956. [PubMed] [Google Scholar]
- 12.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 13.Holcík, M., and V. N. Iyer. 1997. Conditionally lethal genes associated with bacterial plasmids. Microbiology 143:3403-3416. [DOI] [PubMed] [Google Scholar]
- 14.Institute of Medicine. 1998. Workshop summary, p. 1-7. In P. F. Harrison and J. Lederberg (ed.), Antimicrobial resistance: issues and options. National Academy Press, Washington, D.C. [PubMed]
- 15.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, L. B., J. L. Ramos, Z. Kaneva, and S. Molin. 1993. A substrate-dependent biological containment system for Pseudomonas putida based on the Escherichia coli gef gene. Appl. Environ. Microbiol. 59:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasman, L. M., A. Kasman, C. Westwater, J. Dolan, M. G. Schmidt, and J. S. Norris. 2002. Overcoming the phage replication threshold: a mathematical model with implications for phage therapy. J. Virol. 76:5557-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klemm, P., L. B. Jensen, and S. Molin. 1995. A stochastic killing system for biological containment of Escherichia coli. Appl. Environ. Microbiol. 61:481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krylov, V. N. 2001. Phage therapy in terms of bacteriophage genetics: hopes, prospects, safety, limitations. Russ. J. Genet. 37:715-730. [PubMed] [Google Scholar]
- 20.Kudva, I. T., S. Jelacic, P. I. Tarr, P. Youderian, and C. J. Hovde. 1999. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl. Environ. Microbiol. 65:3767-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg, A. A. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205-241. [DOI] [PubMed] [Google Scholar]
- 22.Malmborg, A. C., E. Söderlind, L. Frost, and C. A. K. Borrebaeck. 1997. Selective phage infection mediated by epitope expression on F pilus. J. Mol. Biol. 273:544-551. [DOI] [PubMed] [Google Scholar]
- 23.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merril, C. R., B. Biswas, R. Carlton, N. C. Jensen, G. J. Creed, S. Zullo, and S. Adhya. 1996. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. USA 93:3188-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molenaar, T. J. M., I. Michon, S. A. M. de Haas, T. J. C. van Berkel, J. Kuiper, and E. A. L. Biessen. 2002. Uptake and processing of modified bacteriophage M13 in mice: implications for phage display. Virology 293:182-191. [DOI] [PubMed] [Google Scholar]
- 26.Molin, S., L. Boe, L. B. Jensen, C. S. Kristensen, and M. Givskov. 1993. Suicidal genetic elements and their use in biological containment of bacteria. Annu. Rev. Microbiol. 47:139-166. [DOI] [PubMed] [Google Scholar]
- 27.Molina, L., C. Ramos, M. C. Ronchel, S. Molin, and J. L. Ramos. 1998. Construction of an efficient biologically contained Pseudomonas putida strain and its survival in outdoor assays. Appl. Environ. Microbiol. 64:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norris, J. S., C. Westwater, and D. Schofield. 2000. Prokaryotic gene therapy to combat multidrug resistant bacterial infection. Gene Ther. 7:723-725. [DOI] [PubMed] [Google Scholar]
- 29.Park, S. C., I. Shimamura, M. Fukunaga, K. I. Mori, and T. Nakai. 2000. Isolation of bacteriophages specific to a fish pathogen, Pseudomonas plecoglossicida, as a candidate for disease control. Appl. Environ. Microbiol. 66:1416-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulsen, L. K., N. W. Larsen, S. Molin, and P. Andersson. 1989. A family of genes encoding a cell-killing function may be conserved in all Gram-negative bacteria. Mol. Microbiol. 3:1463-1472. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen, L. K., A. Refn, S. Molin, and P. Andersson. 1991. The gef gene from Escherichia coli is regulated at the level of translation. Mol. Microbiol. 5:1639-1648. [DOI] [PubMed] [Google Scholar]
- 32.Poulsen, L. K., A. Refn, S. Molin, and P. Andersson. 1991. Topographical analysis of the toxic Gef protein from Escherichia coli. Mol. Microbiol. 5:1627-1637. [DOI] [PubMed] [Google Scholar]
- 33.Rao, L., W. Ross, J. A. Appleman, T. Gaal, S. Leirmo, P. J. Schlax, M. T. Record, Jr., and R. L. Gourse. 1994. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 235:1421-1435. [DOI] [PubMed] [Google Scholar]
- 34.Ronchel, M. C., L. Molina, A. Witte, W. Lutbiz, S. Molin, J. L. Ramos, and C. Ramos. 1998. Characterization of cell lysis in Pseudomonas putida induced upon expression of heterologous killing genes. Appl. Environ. Microbiol. 64:4904-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronchel, M. C., and J. L. Ramos. 2001. Dual system to reinforce biological containment of recombinant bacteria designed for rhizoremediation. Appl. Environ. Microbiol. 67:2649-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronchel, M. C., C. Ramos, L. B. Jensen, S. Molin, and J. L. Ramos. 1995. Construction and behavior of biologically contained bacteria for environmental applications in bioremediation. Appl. Environ. Microbiol. 61:2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schofield, D. A., C. Westwater, J. W. Dolan, M. G. Schmidt, and J. S. Norris. 2001. Controlled expression in Klebsiella pneumoniae and Shigella flexneri using a bacteriophage P1-derived C1-regulated promoter system. J. Bacteriol. 183:6947-6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, H. W., and M. B. Huggins. 1982. Successful treatment of experimental Escherichia coli infections in mice with phage: its general superiority over antibiotics. J. Gen. Microbiol. 128:307-318. [DOI] [PubMed] [Google Scholar]
- 40.Smith, H. W., and M. B. Huggins. 1983. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J. Gen. Microbiol. 129:2659-2675. [DOI] [PubMed] [Google Scholar]
- 41.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J. Gen. Microbiol. 133:1111-1126. [DOI] [PubMed] [Google Scholar]
- 42.Soberón-Chávez, G. 1996. Evaluation of the biological containment system based on the Escherichia coli gef gene in Pseudomonas aeruginosa W51D. Appl. Microbiol. Biotechnol. 46:549-553. [DOI] [PubMed] [Google Scholar]
- 43.Sulakvelidze, A., Z. Alavidze, and J. G. Morris. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Summers, W. C. 2001. Bacteriophage therapy. Annu. Rev. Microbiol. 55:437-451. [DOI] [PubMed] [Google Scholar]
- 45.Wright, J. J., A. Kumar, and R. S. Hayward. 1992. Hypersymmetry in a transcriptional terminator of Escherichia coli confers increased efficiency as well as bidirectionality. EMBO J. 11:1957-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yarmolinsky, M. B. 1995. Programmed cell death in bacterial populations. Science 267:836-837. [DOI] [PubMed] [Google Scholar]