Abstract
Oncogenic transformation has been associated with decreased fibronectin (FN) matrix assembly. For example, both the HT-1080 fibrosarcoma and MAT-LyLu cell lines fail to assemble a FN matrix when grown in monolayer culture (2-dimensional [2D] system). In this study, we show that these cells regain the ability to assemble a FN matrix when they are grown as aggregates (3-dimensional [3D] system). FN matrix assembly in 3D correlates with decreased Raf-1 protein expression compared with cells grown in monolayer culture. This effect is associated with reduced Raf-1 mRNA levels as determined by quantitative RT-PCR and not proteasome-mediated degradation of endogenous Raf-1. Interestingly, transient expression of a Raf-1 promoter-reporter construct demonstrates increased Raf-1 promoter activity in 3D, suggesting that the transition to 3D culture may modulate Raf-1 mRNA stability. Finally, to confirm that decreased Raf-1 expression results in increased FN matrix assembly, we used both pharmacological and small interfering RNA knockdown of Raf-1. This restored the ability of cells in 2D culture to assemble a FN matrix. Moreover, overexpression of Raf-1 prevented FN matrix assembly by cells cultured in 3D, resulting in decreased aggregate compaction. This work provides new insight into how the cell microenvironment may influence Raf-1 expression to modulate cell–FN interactions in 3D.
INTRODUCTION
Fibronectin (FN) is a multifunctional, adhesive glycoprotein that has wide tissue distribution and is essential for normal development and tissue repair (Hynes, 1990; Schwarzbauer, 1991; Sottile and Hocking, 2002). Cells secrete FN as a disulfide-bonded dimer that binds principally to integrin cell surface receptors. Integrin–FN interactions allow unfolding of the soluble protein and its assembly into a detergent-insoluble fibrillar matrix that can modulate cell morphology, growth, and tissue architecture (Schwarzbauer and Sechler, 1999; Wierzbicka-Patynowski and Schwarzbauer, 2003). FN matrix assembly can also regulate the subsequent deposition and organization of other extracellular matrix molecules, including fibrinogen, collagen-1, and thrombospondin-1 (Sottile and Hocking, 2002). As a consequence, FN fibrillogenesis initiates the formation of a dynamic protein meshwork that provides important structural and environmental cues required for normal cell behavior.
One hallmark of malignant transformation in vitro is the loss of FN matrix assembly in two-dimensional (2D) culture. For example, transformed cells frequently show decreased FN synthesis, loss of FN receptor expression, or both (Olden and Yamada, 1977; Plantefaber and Hynes, 1989), and in many cases, the loss of surface FN assayed in these cells correlates with malignant transformation, in vivo. Similarly, oncogenic cells can demonstrate loss of normal integrin function, despite adequate receptor expression. For example, human HT-1080 fibrosarcoma cells express the FN-binding integrin α5β1 and adhere to FN-coated substrates, but they lack the ability to assemble a FN matrix even in the presence of excess exogenous FN (Rasheed et al., 1974; Hall et al., 1983; McKeown-Longo and Etzler, 1987; Brenner et al., 2000). In these cells, this aberrant behavior has been correlated with a mutant N-Ras allele that encodes a Ras protein lacking intrinsic GTPase activity (Brown et al., 1984). Such activating mutations in Ras proteins are found in 30% of cancers and are associated with the acquisition of both anchorage- and growth factor-independent growth (Kinbara et al., 2003). Expression of activated variants of both H-Ras and Raf-1 has been shown to suppress integrin function, supporting the idea that cytoplasmic signaling cascades can influence integrin ligand-binding affinity (Hughes et al., 1997; Hughes et al., 2002).
Raf-1 is a serine/threonine kinase that is an important effector of Ras (Katz and McCormick, 1997; Kinbara et al., 2003). Active Ras binds directly to Raf-1, resulting in its activation and recruitment to the plasma membrane. In turn, activated Raf-1 can then bind to critical intermediates in the mitogen-activated protein kinase (MAPK) cascade to initiate intracellular signals required for cell growth and differentiation (Stokoe et al., 1994; Roy et al., 1997). To identify novel targets that disrupt inappropriate Ras signaling, significant research has focused on factors that regulate Raf-1 activity. More recently, investigators have determined that suppression of Raf-1 protein expression can also have therapeutic efficacy (Rudin et al., 2001). Raf-1 protein expression is regulated both by the synthesis of new Raf-1 mRNA and by targeted proteolysis of endogenous Raf-1 protein (Zmuidzinas et al., 1991; Manenti et al., 2002). Although the mechanisms by which Raf-1 protein is stabilized have come under close scrutiny, relatively little work has examined factors that regulate Raf-1 mRNA levels.
Three-dimensional extracellular matrices have been used to model normal cell behavior (Bissell et al., 2003; Grinnell, 2003). Culturing cells in a three-dimensional (3D) context produces distinct cellular morphology and signaling events compared with a rigid two-dimensional (2D) culture system. For example, fibroblast-populated collagen gels demonstrate that fibroblast morphology in 3D is distinct from that observed in 2D (Berry et al., 1998; Grinnell, 2003). Similarly, 3D matrices can induce tissue-specific differentiation of mammary epithelial cells (Li et al., 1987). More recently, 3D culture systems have been used to distinguish between normal and malignant cells and have been shown to support the reversion of transformed cells to a normal phenotype, given the appropriate stimulus (Weaver et al., 1997; Wang et al., 1998; Grinnell, 2003).
Our recent work showed that cells cultured in 3D environments can assemble a FN matrix and that FN matrix formation in this context, contributes significantly to the organization, biomechanical properties, and remodeling of cellular aggregates (Robinson et al., 2003; Robinson et al., 2004). To further explore the role of FN matrix assembly in this process, we used HT-1080 cells that do not assemble a FN matrix in 2D culture. Surprisingly, these cells formed aggregates in 3D culture. Furthermore, spheroid formation correlated with a restored capability for FN matrix assembly. Therefore, we sought to determine the mechanism by which the 2D-to-3D transition could regulate fibrillar matrix formation.
The data presented in this article show that HT-1080 cells grown as 3D aggregates down-regulate Raf-1 protein expression compared with cells grown in 2D, an effect that is also seen in the invasive prostate cancer cell line, MAT-LyLu. Diminished Raf-1 protein expression in 3D is associated primarily with reduced Raf-1 mRNA levels as determined by quantitative RT-PCR and not proteasome-mediated degradation of endogenous Raf-1. Interestingly, transient expression of a Raf-1 promoter-reporter construct demonstrates increased Raf-1 promoter activity in 3D compared with 2D culture, suggesting that the transition to 3D culture may modulate Raf-1 mRNA posttranscriptionally. Finally, Raf-1 knockdown either pharmacologically or by small interfering RNA (siRNA) transfection restored the ability of both HT-1080 and MAT-LyLu cells in 2D culture to assemble a FN matrix, whereas overexpression of Raf-1 prevented FN matrix assembly in 3D, leading to cell dispersal. This work provides new insight into how alterations in Raf-1 expression regulated by the biophysical environment can modulate cell–extracellular matrix (ECM) interactions, suggesting a novel molecular target for the rescue of transformed cells.
MATERIALS AND METHODS
Cell Lines
HT-1080 (CCL-121) human fibrosarcoma and MAT-LyLu rat prostatic cell lines were obtained from the American Type Culture Collection (Manassas, VA) and propagated in Earle’s minimal essential medium containing 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 1.5 g/l sodium bicarbonate (HT-1080 cells), and 10% fetal calf serum (FCS) or in RPMI 1640 medium (Invitrogen, Carlsbad, CA), 10% fetal bovine serum (Hyclone Laboratories, Logan, UT), 0.1 mM nonessential amino acids, and 0.5 μM dexamethasone (MAT-LyLu cells). Cells were tested for mycoplasma contamination using the Mycoalert mycoplasma detection kit (Cambrex Bio Science, Rockland, MD) before use. Cells were cultured under standard tissue culture conditions of 37°C, 5% CO2, and 95% humidity.
Generation of 3D Cell Cultures
Cells were removed from near-confluent plates with trypsin-EDTA, washed, and resuspended at a concentration of 2.5 × 106 cells/ml in complete medium supplemented with 2 mM CaCl2. Fifteen-microliter aliquots of this suspension were deposited on the underside of a 10-cm tissue culture dish lid. The lid was then inverted over 10 ml of phosphate-buffered saline (PBS) for hydration. Hanging drops were incubated under tissue culture conditions for 24 h, allowing the cells to coalesce at the base of the droplets and to form multilayer aggregates (Robinson et al., 2003, 2004).
Assessment of FN Matrix Assembly
The assembly of high-molecular-weight FN multimers (HMWFMs) was assessed using a deoxycholate (DOC) differential solubilization protocol and Western blot analysis as described previously (Sechler et al., 1996; Robinson et al., 2004). Cell monolayers or aggregates were lysed in a DOC lysis buffer (2% sodium deoxycholate, 0.02 M Tris-HCl, pH 8.8, 2 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM EDTA, 2 mM iodoacetic acid, and 2 mM N-ethylmaleimide), passed through a 26-gauge needle, and centrifuged at 15 × g for 20 min at 4°C. The supernatant containing the DOC-soluble components was separated and then pelleted by centrifugation. DOC-insoluble components were solubilized using SDS lysis buffer (1% SDS, 25 mM Tris-HCl, pH 8.0, 2 mM PMSF, 2 mM EDTA, 2 mM iodoacetic acid, and 2 mM N-ethylmaleimide). Reduced lysates were separated on SDS-PAGE gels and probed with a polyclonal anti-FN antibody predicted to cross-react against a variety of mammalian FNs, including human (ab6584; Abcam, Cambridge, United Kingdom). Under reducing conditions, HMWFMs resolve as a 220-kDa band.
