Abstract
The biosynthesis pathways of two anthracyclines, nogalamycin and aclacinomycin, were directed toward angucyclines by using an angucycline-specific cyclase, pgaF, isolated from a silent antibiotic biosynthesis gene cluster. Addition of pgaF to a gene cassette that harbored the early biosynthesis genes of nogalamycin resulted in the production of two known angucyclinone metabolites, rabelomycin and its precursor, UWM6. Substrate flexibility of pgaF was demonstrated by replacement of the nogalamycin minimal polyketide synthase genes in the gene cassette with the equivalent aclacinomycin genes together with aknE2 and aknF, which specify the unusual propionate starter unit in aclacinomycin biosynthesis. This modification led to the production of a novel angucyclinone, MM2002, in which the expected ethyl side chain was incorporated into the fourth ring.
Aromatic polyketides consist of a large group of structurally diverse natural products that exhibit a broad range of biological activities. Despite the structural diversity found within different aromatic polyketides, they all arise from a fundamentally similar biosynthesis pathway which has been elucidated for many representatives of this class of natural products (for a review, see reference 4). Biosynthesis begins with the condensation of a specific number of small carboxylic acids by the iterative action of the minimal polyketide synthase (minPKS) complex, which consists of two ketosynthases (KSα and KSβ) and an acyl carrier protein (ACP) onto which the growing polyketide chain is attached via a flexible phosphopantetheinyl arm. The polyketide chain is subsequently folded into a range of different aromatic compounds by various ketoreductases (KR), cyclases (CYC), and aromatases (ARO). Finally, tailoring reactions catalyzed by diverse oxygenases (OXY), methylases, and reductases, as well as possible noncatalyzed reactions, often further modify the aglycone formed. In this work the latter turned out to be important because of the reactive nature of the isolated intermediates (see below).
The convergent biosynthesis pathways have prompted research into combinatorial biosynthesis, a method that uses genetic engineering of metabolic pathways to provide modified natural products for drug discovery (6, 8). Early research led to the identification of a set of design rules (12) which could be used to engineer novel aromatic polyketides in a predictable manner. However, later research has shown that in many instances different combinations of antibiotic biosynthesis genes have not resulted in the production of the expected compounds. In particular, the jadI cyclase gene from the biosynthesis pathway of the angucycline antibiotic jadomycin has been reported to be incompatible with tetracenomycin (11) and daunorubicin (20) biosynthesis genes. Similarly, the tetracenomycin (tcmI) and daunorubicin (dpsY) cyclase genes were nonfunctional with jadomycin biosynthesis genes (11, 20).
We have recently isolated a silent antibiotic biosynthesis gene cluster that exhibits strong sequence homology to the known angucycline gene clusters (M. Metsä-Ketelä, unpublished results) from soil isolate Streptomyces sp. strain PGA64. The cluster contains a cyclase gene designated pgaF, according to the other homologous angucycline cyclases lanF and urdF from the landomycin (19) and urdamycin (3) biosynthesis pathways, respectively. All of these cyclases also display sequence homology to the jadI cyclase, which has been proven to be responsible for the cyclization of the fourth, angular ring in jadomycin biosynthesis (11).