Assessment of FN Secretion
HT-1080 and MAT-LyLu cells were plated at equal densities in equal volumes of media in 2D or in 3D culture in complete medium containing FN-depleted FCS. Serum was depleted of FN as described previously (Corbett et al., 1997). After 24 h, cells and media were collected together, and cells were pelleted by centrifugation. Then, 100 μl of tissue culture medium from each line was mixed with 100 μl of gelatin-Sepharose beads. This amount of gelatin was calculated to bind in excess of 4 times the amount of FN normally found in serum. Beads and media were rotated for 30 min at room temperature (RT) and then washed five times in ice-cold PBS, followed by boiling in SDS sample buffer containing 5% β-mercaptoethanol. Samples were analyzed by SDS-PAGE gel, followed by immunoblotting with an anti-FN antibody as described above. Medium without cells served as a control.
Assessment of FN Fibril Formation by Fluorescence Microscopy
For 2D cultures, cells were trypsinized from near-confluent plates, suspended at a concentration of 2.5 × 106 cells/ml in complete medium, and plated onto glass coverslips. Cells on coverslips were probed 24 h later with a polyclonal anti-FN antibody (ab6584; Abcam) followed by an Alexa-Fluor 568-conjugated goat anti-rabbit-IgG. Cells were viewed using inverted fluorescence optics. Images were captured using a Spot color camera (Diagnostic Instruments, Sterling Heights, MI) connected to a MacIntosh G4 computer equipped with IPLab image analysis software (Scanalytics, Rockville, MD). For 3D cultures, cells were incubated in the presence of 30 μg/ml rhodamine-labeled FN (Cytoskeleton, Denver, CO) in 15-μl hanging drops for 24 h under tissue culture conditions. Hanging drops were then transferred to glass coverslips and visualized as described above.
Assessment of Integrin Expression in 2D and 3D Culture by Flow Cytometry
For 2D culture, cells were detached from a near-confluent tissue culture plate with trypsin-EDTA (TE; Invitrogen). For 3D cultures, aggregates in hanging drops were pooled, washed three times with ice-cold Hank’s balanced salt solution (HBSS), and then incubated in TE for 20 min at 37°C with agitation. Aggregates were gently triturated to release cells. Single cell suspensions from both 2D and 3D cultures were adjusted to a concentration of 1 × 106 cells/ml and incubated with anti-human integrin α5 (clone VC5; BD Biosciences PharMingen, San Diego, CA), anti-human integrin αvβ3 (clone LM609; Chemicon International, Temecula, CA), anti-human integrin α4 (BD Biosciences PharMingen), or anti-human integrin β1 (HUTS-4; Chemicon International) on ice for 30 min with agitation. Cells were again washed with ice-cold HBSS and incubated on ice for an additional 30 min with an Alexa-Fluor 488-conjugated goat-anti-mouse IgG secondary antibody (Invitrogen). Analysis was performed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA).
Immunoblotting
Protein lysates were prepared from 2D and 3D cultures as described previously (Robinson et al., 2004). Twenty micrograms of protein was separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF). Blots were washed in Tris-buffered saline-Tween 20 (TBS-T) and blocked in 5% nonfat dry milk for 4 h at RT. For assessment of focal adhesion kinase (FAK) expression, blots were probed with an antibody against total FAK (1 μg/ml; Upstate Biotechnology, Lake Placid, NY) and phospho-FAK (pY397-FAK; 0.25 μg/ml; BD Transduction Laboratories, Lexington, KY). For Src expression, blots were probed with antibodies against total Src (0.5 μg/ml; Upstate Biotechnology) and active Src (p-Src; 1.0 μg/ml; Upstate Biotechnology). For extracellular signal-regulated kinase (ERK) expression, blots were probed with antibodies against total ERK and phospho-ERK (Cell Signaling Technology, Beverly, MA). For Ras expression, blots were probed with an antibody against total Ras (t-Ras; 1 μg/ml; Upstate Biotechnology). For the detection of active Ras or active Rho, commercially available assays that detect GTP-Ras (Upstate Biotechnology) or GTP-Rho (Pierce Chemical, Rockford, IL) were used per the manufacturers’ instructions. Cell monolayers or aggregates were lysed in radioimmunoprecipitation assay buffer, and lysates were incubated with a glutathione S-transferase-fusion protein covalently bound to glutathione beads. The beads were recovered by centrifugation and washed three times with PBS. Bound protein was eluted from the beads by boiling in Laemmli sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting with a monoclonal antibody (mAb) specific for Ras or Rho. The amount of bound GTPase is indicative of the amount of the active GTP-bound form of the protein. This was correlated with the amount of Ras or Rho detected in total cell lysates. Raf-1 expression was assessed by probing blots with Raf-1 antibody (8.0 μg/ml; Zymed Laboratories, South San Francisco, CA). A-Raf (C-20) and B-Raf (H-145) rabbit polyclonal antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Goat anti-rabbit IgG-horseradish peroxidase (HRP) and goat anti-mouse IgG-HRP at dilutions of 1:20,000 were then used to probe the FAK/Raf and Src/Ras/ERK/Rho blots, respectively. Blots were washed repeatedly in TBS-T and were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical) and exposed to x-ray film.
Quantification of ERK and Raf-1 Expression in 3D Cultures
At least three separate lysates of 2D and 3D cultures were prepared as described previously and subjected to SDS-PAGE and immunoblotting using anti-Raf-1 or anti-p-ERK antibodies. Images were digitized using a Microtek Scanmaker II digital scanner, and band intensity was quantified using NIH Image gel scanning software (National Institutes of Health, Bethesda, MD). Blots were reprobed using an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (1 μg/ml; Ambion, Austin, TX) to normalize the Raf-1 signal. Means and SEs of band intensities were calculated and compared by a Student’s unpaired t test.
Assessment of Raf-1 Expression in Response to Proteasome Inhibition or Geldanamycin (GA) Treatment
Cells were treated overnight with either proteosome inhibitors MG132 (10 μM), lactacystin (20 μM), N-acetyl-l-leucinyl-l-leucinyl-norleucinal (LLnL; 40 μM), or with GA concentrations ranging from 0.01 to 0.1 μM. For 3D cultures, cells were resuspended at a concentration of 2.5 × 106 cells/ml and plated in 15-μl hanging drops in the presence of the proteosome inhibitor as indicated. Lysates were prepared and protein concentration determined by BCA assay (Pierce Chemical). Twenty micrograms of protein was separated by SDS-PAGE and blotted to PVDF. Immunoblot analysis of Raf-1 expression was performed as described above. GAPDH was used as a loading control. For GA-treated cells, immunohistochemistry for the detection of FN fibrils was performed as described above.
Assessment of Raf-1 mRNA Expression in 2D and 3D Culture
RNA was prepared from 2D and 3D cultures of HT-1080 cells using the Total RNA Purification system (Invitrogen). RNA was treated with DNase I (Invitrogen) for 15 min at RT followed by the addition of EDTA and heated at 65°C for 10 min to remove degraded genomic DNA. One microgram of total RNA was used for cDNA synthesis. cDNA was synthesized using the Superscript First Strand Synthesis system for RT-PCR (Invitrogen). Reverse transcription was performed using random hexamer and Superscript II RT enzyme (Invitrogen) at 42°C for 50 min. The reaction was terminated by heating at 70°C for 15 min. RNase H was added to each reaction and incubated at 37°C for 20 min to remove RNA template from the cDNA:RNA hybrid molecule after first strand synthesis and thereby increase the sensitivity of PCR from cDNA. Standard PCR amplification was performed using 2 μl of cDNA, 45 μl of PCR SuperMix High Fidelity (Invitrogen), and 2 μl each of 20 μM forward and reverse primers. 18s rRNA primers were used as control primers for amplification. Human Raf-1 primer sequences were as follows: 5′-CAG CCC TGT CCA GTA GC-3′ for the forward primer and 5′-GCC TGA CTT TAC TGT TGC-3′ for the reverse primer. Human 18S rRNA primer sequences were 5′-TCA AGA ACG AAA GTC GGA GG-3′ for the forward primer and 5′-GGA CAT CTA AGG GCA TCA CA-3′ for the reverse primer. PCR thermal cycling was performed in a PTC-100 programmable thermal controller (MJ Research, Watertown, MA) with an initial denaturation step for 3 min at 94°C, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min. Cycling was followed by a final extension step for 10 min at 72°C. PCR products were separated on a 1.5% agarose/ethidium bromide gel and photographed under UV light. Real-time PCR amplification was performed using 50 ng of cDNA, 25 μl of PCR SYBR Green Master Mix (Applied Biosystems, Foster City, CA), 0.5 μl each of 5 μM Raf-1 forward primer 5′-TTT CCT GGA TCA TGT TCC CCT-3′, and Raf-1 reverse primer 5′-ACT TTG GTG CTA CAG TGC TCA-3′. 18s rRNA primers (18s forward primer, GAT GGG CGG CGG AAA ATA G; and 18s reverse primer, GCG TGG ATT CTG CAT AAT GGT) were used as control primers for amplification. Real-time PCR amplification was performed in a 7900 HT sequence detection system (Applied Biosystems) with initial heating hold for 10 min at 95°C, followed by 40 cycles of 15-s denaturation at 95°C and annealing/extension at 60°C (1 min). Data analyses of threshold cycle (CT) values of samples were performed using SDS 2.2 software (Applied Biosystems).
Cloning of the Raf-1 Promoter
Raf-1 promoter forward primer (5′-AAC TAT CTA GTTCAT TCT TGG ATG GAT GAC AAC-3′) and reverse primer (5′-GCC CGC CGC CGG CTC CCC CGG CAT CCA CGA C-3′) were designed based on published sequence of the human Raf-1 promoter (Beck et al., 1990). The above-mentioned oligonucleotides were synthesized by the University of Medicine and Dentistry of New Jersey DNA core facility (New Brunswick, NJ). The primers were then used to amplify a 1.2-kb PCR product from human genomic DNA. PCR thermal cycling was performed in a programmable thermal controller (Biometra, Göttingen, Germany) with initial heating for 3 min at 94°C, followed by 35 cycles of 45-s denaturation at 94°C, annealing at 50°C (for 30 s), extension at 72°C (for 1 min), and a final extension for 10 min at 72°C. The 1.2-kb PCR product was gel purified using the QIAGEN gel extraction kit (QIAGEN, Valencia, CA) and then cloned into pCR 2.1-TOPO vector by TA cloning (Invitrogen). TOPO-TA clones containing the human Raf-1 promoter were then digested with HindIII and SalI restriction enzymes to confirm 5′-to-3′ orientation. The clone that contained the human Raf-1 promoter in the correct orientation was digested with SacI and XhoI to release the 1.2-kb human Raf-1 promoter insert from the TOPO vector, which was then cloned in frame into the pGL3 basic luciferase promoter vector (Promega, Madison, WI) that had been previously linearized with the same restriction enzymes. Both strands of the pGL3 clone with the human Raf-1 promoter insert were sequenced. pRL-TK vector containing cDNA encoding Renilla luciferase was used as an internal control for cotransfection into HT1080 cells in combination with our experimental reporter vector construct pGL3-humraf1PR.
Transfection of the Raf-1 Promoter into HT1080 Cells
Both the experimental reporter vector pGL3 containing the Raf-1 promoter and the control reporter vector pRL-TK were cotransfected into HT1080 cells by electroporation. HT-1080 cells (5 × 106) were resuspended in 0.4 ml of transfection medium (serum-free RPMI 1640 medium, 10 mM dextrose, and 0.1 mM dithiothreitol [DTT]) and pipetted into an electroporation cuvette (Bio-Rad, Hercules, CA). Thirty micrograms of pGL3/humraf1 plasmid DNA and 6 μg of pRl-TK plasmid DNA were added to the cell suspension in the cuvette and electroporated at 200 V and 0.975 μF using a Gene Pulser II (Bio-Rad) electroporation instrument. Transfected cells were replated in 10-cm tissue culture dish. The next day, cells were trypsinized, washed with 1× PBS, and passed through a Dead Cell Removal microbead column (Miltenyi Biotec, Auburn, CA) to eliminate dead cells. The remaining live cells were used to make either hanging drops for 3D culture or plated in 2D culture. After 24 h, cells from both 2D and 3D cultures were harvested and assayed for Raf-1 promoter activity.
Raf-1 Promoter Assay
Dual luciferase reporter assay for the expression of Raf-1 promoter was performed using the Dual-Luciferase Reporter Assay system (Promega). Growth medium from the Raf-1 promoter transfected HT-1080 cells in 2D and 3D culture was removed. Then, 1 ml of 1× passive lysis buffer (PLB) was added to the cells in 2D culture. The cells were then manually scraped from the culture dish in the presence of 1× PLB. The hanging drops were pooled, gently rinsed with 1× PBS, centrifuged, and the pellet was resuspended in a suitable volume of 1× PLB. The lysates were then subjected to a freeze-thaw cycle to accomplish complete lysis and then Qiashredded (QIAGEN) to residual cell debris. The protein concentrations of the lysates were determined using a standard BCA protein assay. One hundred microliters of Luciferase Assay Reagent II was added to a tube containing 20 μl of cell lysate (0.5–1.0 μg of total protein), mixed by pipetting, and placed in a luminometer programmed to perform a 2-s premeasurement delay, followed by a 10-s measurement period for each reporter assay. The reaction was then stopped by the addition of 100 μl of Stop & Glo Reagent, which activated the Renilla luciferase. The ratio of luminescence from the experimental reporter (firefly luminescence) to luminescence from the control vector (Renilla luminescence) was calculated to measure Raf-1 promoter activity.
Raf-1 siRNA Transfection
HT1080 cells in antibiotic-free tissue culture medium were seeded into a 24-well plate at densities empirically determined to yield 50 and 70% confluence within 24 h. Commercially available Raf-1-specific siRNA that is effective against human Raf-1 (Ambion) was transfected into HT-1080 cells using the Silencer siRNA transfection kit (Ambion). Briefly, 1.5 μl of siPORT lipid was added dropwise into 6 μl of Opti-MEM I medium. The mixture was vortexed and incubated at RT for 30 min. Then, 2 μl of 20 mM Raf-1 siRNA was added to 40 μl of Opti-MEM medium. The diluted siRNA was added to the siPORT lipid mixture, mixed gently, and incubated for 20 min at RT. Cells were washed once with fresh Opti-MEM and 200 μl of Opti-MEM were added to each well. The transfection agent/siRNA complex was added dropwise onto the cells in each well and incubated for 4 h at 37°C under tissue culture conditions. After 4 h, 2 ml of normal growth medium was added, and the cells were incubated for a further 24 h. Controls included untransfected HT-1080 cells as well as cells transfected with the siPORT lipid plus GAPDH siRNA. Alternately, the mammalian Raf-1 siRNA expression plasmid pKD-Raf-1-v4 or the pKD-NegCon-v1 that generates an siRNA that is effective against human, rat, and mouse Raf-1 were used to transfect both HT-1080 and MAT-LyLu cells according to the manufacturer’s protocol. After 24 h, FN fibril formation was assessed by immunohistochemistry and Western blotting as described above.
Transfection with Raf-1 Plasmid
A plasmid encoding wild-type human Raf-1 is commercially available (Upstate Biotechnology). Approximately 5 × 106 cells/ml in 400 μl of transfection medium (RPMI 1640 medium, 0.1 mM DTT, and 10 mM dextrose) and 20 μg of plasmid DNA were transfected by electroporation using a Gene Pulser II apparatus (Bio-Rad) at 200 V and 960 μF. Transfected cells were replated and either used the next day or cultured for 48 h whereupon 800 μg/ml G418 was added to the cultures. Resistant cells were pooled and grown to confluence. Cells were maintained in 200 μg/ml G418. Raf-1-expressing cells were replated onto coverslips and tested for FN fibril formation as described above.
RESULTS
Cells in 3D Aggregate Culture Assemble a FN Matrix
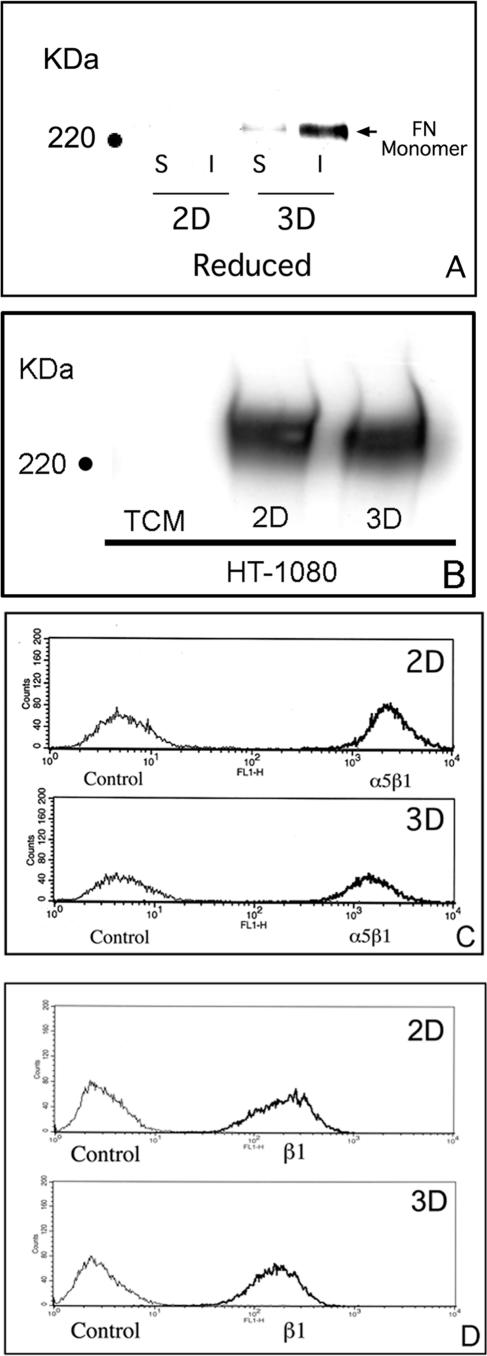
We previously reported that the α5β1 integrin can mediate strong intercellular cohesion between cells cultured as multilayered aggregates through a process that is dependent on FN matrix assembly (Robinson et al., 2003, 2004). To further explore the role of FN fibril formation in cell–cell cohesion, we used the human fibrosarcoma cell line HT-1080. These cells are unable to assemble a FN matrix in 2D culture despite adequate expression of the FN receptor α5β1 and the presence of excess exogenous FN (Brenner et al., 2000). When grown as 3D cultures, aggregates of HT-1080 cells become more compact in the presence of FN (Robinson et al., 2004). To determine whether FN matrix assembly may play a role in this process, HT-1080 cells in complete medium were grown either as conventional 2D culture or as multilayer aggregates. A DOC differential solubilization assay was used to assess the assembly of FN dimers into HMWFMs indicative of FN matrix assembly. HT-1080 cells cultured as aggregates incorporated FN into both DOC-soluble and DOC-insoluble fractions, indicative of surface-bound and matrix FN, respectively. The detection of DOC-insoluble FN by immunoblotting correlated with the detection of fibrillar FN by fluorescence microscopy in HT-1080 cell aggregates (Supplemental Figure 1S). These data demonstrate that the transition from a rigid, planar surface to a multilayer architecture restores the capacity for FN matrix assembly to HT-1080 cells and suggest that the biophysical environment can influence fibrillar matrix formation.