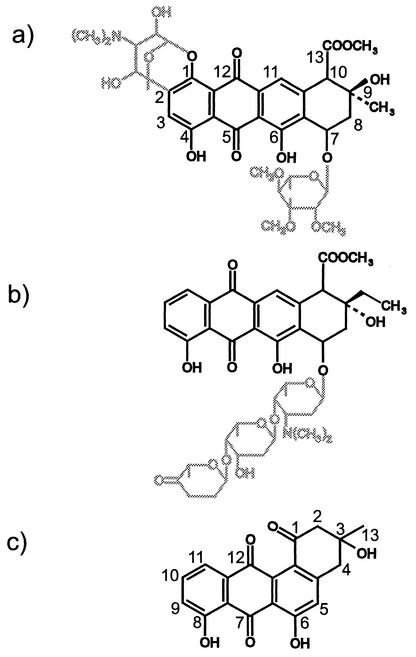
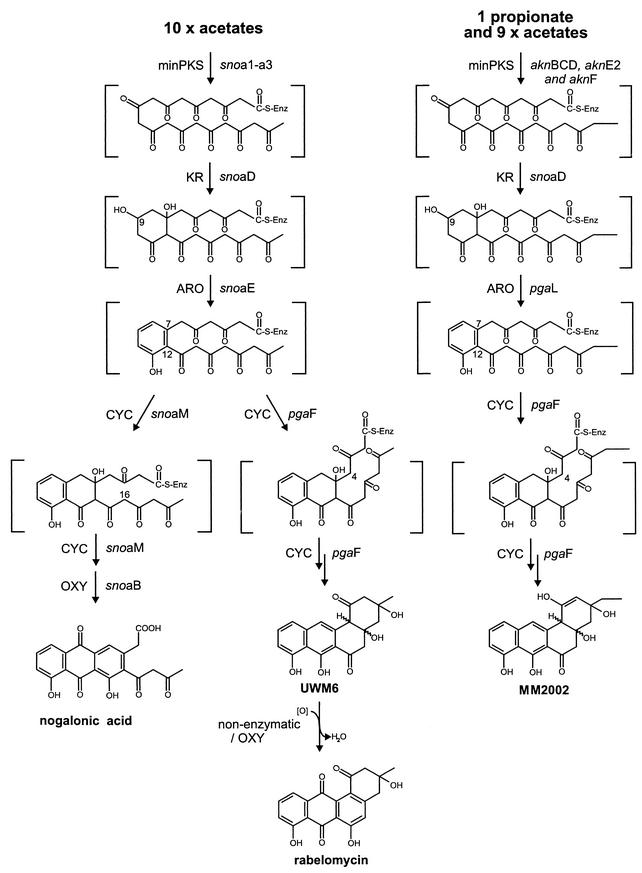
Our previous investigations on the biosynthesis pathways of the anthracycline antibiotics nogalamycin (17, 24) and aclacinomycins (14) (Fig. 1) combined with the genes from the silent angucycline gene cluster presented us an opportunity to study the interface of these two classes of aromatic polyketides and to assess the extent to which these pathways are compatible. In particular, the nogalamycin biosynthesis genes offered a good starting point for this study, since nogalamycin biosynthesis proceeds identically, up to a point, to the proposed biosynthesis pathways of angucyclines (Fig. 2); in both pathways the polyketide chain synthesized is composed of 10 acetates and the C-9 ketone is reduced to hydroxyl, followed by cyclization between C-7 and C-12 and aromatization of the first ring. The second ring is also closed in a similar manner, and differences start to appear only in the third cyclization event, in which opposite carbons (C-16 in nogalamycin and C-4 in angucyclines) carry out the nucleophilic attack.
FIG. 1.
Structures of anthracycline antibiotics nogalamycin (a), produced by S. nogalater, and aclacinomycin A, produced by S. galilaeus (b), and an example of angucycline antibiotic rabelomycin (c). Black and gray lines indicate atoms derived from PKS and deoxysugar biosynthesis pathways, respectively.
FIG. 2.
Structures and hypothetical biosynthesis pathways of the compounds produced in this study. The pathway resulting in the production of nogalonic acid is described elsewhere in detail (14). Structures within brackets indicate hypothetical intermediates.
The biosynthesis of aklavinone, the aglycone moiety of aclacinomycin, progresses highly similarly to that of nogalamycin, with the exception of the use of a propionate starter unit, which results in an ethyl side chain in the fourth ring (14), and the stereochemistry in the cyclization of the fourth ring, which is opposite in the nogalamycin and aclacinomycin biosynthesis pathways (16). In this study, the differences in anthracycline starter unit utilization could be used to investigate the substrate flexibility of PgaF, as priming of the minPKS complex with propionate results in an unnatural substrate for PgaF that is composed of a longer carbon chain (Fig. 2). Here we report on the use of the pgaF cyclase to divert the biosynthetic pathways of two anthracyclines, nogalamycin and aclacinomycin, toward angucyclines.
MATERIALS AND METHODS
Bacterial strains and cultivation of bacteria.