Two possible explanations for the increased matrix assembly observed when HT-1080 cells are cultured as aggregates is that it reflects either increased synthesis of cellular FN or increased expression of the integrin α5β1, the major FN receptor on these cells (Brenner et al., 2000). To determine whether the culture environment influences secretion of cellular FN, cells were grown in 2D culture or as aggregates in culture media depleted of exogenous FN. After 24 h, the media were recovered and incubated with gelatin-Sepharose to bind secreted FN. As demonstrated in Figure 1B, there is no increase in the level of secreted FN when cells are grown in 3D culture. The surface expression of α5β1 was also examined by flow cytometry using an α5-specific mAb. As indicated in Figure 1C, surface α5β1 expression, rather than increasing, actually diminished when cells were cultured as aggregates. It is also possible that the transition to 3D culture may modulate the activation status of the integrin. To evaluate the effect of 3D culture on β1 integrin conformation, flow cytometric analysis was performed using the mAb HUTS-4, which specifically recognizes an active conformation of the β1 integrin (Luque et al., 1996). No change in β1 integrin conformation, as assessed by HUTS-4, was detected (Figure 1D). HT-1080 cells also express the FN-binding integrins α4β1 and αvβ3, which can support FN matrix assembly with activation. There was no significant difference in the surface expression of these integrins when the cells were grown in 2D or 3D conditions (Supplemental Figure 2S). Although the activation status of αvβ3 was not examined directly, previous studies have demonstrated that αvβ3, when coexpressed with α5β1, is in a low affinity state (Ly et al., 2003). Therefore, αvβ3 is unlikely to significantly contribute to FN matrix assembly in these cells. Thus, increased FN matrix assembly by HT-1080 cells cultured as multilayered aggregates does not occur as a consequence of a change in integrin expression.
Figure 1.
Induction of FN matrix assembly in 3D culture does not correlate with changes in secreted FN or surface integrin expression. The assembly of FN into HMWFMs was assessed using a DOC differential solubilization assay, SDS-PAGE under reducing conditions, and immunoblot analysis. (A) FN was detected in the soluble and insoluble fractions of the 3D samples only. (B) Secreted FN by cells in 2D or 3D culture was determined by immunoblotting. Surface expression of α5β1 (C) or of activated β1 integrin (D) was determined by staining with monoclonal mAbs recognizing the α5 integrin subunit or activated β1, respectively. Control cells were incubated in a preimmune IgG to establish baseline mean fluorescence. No significant change in FN secretion or surface integrin expression was detected.
FN Matrix Assembly in 3D Culture Correlates with Down-Regulation of Raf-1 Expression
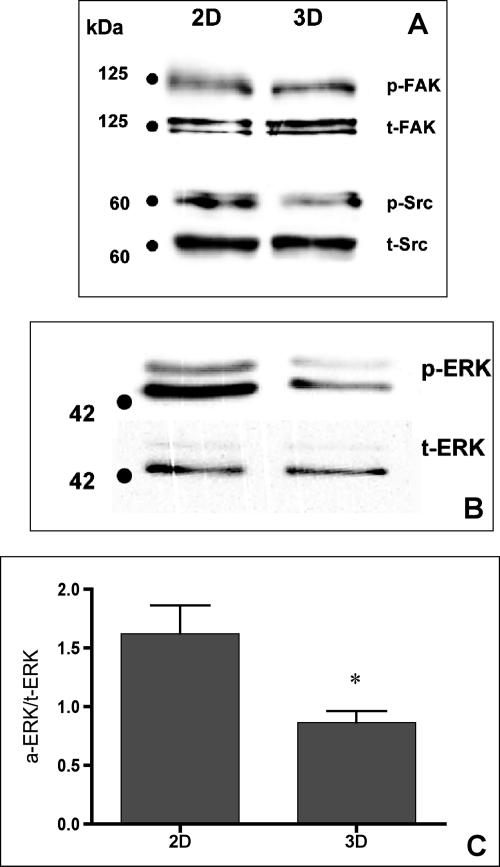
Intracellular signaling pathways influence FN matrix assembly (Wierzbicka-Patynowski and Schwarzbauer, 2003). For example, cells lacking FAK demonstrate altered FN fibril organization (Ilic et al., 2004). Similarly, mutant fibroblasts lacking three Src-family kinases (Src, Yes, and Fyn) display decreased FN fibril assembly compared with wild-type cells (Wierzbicka-Patynowski and Schwarzbauer, 2002). Therefore, we explored whether HT-1080 cells cultured as multilayer aggregates demonstrate an alteration in FAK or Src expression that could account for the increased FN matrix assembly observed in 3D. As indicated in Figure 2A, there was no significant difference in FAK or Src protein levels when cells cultured in 2D versus aggregates were compared (p ≥ 0.05; Student’s unpaired t test). Furthermore, the basal activity of both FAK and Src, as determined by their phosphorylation status, was not significantly altered by the difference in culture conditions.
Figure 2.
Assessment of FAK, Src, and ERK expression in 2D and 3D culture. Antibodies against total and phosphorylated FAK, Src, or ERK were used to assess whether culture conditions influenced expression or function of these known modulators of FN matrix assembly. No qualitative difference in the expression FAK, Src, or ERK was detected. A decrease in p-ERK, but not p-FAK or p-Src, was noted. This decrease was quantified by densitometric analysis of three separate experiments comparing p-ERK and t-ERK expression as a function of culture condition. An unpaired Student’s t test detected a significant difference (*p ≤ 0.05) in p-ERK-1 expression between cells grown as 2D and 3D cultures (C).
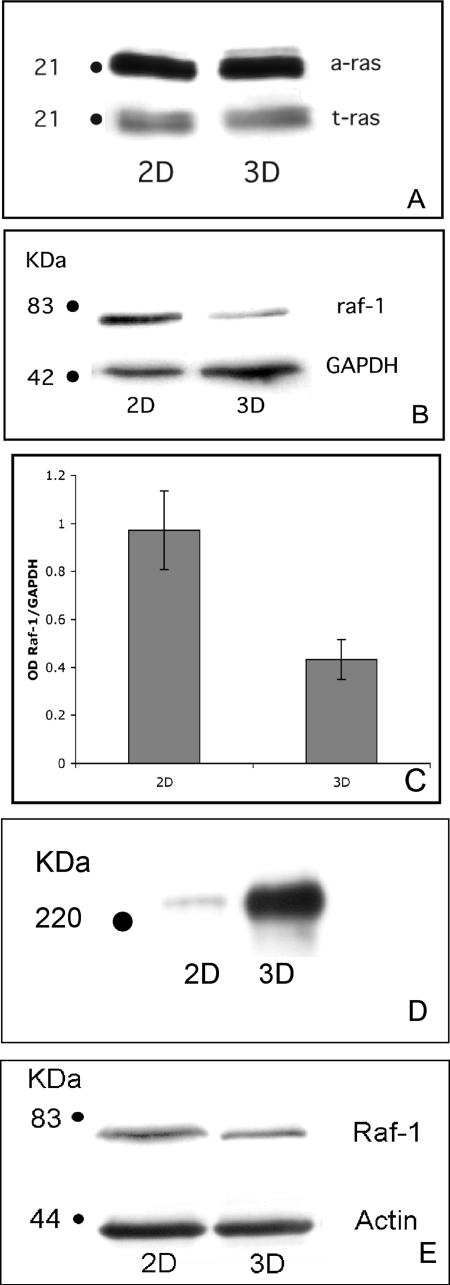
Previous work has shown that treatment of HT-1080 cells with the mitogen-activated protein kinase kinase (MEK) inhibitor PD98059 could induce matrix assembly in 2D culture (Brenner et al., 2000). Therefore, we determined whether 3D culture conditions modulate ERK activity. Although no change in total ERK was noted, a significant decrease in phospho-ERK was observed (Figure 2, B and C). HT-1080 cells possess one activated allele of N-ras (Brown et al., 1984). Because active Ras is located upstream of ERK, we examined whether culture environment could modulate Ras activity. HT-1080 cells cultured in 2D or as aggregates were lysed, and Ras activation was determined using a pull-down assay specific for Ras-GTP. No difference between active Ras-GTP and t-Ras was detected (Figure 3A). The immediate downstream effector of Ras is the serine-threonine kinase Raf-1. Activated Raf-1 has also been shown to suppress α5β1-mediated FN fibrillogenesis (Hughes et al., 1997). Therefore, to test whether cell culture conditions alter Raf-1 expression, HT-1080 cells in 2D or 3D culture were assessed for Raf-1 protein by immunoblotting. Interestingly, HT-1080 cells grown for 24 h as aggregates demonstrated a significant decrease in Raf-1 protein expression compared with cells cultured in 2D (Figure 3, B and C). Diminished Raf-1 protein levels are evident within the same time frame that FN fibril assembly occurs, suggesting that these two events may be linked. Because B-Raf signaling has also been strongly linked to ERK activation, we examined the expression of B-Raf in this system. No change in B-Raf protein level was detected when HT-1080 cells were grown in 3D compared with 2D (Supplemental Figure 3S).