The bacterial strain designated PGA64 was obtained from a soil sample collected in Barcelona, Spain. One gram of the dried soil sample was suspended in 5 ml of H2O and centrifuged to remove solid substances. The supernatant containing bacterial strains was spread as 100-μl aliquots on International Streptomyces Project (ISP) medium 4-related (ISP4) agar plates in which fructose replaced starch as a carbon source and that were supplemented with heximide (25 μg/ml). Soil isolate PGA64 was purified on agar plates by repeated plating. Cultures were maintained on ISP4 agar (Difco, Detroit, Mich.), which is a medium commonly used to isolate Streptomyces and closely related genera from soil. Manipulations of Streptomyces DNA were carried out with Escherichia coli XL2-Blue (Stratagene, La Jolla, Calif.). The Streptomyces strains used were Streptomyces nogalater ATCC 27451 (2), Streptomyces galilaeus ATCC 31615 (13), and Streptomyces lividans TK24 (9). E. coli XL2-Blue and Streptomyces strains were grown at 37 and 30°C in Luria-Bertani medium (15) and E1 medium (23), respectively.
PCR amplifications and cloning procedures.
Amplification of DNA fragments by PCR was performed with 100 pmol of each oligonucleotide primer, 0.1 to 100 ng of plasmid or chromosomal DNA template, 5 mmol of each deoxynucleoside triphosphate, and 2 U of DynaZyme II DNA polymerase (Finnzymes Oy, Espoo, Finland). The template was initially denaturated by heating at 99°C for 5 min, followed by 30 cycles of amplification, i.e., denaturation at 96°C for 1 min, annealing at 55 to 62°C (depending on the primers) for 2 min, and extension at 73°C for 1 min 30 s. The primers that were used are shown in Table 1. The PCR products were cloned with the TOPO-TA cloning system (Invitrogen, Carlsbad, Calif.) and sequenced with an automatic ABI310 DNA sequencer (Applied Biosystems, Foster City, Calif.). DNA isolations and manipulations were carried out by standard procedures (9, 15). DNA propagated in E. coli was introduced by standard protoplast transformation into S. lividans TK24 (9).
TABLE 1.
Sequences of oligonucleotide primers used in amplification of the genes used in this studya
| Gene | Forward and reverse primer sequences | Restriction site | Reference |
|---|---|---|---|
| snoa1 and snoa2 | GATTCTAGAACTTCAAGACGATCAAAGC | XbaI | Kantola, unpublished |
| GATAAGCTTTTAATTAACTTACCGCGGCTCCGG | HindIII and PacI | Kantola, unpublished | |
| snoa3 | GATTTAATTAACGGTAAGAGGCCACGGAA | PacI | Kantola, unpublished |
| GATAAGCTTGCTAGCAGTTCCGTGCCGTACACA | HindIII and NheI | Kantola, unpublished | |
| snoaD | GATGCTAGCTTGTATTCCGCCGTGTC | NheI | Kantola, unpublished |
| GATAAGCTTGTTAACTGTCATGGTCCTCCGTCC | HindIII and HpaI | Kantola, unpublished | |
| snoaE | GATGTTAACGGCCTGGGAAACTACTGA | HpaI | Kantola, unpublished |
| GATAAGCTTATGCATGAACCGTCCTCCGCTGT | HindIII and NsiI | Kantola, unpublished | |
| pgaF | CTTAAGCCCGCTCACCCGAAGAAGAG | AflII | This work |
| TACGTACTTCACTGTGTGGCCGTCCA | SnaBI | This work | |
| pgaL | GTTAACGACCTGAAACCGACCCCTGA | HpaI | This work |
| ATGCATGTGTGGTCCTCGCTCATGTC | NsiI | This work | |
| aknBCDE2F | GATTCTAGAGCCCGAACTGTATCTGGTA | XbaI | 12 |
| GATAAGCTTGCTAGCAGGACAAACGTCAGCAGCT | NheI | 12 |
The HindIII sites at the ends of the sno reverse primers were used as insertion sites for the indicated gene.
Expression constructs.
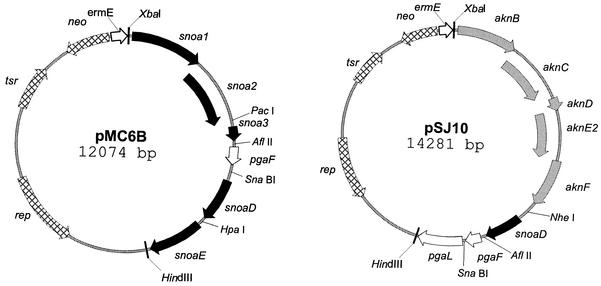
The nogalamycin biosynthesis genes were previously amplified (J. Kantola, unpublished results) by PCR with oligonucleotide primers that contain unique restriction sites at their ends (Table 1) to facilitate the construction of the desired expression vectors. The genes snoa1-a3, snoaD, and snoaE were combined in pUC19 (22) one at the time by using standard molecular biology methods (15) to give plasmid pMC5. The angucycline cyclase pgaF was inserted as a Klenow fragment-treated EcoRI fragment to a similarly blunt-ended NheI site of the pMC5 cassette, resulting in plasmid pMC6B (Fig. 3). To investigate the effect of the aclacinomycin minPKS genes, the sno minPKS genes were first removed from plasmid pMC6BD by XbaI-NheI digestion, and the akn genes were inserted as an XbaI-NheI fragment to give plasmid pSJ10 (Fig. 3). Plasmid pMC6BD was used in this case, because the NheI restriction site in pMC6B was no longer intact, as it was used for insertion of the pgaF cyclase gene. Plasmid pMC6BD, which causes the production of the same compounds as pMC6B in S. lividans TK24, differs from pMC6B in that it contains an aromatase (the pgaL gene from the silent angucycline gene cluster) different from the homologous nogalamycin snoaE aromatase. Both constructs were ligated as XbaI-HindIII fragments to similarly digested vector Streptomyces(pIJE486) and introduced into S. lividans TK24. In this system, all of the genes are expressed from the constitutive ermE promoter from Saccharopolyspora erythraea (9), which has been cloned into the polylinker of Streptomyces cloning vector pIJ486 (18).
FIG. 3.
Diagrams of expression constructs used in this work. The genes have been obtained from S. nogalater (sno; black), S. galilaeus (akn; gray), and Streptomyces PGA64 (pga; white); and they encode the following proteins: KSα (snoa1, aknB), KSβ (snoa2, aknC), ACP (snoa3, aknD), starter unit selection (aknE2, aknF), KR (snoaD), ARO (snoaE, pgaL), and CYC (pgaF). The antibiotic biosynthesis genes were cloned in the pIJ486 vector under the constitutive ermE promoter (8).
Isolation and purification of products.
For structure elucidation the strains were grown in 250-ml flasks (a total of 2 liters of E1 medium supplemented with thiostrepton [50 μg/ml] and XAD-7 absorbent [20 g/liter] at 30°C and 330 rpm) for 7 days. Compounds were extracted with acidic methanol-chloroform (1:3) and evaporated to dryness. The crude extract was subjected to silica gel liquid chromatography and was further separated by reversed-phase high-pressure liquid chromatography on a Merck Hitachi instrument (L-6200A/L-4250).
Instrumental analysis.
Structural elucidation of the compounds by nuclear magnetic resonance (NMR) was performed with a JEOL JNM-GX 400 spectrometer in dimethyl sulfoxide (DMSOd6). The spectra were run at ambient temperatures in the solvent, and both the 1H and 13C NMR spectra were referenced internally to tetramethylsilane, which was assigned a value of 0 ppm.
Nucleotide sequence accession number.
The 16.5-kb DNA fragment sequenced from Streptomyces sp. strain PGA64 has been submitted to GenBank and given accession number AY034378.
RESULTS AND DISCUSSION
Deduced function of pgaF gene product.
The gene cluster designated pga was cloned from a rubromycin-producing soil isolate, Streptomyces sp. strain PGA64. Analysis of the 16.5-kb DNA fragment sequenced revealed that the region was most similar to gene clusters involved in the biosynthesis of angucycline metabolites (Metsä-Ketelä, unpublished). In particular, the cluster harbors a gene designated pgaF, which is homologous to various cyclases found in angucycline biosynthesis gene clusters. These cyclases are distinct from other cyclases found in databases, and although they do show a lower degree of similarity to other cyclases involved in secondary metabolite production, they are indicative of angucycline biosynthesis. pgaF encodes a short 109-amino-acid peptide which is most related to the lanF gene product from the landomycin biosynthesis (19) pathway (82% identity, 92% similarity), followed by the jadI gene product (79% identity, 86% similarity), which has been shown to cyclize the last angular ring in jadomycin biosynthesis (11).