Figure 3.
Induction of FN matrix assembly correlates with down-regulation of Raf-1 protein levels. Ras-GTP levels in HT-1080 cells were determined in 2D or 3D culture using a commercially available assay. (A) No difference in active or total Ras was noted. (B) Immunoblot analysis of Raf-1 expression in lysates generated from 3D cultures show a marked decrease in Raf-1 expression compared with cells cultured as monolayers. Raf-1 expression was normalized against the expression of GAPDH. (C) This decrease was quantified by densitometric analysis of five separate experiments comparing Raf-1 expression as a function of culture condition. GAPDH was used to normalize the data. An unpaired Student’s t test detected a significant difference (p < 0.05) in Raf-1 expression between cells grown as 2D and 3D cultures. (D) Transition to 3D culture induces FN matrix assembly in the rat prostatic carcinoma cell line MAT-LyLu. (E) This correlates with a decrease in Raf-1 expression in these cells.
In addition to MEK inhibition, both serum starvation and incubation with dexamethasone have been shown to induce FN matrix assembly in HT-1080 cells in 2D culture (Brenner et al., 2000). We therefore determined whether any of these treatments were associated with a down-regulation of Raf-1 protein. Immunoblot analysis of lysates from treated cells revealed no change in Raf-1 protein levels (Supplemental Figure 4S). These data suggest that the induction of FN matrix assembly by a decrease in Raf-1 protein levels is associated specifically with 3D culture conditions.
To determine whether a 2D to 3D transition could influence FN matrix assembly in other cell types, we examined the rat prostatic carcinoma cell line MAT-LyLu. These cells also express the integrin α5β1 in 2D culture, but they do not efficiently assemble a FN matrix. As demonstrated in Figure 3D, FN matrix assembly is induced in MAT-LyLu cells cultured as aggregates. We examined secretion of cellular FN by MAT-LyLu cells grown as monolayer culture or as aggregates using gelatin-Sepharose to bind secreted FN, as described above. Similar to the result obtained for HT-1080 cells, no difference in FN secretion was noted. Rather, FN matrix assembly correlated with a decrease in Raf-1 protein expression (Figure 3E).
Effect of Proteasome Inhibition on Raf-1 Protein Levels
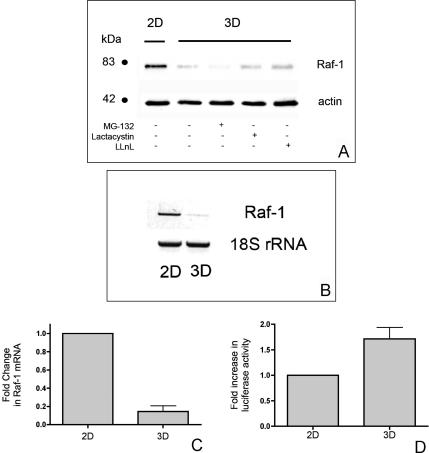
One possible mechanism for down-regulation of Raf-1 protein expression by the transition to 3D culture could be via the proteolysis of Raf-1 protein. Endogenous Raf-1 exists in a complex with heat-shock protein of 90 kDa (Hsp90) (Stancato et al., 1993). The release of Raf-1 from this complex leads to a rapid decrease in the half-life of the Raf-1 protein because of accelerated proteasome-mediated degradation (Schulte et al., 1995). Therefore, to determine whether 3D culture induces the targeted degradation of Raf-1, HT-1080 cells were cultured as aggregates for 24 h in the presence or absence of the cell-permeable proteasome inhibitors MG132, lactacystin, or LLnL. Our results show that proteasome inhibition does not rescue the diminished Raf-1 protein in 3D to the levels observed when cells are cultured in 2D (Figure 4A and Supplemental Figure 5S).
Figure 4.
Down-regulation of Raf-1 protein in 3D culture correlates with decreased Raf-1 mRNA levels. To explore potential mechanisms associated with Raf-1 down-regulation, aggregates of HT-1080 cells were incubated in the presence or absence of the proteosome inhibitors MG132 (10 μM), lactacystin (20 μM), or LLnL (40 μM). (A) Raf-1 expression was detected by immunoblotting. In separate experiments, cells were cultured in 2D or 3D overnight as indicated. Cells were lysed and total RNA extracted by TRIzol. (B) RT-PCR was performed using primers specific for Raf-1 or for the 18S ribosomal subunit. Alternately, Raf-1 mRNA was quantified using real-time PCR. (C) Data analysis of CT values of samples was performed using SDS 2.2 software to determine the -fold change in mRNA. To determine whether the decrease in Raf-1 protein is mediated by transcriptional regulation of Raf-1 mRNA, a 1194-base pair fragment containing the promoter region of the Raf-1 gene was amplified by PCR and cloned into the pGL3-basic luciferase expression vector. The pGL3-Raf luciferase construct was then used to transiently transfect HT-1080 cells. The pRL-SV40 vector containing the Renilla luciferase gene was cotransfected and served as an internal control. At 24 h after transfection, cells were harvested by trypsinization and replated in 2D or in 3D aggregates as described previously for an additional 24 h. (D) The Dual-Luciferase Reporter assay was used to determine the –fold increase in Raf-1 promoter activity.
Down-Regulation of Raf-1 Protein Correlates with Decreased Raf-1 mRNA Levels
An alternate mechanism for the change in Raf-1 protein levels could be that it reflects a decrease in Raf-1 mRNA levels. To test this hypothesis, HT-1080 cells were cultured in 2D or as aggregates, and Raf-1 mRNA levels were analyzed by both semiquantitative (Figure 4B) and quantitative RT-PCR (Figure 4C). We found that mRNA levels of Raf-1 were significantly reduced when cells were cultured as aggregates compared with cells cultured in 2D. Although these data do not exclude a contribution of protease activation to diminished Raf-1 protein, they show the novel result that the biophysical environment modulates Raf-1 mRNA levels. The decrease in Raf-1 protein and mRNA correlates with increased FN matrix assembly in these cells.
To determine whether the decrease in Raf-1 protein is mediated by transcriptional regulation of Raf-1 mRNA, a 1194-base pair fragment containing the promoter region of the Raf-1 gene was amplified by PCR and cloned into the pGL3-basic luciferase expression vector. The pGL3-Raf luciferase construct was then used to transiently transfect HT-1080 cells by electroporation. The pRL-SV40 vector containing the Renilla luciferase gene was cotransfected and served as an internal control. At 24 h after transfection, cells were harvested by trypsinization and replated in 2D or in 3D aggregates as described previously for an additional 24 h. As demonstrated in Figure 4D, Raf-1 promoter activity was not diminished by culture in 3D. On the contrary, Raf-1 promoter activity was consistently increased compared with cells cultured in 2D. These data suggest the novel idea that the transition to 3D culture may lead to posttranscriptional regulation of Raf-1 mRNA levels.
Knockdown of Raf-1 Expression Induces FN Matrix Assembly by HT-1080 Cells in 2D Culture
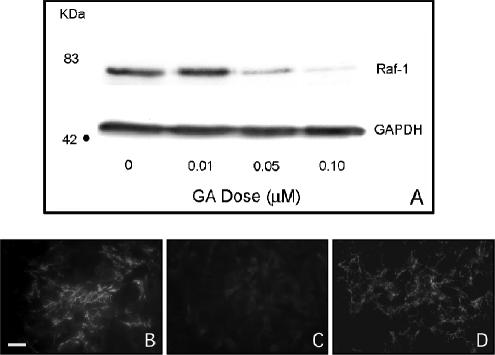
To confirm that down-regulation of Raf-1 expression can induce FN matrix assembly in cells cultured in 2D, we first used the benzoquinone anisanmycin antibiotic GA. The primary target of GA is Hsp90 that acts as a chaperone to a variety of important signaling molecules, including Raf-1 (Neckers et al., 1999). Treatment of cells with GA leads to destabilization of nonchaperoned proteins, resulting in their degradation. As demonstrated in Figure 5A, GA treatment of HT-1080 cells in 2D culture results in a dose-dependent decrease in Raf-1 protein expression after 24 h. When FN matrix assembly is examined by immunofluorescence microscopy, an intermediate dose of GA (0.05 μM) induces FN fibril assembly in 2D compared with vehicle control (Figure 5, B and C, respectively). Increasing the dose of GA (0.10 μM) produces a corresponding stimulation of fibrillar FN deposition. FN matrix assembly in response to GA treatment is similar to that observed for dexamethasone, an agent that has been shown to restore α5β1-mediated FN fibrillogenesis to HT-1080 cells (Figure 5D).
Figure 5.
GA treatment decreases Raf-1 expression and induces FN matrix assembly in 2D culture. (A) GA treatment results in a dose-dependent decrease in Raf-1 protein levels. GA also stimulated FN matrix assembly in 2D culture (B) compared with a vehicle control (C). (D) Matrix assembly in GA-treated cultures was similar to that observed for dexamethasone-treated cells. Bar, 20 μm.