Conversion of nogalamycin biosynthetic pathway toward angucyclines.
We have recently elucidated the biosynthesis pathway of auramycinone by stepwise cloning of genes and isolation of the compounds produced (7; Kantola, unpublished). From these studies we acquired plasmid pMC5, which contains the genes that are responsible for the putatively mutual part of auramycinone and angucyclinone biosynthesis, namely, snoa1-a3 (minPKS), snoaD (KR), and snoaE (ARO). The putative angucycline cyclase pgaF was amplified from Streptomyces sp. strain PGA64 by PCR; inserted into the pMC5 cassette, resulting in plasmid pMC6B (Fig. 3); and introduced into S. lividans TK24 by standard protoplast transformation methods (9).
Fermentation of S. lividans TK24/pMC6B resulted in the production of two previously undetected compounds that appeared in different ratios in separate flask culture fermentations after 7 days of cultivation. The two compounds were isolated from the culture broth, and structural elucidation of the compounds by NMR (Table 2) revealed that the compounds were UWM6 and rabelomycin, which have previously been described as intermediates in the jadomycin biosynthesis pathway (11). Additionally, during the NMR work the instability of UWM6 under the conditions used was observed, as the sample slowly turned red; the maximum of the visible spectrum changed from 440 to 510 nm. This indicates additional conjugation within the molecule, which could result from the elimination of two water molecules and the formation of a fully aromatized derivative of UWM6 that is aromatized similarly to 6-hydroxytetrangulol (21). Furthermore, this transformation was accelerated in acidic conditions, which are favorable to the elimination reaction.
TABLE 2.
1H and 13C NMR spectral data for compounds MM2002 and UWM6
| Site | MM2002
|
UWM6
|
||
|---|---|---|---|---|
| 1H NMR spectrum (400 MHz) | 13C NMR spectrum (100 MHz) | 1H NMR spectrum (400 MHz) | 13C NMR spectrum (100 MHz) | |
| 1 | 166.1 (s) | 203.9 (s) | ||
| 1-OH | brda, exchangeable | |||
| 2A | 5.95, 1H, s | 123.5 (d) | 3.00, 1H, d, 18.0 | 57.5 (t) |
| 2B | 2.58, 1H, d, 18.0 | |||
| 3 | 72.4 (s) | 72.3 (s) | ||
| 3-OH | brd, exchangeable | brd, exchangeable | ||
| 4A | 2.72, 1H, d, 17.6 | 42.6 (t) | 2.90, 1H, d, 17.8 | 46.9 (t) |
| 4B | 2.50, 1H, d, 17.6 | 2.56, 1H, d, 17.8 | ||
| 4a | 71.1 (s) | 70.0 (s) | ||
| 4a-OH | brd, exchangeable | brd, exchangeable | ||
| 5A | 2.92, 1H, d, 17.8 | 48.2 (t) | 3.18, 1H, d, 17.9 | 53.1 (t) |
| 5B | 2.78, 1H, d, 17.8 | 2.87, 1H, d, 17.9 | ||
| 6 | 202.0 (s) | 202.7 (s) | ||
| 6a | 113.0 (s) | 110.0 (s) | ||
| 7 | 163.4 (s) | 164.6 (s) | ||
| 7a | 108.9 (s) | 108.7 (s) | ||
| 7-OH | 16.0, 1H, s | 15.9, 1H, brs | ||
| 8 | 157.5 (s) | 157.2 (s) | ||
| 8-OH | 9.66, 1H, s | 9,67, 1H, s | ||
| 9 | 6.84, 1H, d, 7.8 | 110.7 (d) | 6.78, 1H, dd, 7.8, 0.9 | 110.8 (d) |
| 10 | 7.47, 1H, dd, 8.2, 7.8 | 132.2 (d) | 7.45, 1H, dd, 8.1, 7.8 | 132.2 (d) |
| 11 | 7.17, 1H, d, 8.2 | 118.2 (d) | 7.18 1H, dd, 8.0, 0.9 | 118.0 (d) |
| 11a | 139.1 (s) | 138.6 (s) | ||
| 12 | 7.01, 1H, s | 121.1 (d) | 7.14, 1H, s | 124.8 (d) |
| 12a | 133.2 (s) | 132.9 (s) | ||
| 12b | 3.87, 1H, s | 58.2 (s) | 3.80, 1H, s | 62.0 (d) |
| 13 | 2.26, 2H, m | 29.9 (t) | 1.31, 3H, s | 23.6 (q) |
| 14 | 1.11, 3H, t, 7.2 | 10.5 (q) | ||
brd, broadened.
Of the two compounds identified, UWM6 has been suggested to be the compound that results from the direct action of the corresponding jadomycin biosynthesis genes and rabelomycin has been suggested to emerge from oxidation of UWM6 in flask cultures or from the concomitant action of endogenous genes of the host strain, S. lividans (11). These suggestions were based on the fact that the principal product was UWM6 on R2YENG agar plates. Our results coincide with the nonenzymatic model for UWM6 oxidation, since addition of an absorbent, XAD-7, to liquid cultures shifted the production greatly toward UWM6 and only minor amounts of rabelomycin were produced. This can be interpreted to mean that the UWM6 produced by the strain binds to XAD-7 after export to the culture medium and is subsequently protected from oxidation.
Interestingly, studies on the total synthesis of angucyclinones present an attractive analogy and a possible explanation for the nonenzymatic oxidation of the C-12 carbon; Krohn et al. (10) have reported a method for the total synthesis of a rabelomycin derivative in which the C-1 ketone is synthesized by photoinduced oxygenation upon exposure to daylight. They suggested that the reaction could proceed through a diradical intermediate derived from photoinduced hydrogen abstraction at C-1 by the neighboring C-12 carbonyl group. In the biosynthetic route presented here, a similar diradical could conceivably be formed after dehydration of UWM6 and photoinduced hydrogen abstraction at C-12 by the C-1 carbonyl group. The hydrogen abstraction can then be followed by reaction with oxygen and subsequent ketone formation.
Conversion of aclacinomycin biosynthetic pathway toward angucyclines.
To further test the ability of pgaF to act in conjunction with unnatural biosynthesis genes, we replaced the sno minPKS genes with the corresponding genes aknB, aknC, and aknD as well as genes aknE2 and aknF from the aclacinomycin pathway. The aknE2 and aknF genes, which are homologous to the dpsC and dpsD genes from the daunorubicin biosynthesis pathway (1), respectively, are responsible for the initiation of biosynthesis from a longer propionate starter unit (14). This expression construct, which was designated pSJ10 (Fig. 3) and which was transformed into S. lividans TK24, was cultivated in E1 medium with XAD-7. The compounds produced were purified as described above for S. lividans TK24/pMC6B. According to the NMR results (Table 2), the strain produced the expected UWM6 derivative, MM2002, which, however, appeared in the NMR solvent used as an enol tautomer (Fig. 2). The accumulation of MM2002 in the culture medium demonstrated that the pgaF cyclase is capable of processing a C21 polyketide chain and cyclizing the last ring correctly.
The jadI jadomycin cyclase, unlike the pgaF cyclase described here, has been reported to exhibit strict context-dependent behavior and nonfunctionality with the tcm and dps biosynthesis genes. Tetracenomycin biosynthesis proceeds through an unreduced intermediate of the first ring, which subsequently leads to cyclization between C-9 and C-14 that is different from the folding pattern of reduced polyketides like anthracyclines and angucyclines. The differences in the early part of the biosynthesis of unreduced and reduced polyketides are the most likely reasons why these two pathways were incompatible, as investigators acknowledged previously (11).
On the contrary, the biosynthesis of daunorubicin proceeds in a manner highly similar to the biosynthesis of aclacinomycins, as described above, and no clear explanation for the incompatibility of the jad and dps biosynthesis cyclases could be presented (20). However, it is interesting that the expression constructs used in the jad and dps biosynthesis experiments also included the tcmJ gene from the biosynthesis pathway of tetracenomycin. The exact function of tcmJ has not been elucidated, but it has been reported to improve production of tetracenomycin in its native cluster (5). Nonetheless, in light of the fact that the pgaF cyclase is functional with two different sets of anthracycline biosynthesis genes, it would be interesting to see if the inclusion of tcmJ in the expression constructs has an effect on the nonfunctionality of the jad and dnr genes.