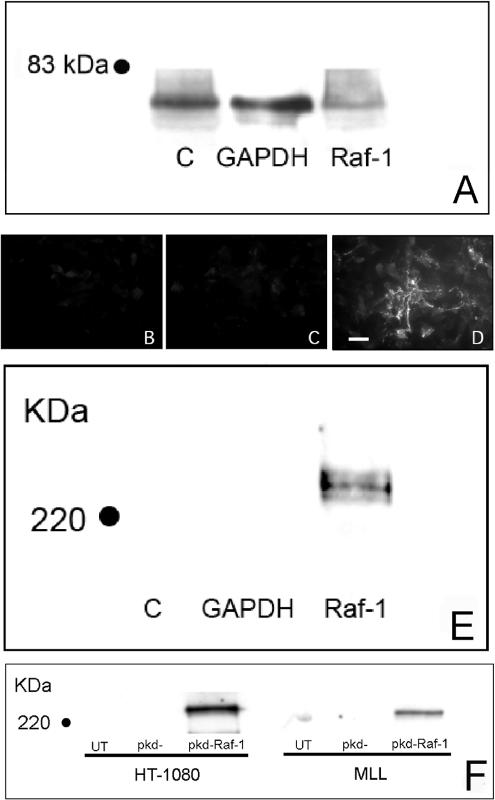
Geldanamycin supports the stable conformation of several signaling molecules that have been implicated in integrin function. Therefore, to determine whether the effect of GA on FN matrix assembly occurs specifically as a result of Raf-1 down-regulation, we used siRNA-mediated RNA interference to specifically target Raf-1, without affecting A-Raf or B-Raf, as confirmed by immunoblotting. Raf-1 siRNA transfection of HT-1080 cells resulted in a ∼50% decrease in Raf-1 protein expression after 24 h (Figure 6A). As demonstrated in Figure 6, D and E, Raf-1 knockdown rescues the ability of HT-1080 cells to assemble a FN matrix compared with control cells (Figure 6, B, C, and E). An siRNA targeting an alternate region of Raf-1 also induced FN matrix assembly in both HT-1080 and MAT-LyLu cells (Figure 6F). Together, these data indicate that specific down-regulation of Raf-1 protein restores FN fibrillogenesis to these cells.
Figure 6.
siRNA transfection of cells in 2D culture down-regulates Raf-1 expression and stimulates FN matrix assembly. HT-1080 cells were transfected with a commercially available and validated Raf-1 siRNA (A–E) or an expression plasmid (pKD-Raf-1) containing human Raf-1 sequence (F). When expressed, this sequence forms an shRNA that is processed to a Raf-1 siRNA that targets a distinct Raf-1 exon in both human and rat. (A) The Raf-1 siRNA down-regulated Raf-1 expression as assessed by immunoblot analysis. Raf-1 knockdown stimulated FN matrix assembly (D) compared with untransfected (B) and siRNA controls (C), as detected by immunofluorescence microscopy. (E and F) These results were confirmed by assessing the DOC-insoluble FN fraction by immunoblotting. (F) MAT-LyLu cells cultured in 2D were also transfected with pKD-Raf-1. Raf-1 siRNA transfection also resulted in an increased fraction of DOC-insoluble FN typical of FN matrix. Bar, 20 μm.
Maintenance of Raf-1 Expression Prevents FN Matrix Assembly by Cells Cultured in 3D
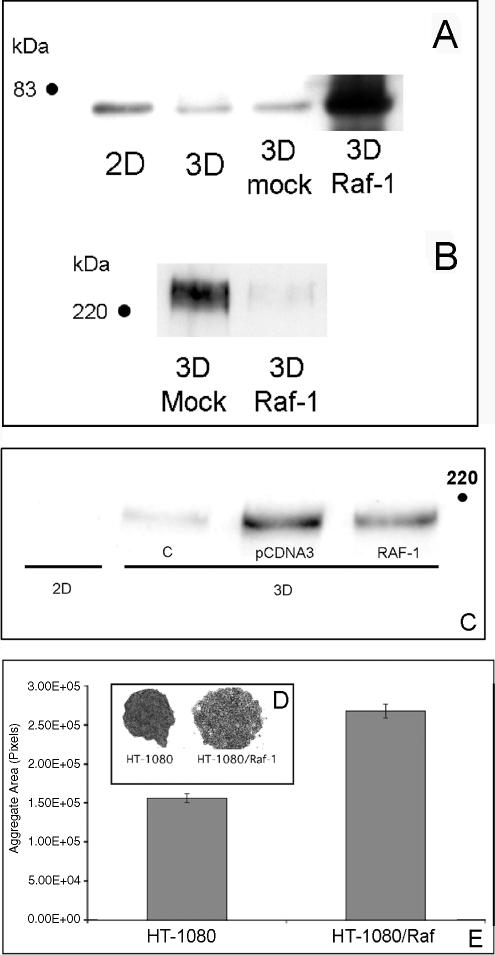
Our data suggest that increased FN matrix assembly by HT-1080 cells cultured as aggregates occurs as a consequence of diminished Raf-1 expression. Therefore, we reasoned that overexpression of Raf-1 would prevent HT-1080 cells in aggregate culture from regaining their ability to assemble FN fibrils. To test this, HT-1080 cells were transfected with a cDNA for human Raf-1 or an empty vector control. The level of Raf-1 in transfected cells (in 3D) ranged from 10- to 12-fold greater than vector-transfected cells in 3D (5- to 6-fold greater than cells cultured in 2D). As demonstrated in Figure 7, A and B, the increase in Raf-1 expression reduces FN fibril formation, compared with control cells. A decrease in FN matrix of ∼40% was also seen in MAT-LyLu cells overexpressing Raf-1 (Figure 7C). To determine whether the decreased FN matrix formation correlates with a difference in aggregate morphology, transfected HT-1080 cells were bulk-selected in G418. Figure 7, D and E, shows that the overexpression of Raf-1 in 3D results in loosely associated cells that exhibit increased aggregate size. Together, these data indicate that the regulation Raf-1 protein level by the physical environment modulates FN fibrillogenesis to influence integrin-dependent cell–cell interactions.
Figure 7.
Overexpression of Raf-1 inhibits FN matrix assembly in 3D. HT-1080 or MAT-LyLu cells were transfected by electroporation with a Raf-1 cDNA plasmid or vector control. Cells were then plated as 2D cultures or used to generate 3D hanging drops. Raf-1 expression was assessed by Western blot analysis. When grown as hanging drops, HT-1080 cells expressing exogenous Raf-1 (A) exhibit decreased FN matrix assembly as assessed by determining the DOC-insoluble FN fraction by immunoblotting (B). (C) A similar effect is seen with MAT-LyLu cells. When grown as hanging drops, HT-1080/Raf-1 cells did not compact to the same extent as did HT-1080 cells (C and D) as reflected by their significantly increased aggregate surface area (Student’s unpaired t test; p < 0.05).
DISCUSSION
In this article, we further investigate the role of FN matrix assembly in integrin-mediated cell–cell cohesion. We demonstrate that both HT-1080 and MAT-LyLu cells cultured as 3D aggregates regain their ability to assemble a FN matrix, which promotes increased intracellular cohesion. The induction of FN matrix assembly correlates with diminished Raf-1 protein expression in these cells that occurs primarily as a result of decreased Raf-1 mRNA levels. Interestingly, HT-1080 cells cultured in 3D demonstrate increased Raf-1 promoter activity, suggesting that the regulation of Raf-1 mRNA levels can occur posttranscriptionally. To confirm that Raf-1protein levels regulate FN matrix assembly in this context, we show that both pharmacological and siRNA knockdown of Raf-1 induces FN matrix assembly in 2D culture. Moreover, overexpression of Raf-1 inhibits FN matrix assembly in 3D culture, leading to diminished intercellular cohesion. Together, these data show that the biophysical environment can regulate Raf-1 expression to modulate fibrillar matrix formation and integrin-dependent cell–cell interactions.
Various studies support the concept that the morphology and function of cells grown as 3D aggregates can differ significantly from those of cells grown as conventional 2D monolayers (Li et al., 1987; Eckes et al., 1993; Cukierman et al., 2002; Bissell et al., 2003). For example, the studies of Bissell and colleagues have shown that changes in dimensionality are critical for the expression of the malignant phenotype of HMT-3522 mammary epithelial cells (Weaver et al., 1997; Wang et al., 1998; Bissell et al., 1999). Cells of the HMT-3522 series are indistinguishable when cultured as 2D monolayers. Phenotypic differences between the various lines become apparent only when these cells are cultured in a 3D-reconstituted basement membrane. Furthermore, cells can be reverted to near-normal phenotype by the inhibition of β1 integrin-mediated signaling (Weaver et al., 1997). This effect, however, is manifested only when cells are grown as 3D spheroids. Acquired multidrug resistance (MDR) has also been shown to be associated with the culture microenvironment. Kerbel and colleagues showed that MDR in mouse EMT-6 mammary carcinoma cells is manifested in vitro only if cells were grown in 3D configurations and not in conventional monolayer cultures (Kobayashi et al., 1993).
Our work shows that the transition from 2D culture to multilayer aggregate induces FN matrix assembly by both HT-1080 and MAT-LyLu cells. This correlates with a significant decrease in Raf-1 protein. The effect of 3D culture is largely specific to Raf-1, because comparable effects were not seen for two other signaling molecules implicated in FN matrix assembly, FAK and Src. These data suggest that the ability of the 3D microenvironment to influence Raf-1 expression in certain cell types could provide one explanation for phenotypic alterations observed with 2D-to-3D transition, because alterations in Raf-1 expression can modulate integrin function. For example, changes in Raf-1 protein levels could impact fibrillar matrix assembly to produce altered FN fibril length or fibril deposition. Because the characteristics of cell interactions with fibrillar matrix proteins are distinct from those of two-dimensional substrates (Cukierman et al., 2001, 2002), such changes in matrix architecture and density could impact the cell–matrix contacts, leading to changes in cell growth, differentiation, and survival. Thus, Raf-1 expression, in addition to activation, is likely to be a critical factor controlling integrin-mediated cell behaviors.
Activating mutants of Ras have been associated with malignant transformation and suppression of normal integrin function. For example, the expression of activated variants of both H-Ras and Raf-1 blocks integrin activation (Hughes et al., 1997, 2002). In these experiments, the effect of the mutant H-Ras required binding to Raf-1. Additionally, cells expressing Raf-1:ER, a conditionally active form of Raf-1, lose α5β1 integrin function upon induction with 4′-hydroxytamoxifen. Our data support previous work demonstrating a pivotal role for Raf-1 in the regulation of FN matrix assembly by showing that the 3D microenvironment, through its effect on Raf-1 mRNA and protein levels, restores the capacity for FN matrix assembly in two transformed cell types. This effect of 3D culture is independent of both FN secretion and integrin receptor expression. Rather, it suggests a mechanism dependent on postsecretory events associated with fibrillogenesis. The exact mechanism for the effect of Raf-1 on this process is unclear; however, one possible explanation may be the effect of the Ras-Raf–ERK signaling pathway on the posttranslational modification of integrin subunits.
Factors that influence Raf-1 expression are likely to become important therapeutic targets. Recently, depletion of Raf-1 in a variety of transformed cells has been achieved with GA, which binds to and maintains the correct conformation of important signaling molecules, including Raf-1 (Stancato et al., 1993). Inhibition of Raf-1–Hsp90 interactions leads to destabilization of Raf-1 and its targeted degradation via the proteasome pathway (Schulte et al., 1995, 1997). We used GA to pharmacologically deplete Raf-1 in HT-1080 cells, confirming that Raf-1 can undergo ubiquitin-dependent proteolysis in these cells. Recent work has suggested that cell–ECM interactions can regulate Raf-1 protein stability. Manenti et al. (2002) demonstrated that proteasome-dependent degradation of Raf-1 occurs in NIH-3T3 cells when they are cultured in suspension for an extended time. Furthermore, they showed that cell suspension induced ubiquitylation of Raf-1 and that the down-regulation of Raf-1 in suspended cells could be blocked by proteasome inhibition. Clearly, there is precedence for cell attachment to influence Raf-1 expression by proteolysis. However, we saw only a modest increase in Raf-1 expression when HT-1080 cells were treated with a proteasome inhibitor in 3D, suggesting that the effect of culture conditions on Raf-1 expression is largely independent of proteasome function. This is not surprising because, in hanging drop culture, the cells are not kept in suspension. Rather, the culture system allows the formation of multilayered cellular aggregates in which fibrillar matrix formation supports cellular rearrangement, aggregate compaction, and spheroid formation (Robinson et al., 2004). Additionally, cell viability is maintained over time. Although we cannot rule out that Raf-1 expression is influenced by proteolysis in our system, the effect seems largely mediated by a decrease in Raf-1 mRNA levels, which may reflect an alteration in mRNA stability. Few studies have examined the stability of the Raf-1 mRNA transcript. However, previous work suggests that the mRNA half-life, at least in monocytes, can be regulated by adhesion (Colotta et al., 1991). Interestingly, the 3′ untranslated region of Raf-1 is highly conserved among species (Duret et al., 1993; Duret and Bucher, 1997).
The transition from 2D culture to 3D matrices can modulate not only cell signaling events but also transcriptional and posttranscriptional processes. This has been best described for cells grown in collagen gels. For example, human fibroblasts transferred from 2D to 3D down-regulate collagen I synthesis due to decreased transcription of the collagen I gene and to decreased collagen I mRNA half-life (Eckes et al., 1993). In contrast, up-regulation of the α2 integrin subunit occurs with transition of fibroblasts to 3D culture, as a consequence of activation of the transcription factor nuclear factor-κB (Xu and Clark, 1997; Xu et al., 1998). Similarly, when hepatic stellate cells are cultured within collagen gels, matrix metalloproteinase expression is strongly induced compared with 2D culture as a consequence of transcriptional up-regulation (Takahra et al., 2004). Mechanical loading can also have an effect on cell signaling and transcriptional regulation. Breast epithelial cells differentiate into tubules when cultured in floating collagen gels but not when the same collagen gels are anchored to a dish (Parry et al., 1985; Keely et al., 1995). In this instance, the mechanical sensing of the biophysical environment led to the down-regulation of Rho activity, an event required for tubulogenesis in this system (Wozniak et al., 2003). Similarly, the mRNA and protein level of FN and large and small collagen XII variants was shown to be higher when skin fibroblasts are maintained in attached collagen gels than when they are cultured in a floating lattice relieved of tension (Fluck et al., 2003). Cells in hanging drop culture aggregate at the air–water interface and therefore lack tension associated with a rigid substrate. The formation of FN matrices in this context is completely dependent on cell–cell interactions and cell-directed force generation as the typical associations between FN and the planar surface of the tissue culture plate are absent. Although the mechanics of this system are not well understood, cell-generated tension allows for fibrillar matrix formation, aggregate compaction, and cell rearrangement (Robinson et al., 2003, 2004). Indeed, it represents a unique culture system in that the only supporting stromal environment is that generated by the cells de novo. The forces that are generated in this context are likely to be more reflective of those generated in vivo. Matrix reorganization and remodeling occurs in conjunction with alterations in fibrillar ECM components, their density, and their structure. It follows that, as ECM content and architecture change, the generated forces will be equally dynamic. Our data suggest that such changes in the biophysical environment can influence Raf-1 protein levels and in turn, integrin-dependent cell behaviors.
In conclusion, we show for the first time that transformed cells cultured as multilayer aggregates regain their ability to assemble a FN matrix due to a down-regulation of Raf-1 mRNA and protein levels. Regulation of Raf-1 expression by the biophysical environment may act in concert with Raf-1 activation to fine-tune a variety of integrin-dependent cellular activities.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health Grant GM-061487 (to S.A.C.) and Department of Defense Grant W81XWH 04-1-0265 (to R.A.F.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0849) on May 17, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Beck T. W., Brennscheidt U., Sithanandam G., Cleveland J., Rapp U. R. Molecular organization of the human Raf-1 promoter region. Mol. Cell. Biol. 1990;10:3325–3333. doi: 10.1128/mcb.10.7.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. P., Harding K. G., Stanton M. R., Jasani B., Ehrlich H. P. Human wound contraction: collagen organization, fibroblasts, and myofibroblasts. Plast. Reconstr. Surg. 1998;102:124–131. doi: 10.1097/00006534-199807000-00019. discussion 132–124. [DOI] [PubMed] [Google Scholar]
- Bissell M. J., Rizki A., Mian I. S. Tissue architecture: the ultimate regulator of breast epithelial function. Curr. Opin. Cell Biol. 2003;15:753–762. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell M. J., Weaver V. M., Lelievre S. A., Wang F., Petersen O. W., Schmeichel K. L. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757s–1763s. discussion 1763s–1764s. [PubMed] [Google Scholar]
- Brenner K. A., Corbett S. A., Schwarzbauer J. E. Regulation of fibronectin matrix assembly by activated Ras in transformed cells. Oncogene. 2000;19:3156–3163. doi: 10.1038/sj.onc.1203626. [DOI] [PubMed] [Google Scholar]
- Brown R., Marshall C. J., Pennie S. G., Hall A. Mechanism of activation of an N-ras gene in the human fibrosarcoma cell line HT1080. EMBO J. 1984;3:1321–1326. doi: 10.1002/j.1460-2075.1984.tb01970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotta F., Polentarutti N., Mantovani A. Differential expression of Raf-1 protooncogene in resting and activated human leukocyte populations. Exp. Cell Res. 1991;194:284–288. doi: 10.1016/0014-4827(91)90366-3. [DOI] [PubMed] [Google Scholar]
- Corbett S. A., Lee L., Wilson C. L., Schwarzbauer J. E. Covalent cross-linking of fibronectin to fibrin is required for maximal cell adhesion to a fibronectin-fibrin matrix. J. Biol. Chem. 1997;272:24999–25005. doi: 10.1074/jbc.272.40.24999. [DOI] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D. R., Yamada K. M. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Yamada K. M. Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- Duret L., Bucher P. Searching for regulatory elements in human noncoding sequences. Curr. Opin. Struct. Biol. 1997;7:399–406. doi: 10.1016/s0959-440x(97)80058-9. [DOI] [PubMed] [Google Scholar]
- Duret L., Dorkeld F., Gautier C. Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 1993;21:2315–2322. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckes B., Mauch C., Huppe G., Krieg T. Downregulation of collagen synthesis in fibroblasts within three-dimensional collagen lattices involves transcriptional and posttranscriptional mechanisms. FEBS Lett. 1993;318:129–133. doi: 10.1016/0014-5793(93)80006-g. [DOI] [PubMed] [Google Scholar]
- Fluck M., Giraud M. N., Tunc V., Chiquet M. Tensile stress-dependent collagen XII and fibronectin production by fibroblasts requires separate pathways. Biochim. Biophys. Acta. 2003;1593:239–248. doi: 10.1016/s0167-4889(02)00394-4. [DOI] [PubMed] [Google Scholar]
- Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;13:264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Hall A., Marshall C. J., Spurr N. K., Weiss R. A. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature. 1983;303:396–400. doi: 10.1038/303396a0. [DOI] [PubMed] [Google Scholar]
- Hughes P. E., Oertli B., Hansen M., Chou F. L., Willumsen B. M., Ginsberg M. H. Suppression of integrin activation by activated Ras or Raf does not correlate with bulk activation of ERK MAP kinase. Mol. Biol. Cell. 2002;13:2256–2265. doi: 10.1091/mbc.01-10-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P. E., Renshaw M. W., Pfaff M., Forsyth J., Keivens V. M., Schwartz M. A., Ginsberg M. H. Suppression of integrin activation: a novel function of a Ras/Raf-initiated MAP kinase pathway. Cell. 1997;88:521–530. doi: 10.1016/s0092-8674(00)81892-9. [DOI] [PubMed] [Google Scholar]
- Hynes R. O. Fibronectins. New York: Springer-Verlag; 1990. [Google Scholar]
- Ilic D. FAK promotes organization of fibronectin matrix and fibrillar adhesions. J. Cell Sci. 2004;117:177–187. doi: 10.1242/jcs.00845. [DOI] [PubMed] [Google Scholar]
- Katz M. E., McCormick F. Signal transduction from multiple Ras effectors. Curr. Opin. Genet. Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- Keely P. J., Fong A. M., Zutter M. M., Santoro S. A. Alteration of collagen-dependent adhesion, motility, and morphogenesis by the expression of antisense alpha 2 integrin mRNA in mammary cells. J. Cell Sci. 1995;108:595–607. doi: 10.1242/jcs.108.2.595. [DOI] [PubMed] [Google Scholar]
- Kinbara K., Goldfinger L. E., Hansen M., Chou F. L., Ginsberg M. H. Ras GTPases: integrins’ friends or foes? Nat. Rev. Mol. Cell Biol. 2003;4:767–776. doi: 10.1038/nrm1229. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Man S., Graham C. H., Kapitain S. J., Teicher B. A., Kerbel R. S. Acquired multicellular-mediated resistance to alkylating agents in cancer. Proc. Natl. Acad. Sci. USA. 1993;90:3294–3298. doi: 10.1073/pnas.90.8.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. L., Aggeler J., Farson D. A., Hatier C., Hassell J., Bissell M. J. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl. Acad. Sci. USA. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque A., Gomez M., Puzon W., Takada Y., Sanchez-Madrid F., Cabanas C. Activated conformations of very late activation integrins detected by a group of antibodies (HUTS) specific for a novel regulatory region (355–425) of the common beta 1 chain. J. Biol. Chem. 1996;271:11067–11075. doi: 10.1074/jbc.271.19.11067. [DOI] [PubMed] [Google Scholar]
- Ly D. P., Zazzali K. M., Corbett S. A. De novo expression of the integrin alpha5beta1 regulates alphavbeta3-mediated adhesion and migration on fibrinogen. J. Biol. Chem. 2003;278:21878–21885. doi: 10.1074/jbc.M212538200. [DOI] [PubMed] [Google Scholar]
- Manenti S., Delmas C., Darbon J. M. Cell adhesion protects c-Raf-1 against ubiquitin-dependent degradation by the proteasome. Biochem. Biophys. Res. Commun. 2002;294:976–980. doi: 10.1016/S0006-291X(02)00594-6. [DOI] [PubMed] [Google Scholar]
- McKeown-Longo P. J., Etzler C. A. Induction of fibronectin matrix assembly in human fibrosarcoma cells by dexamethasone. J. Cell Biol. 1987;104:601–610. doi: 10.1083/jcb.104.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L., Schulte T. W., Mimnaugh E. Geldanamycin as a potential anti-cancer agent: its molecular target and biochemical activity. Investig. New Drugs. 1999;17:361–373. doi: 10.1023/a:1006382320697. [DOI] [PubMed] [Google Scholar]
- Olden K., Yamada K. M. Mechanism of the decrease in the major cell surface protein of chick embryo fibroblasts after transformation. Cell. 1977;11:957–969. doi: 10.1016/0092-8674(77)90307-5. [DOI] [PubMed] [Google Scholar]
- Parry G., Lee E. Y., Farson D., Koval M., Bissell M. J. Collagenous substrata regulate the nature and distribution of glycosaminoglycans produced by differentiated cultures of mouse mammary epithelial cells. Exp. Cell Res. 1985;156:487–499. doi: 10.1016/0014-4827(85)90556-7. [DOI] [PubMed] [Google Scholar]
- Plantefaber L. C., Hynes R. O. Changes in integrin receptors on oncogenically transformed cells. Cell. 1989;56:281–290. doi: 10.1016/0092-8674(89)90902-1. [DOI] [PubMed] [Google Scholar]
- Rasheed S., Nelson-Rees W. A., Toth E. M., Arnstein P., Gardner M. B. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Robinson E. E., Foty R. A., Corbett S. A. Fibronectin matrix assembly regulates alpha5beta1-mediated cell cohesion. Mol. Biol. Cell. 2004;15:973–981. doi: 10.1091/mbc.E03-07-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson E. E., Zazzali K. M., Corbett S. A., Foty R. A. alpha5beta1 integrin mediates strong tissue cohesion. J. Cell Sci. 2003;116:377–386. doi: 10.1242/jcs.00231. [DOI] [PubMed] [Google Scholar]
- Roy S., Lane A., Yan J., McPherson R., Hancock J. F. Activity of plasma membrane-recruited Raf-1 is regulated by Ras via the Raf zinc finger. J. Biol. Chem. 1997;272:20139–20145. doi: 10.1074/jbc.272.32.20139. [DOI] [PubMed] [Google Scholar]
- Rudin C. M., et al. Phase I Trial of ISIS 5132, an antisense oligonucleotide inhibitor of c-raf-1, administered by 24-hour weekly infusion to patients with advanced cancer. Clin. Cancer Res. 2001;7:1214–1220. [PubMed] [Google Scholar]
- Schulte T. W., An W. G., Neckers L. M. Geldanamycin-induced destabilization of Raf-1 involves the proteasome. Biochem. Biophys. Res. Commun. 1997;239:655–659. doi: 10.1006/bbrc.1997.7527. [DOI] [PubMed] [Google Scholar]
- Schulte T. W., Blagosklonny M. V., Ingui C., Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J. Biol. Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer J. E. Fibronectin: from gene to protein. Curr. Opin. Cell Biol. 1991;3:786–791. doi: 10.1016/0955-0674(91)90051-y. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Sechler J. L. Fibronectin fibrillogenesis: a paradigm for extracellular matrix assembly. Curr. Opin. Cell Biol. 1999;11:622–627. doi: 10.1016/s0955-0674(99)00017-4. [DOI] [PubMed] [Google Scholar]
- Sechler J. L., Takada Y., Schwarzbauer J. E. Altered rate of fibronectin matrix assembly by deletion of the first type III repeats. J. Cell Biol. 1996;134:573–583. doi: 10.1083/jcb.134.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile J., Hocking D. C. Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol. Biol. Cell. 2002;13:3546–3559. doi: 10.1091/mbc.E02-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancato L. F., Chow Y. H., Hutchison K. A., Perdew G. H., Jove R., Pratt W. B. Raf exists in a native heterocomplex with hsp90 and p50 that can be reconstituted in a cell-free system. J. Biol. Chem. 1993;268:21711–21716. [PubMed] [Google Scholar]
- Stokoe D., Macdonald S. G., Cadwallader K., Symons M., Hancock J. F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- Takahra T., Smart D. E., Oakley F., Mann D. A. Induction of myofibroblast MMP-9 transcription in three-dimensional collagen I gel cultures: regulation by NF-kappaB, AP-1 and Sp1. Int. J. Biochem. Cell Biol. 2004;36:353–363. doi: 10.1016/s1357-2725(03)00260-7. [DOI] [PubMed] [Google Scholar]
- Wang F., Weaver V. M., Petersen O. W., Larabell C. A., Dedhar S., Briand P., Lupu R., Bissell M. J. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc. Natl. Acad. Sci. USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver V. M., Petersen O. W., Wang F., Larabell C. A., Briand P., Damsky C., Bissell M. J. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I., Schwarzbauer J. E. Regulatory role for SRC and phosphatidylinositol 3-kinase in initiation of fibronectin matrix assembly. J. Biol. Chem. 2002;277:19703–19708. doi: 10.1074/jbc.M200270200. [DOI] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I., Schwarzbauer J. E. The ins and outs of fibronectin matrix assembly. J. Cell Sci. 2003;116:3269–3276. doi: 10.1242/jcs.00670. [DOI] [PubMed] [Google Scholar]
- Wozniak M. A., Desai R., Solski P. A., Der C. J., Keely P. J. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Clark R. A. A three-dimensional collagen lattice induces protein kinase C-zeta activity: role in alpha2 integrin and collagenase mRNA expression. J. Cell Biol. 1997;136:473–483. doi: 10.1083/jcb.136.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zutter M. M., Santoro S. A., Clark R. A. A three-dimensional collagen lattice activates NF-kappaB in human fibroblasts: role in integrin alpha2 gene expression and tissue remodeling. J. Cell Biol. 1998;140:709–719. doi: 10.1083/jcb.140.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmuidzinas A., Mamon H. J., Roberts T. M., Smith K. A. Interleukin-2-triggered Raf-1 expression, phosphorylation, and associated kinase activity increase through G1 and S in CD3-stimulated primary human T cells. Mol. Cell. Biol. 1991;11:2794–2803. doi: 10.1128/mcb.11.5.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.