Acknowledgments
This research was supported by a grant from the Jenni and Antti Wihuri Foundation to M.M-K.
REFERENCES
- 1.Bao, W., P. J. Sheldon, E. Wendt-Pienkowski, and C. R. Hutchinson. 1999. The Streptomyces peucetius dpsC gene determines the choice of starter unit in biosynthesis of the daunorubicin polyketide. J. Bacteriol. 181:4690-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhuyan, B. K., and A. Dietz. 1965. Fermentation, taxonomic, and biological studies of nogalamycin. J. Antimicrob. Chemother. 5:836-844. [PubMed] [Google Scholar]
- 3.Decker, H., and S. Haag. 1995. Cloning and characterization of a polyketide synthase gene from Streptomyces fradiae Tu2717, which carries the genes for biosynthesis of the angucycline antibiotic urdamycin A and a gene probably involved in its oxygenation. J. Bacteriol. 177:6126-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2497. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson, C. R. 1997. Biosynthetic studies of daunorubicin and tetracenomycin C. Chem. Rev. 97:2525-2535. [DOI] [PubMed] [Google Scholar]
- 6.Hutchinson, C. R. 1998. Combinatorial biosynthesis for new drug discovery. Curr. Opin. Microbiol. 1:319-329. [DOI] [PubMed] [Google Scholar]
- 7.Kantola, J., T. Kunnari, A. Hautala, J. Hakala, K. Ylihonko, and P. Mäntsälä. 2000. Elucidation of anthracyclinone biosynthesis by stepwise cloning of genes for anthracyclines from three different Streptomyces spp. Microbiology 146:155-163 [DOI] [PubMed] [Google Scholar]
- 8.Khosla, C., and R. J. X. Zawada. 1996. Generation of polyketide libraries via combinatorial biosynthesis. Trends Biotechnol. 14:335-341. [DOI] [PubMed] [Google Scholar]
- 9.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, John Innes Centre, Norwich, United Kingdom.
- 10.Krohn, K., F. Ballwanz, and W. Baltus. 1993. Total synthesis of angucyclines. 1. Synthesis of a daunomycin-rabelomycin hybrid. Liebigs. Ann. Chem. 1993:911-913.
- 11.Kulowski, K., E. Wendt-Pienkowski, L. Han, K. Yang, L. C. Vining, and C. R. Hutchinson. 1999. Functional characterization of the jadI gene as a cyclase forming angucyclinones. J. Am. Chem. Soc. 121:1786-1794. [Google Scholar]
- 12.McDaniel, R., S. Ebert-Khosla, D. A. Hopwood, and C. Khosla. 1995. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature 375:549-554. [DOI] [PubMed] [Google Scholar]
- 13.Oki, T., Y. Matsuzawa, A. Yoshimoto, K. Numata, I. Kitamura, S. Hori, A. Takamatsu, H. Umezawa, M. Ishizuka, H. Naganawa, H. Suda, M. Hamada, and T. Takeuchi. 1975. New antitumor antibiotics, aclacinomycins A and B. J. Antibiot. 28:830-834. [DOI] [PubMed] [Google Scholar]
- 14.Räty, K., J. Kantola, A. Hautala, J. Hakala, K. Ylihonko, and P. Mäntsälä. 2002. Cloning and characterization of Streptomyces galilaeus aclacinomycins PKS cluster. Gene 293:115-122. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Torkkell, S., T. Kunnari, K. Palmu, J. Hakala, P. Mäntsälä, and K. Ylihonko. 2000. Identification of a cyclase gene dictating the C-9 stereochemistry of anthracyclines from Streptomyces nogalater. Antimicrob. Agents Chemother. 44:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torkkell, S., T. Kunnari, K. Palmu, P. Mäntsälä, J. Hakala, and K. Ylihonko. 2001. The entire nogalamycin biosynthetic gene cluster of Streptomyces nogalater: characterization of a 20-kb DNA region and generation of hybrid structures. Mol. Genet. Genomics 266:276-288. [DOI] [PubMed] [Google Scholar]
- 18.Ward, J. N., G. R. Janssen, T. Kieser, M. J. Bibb, M. J. Buttner, and M. J. Bibb. 1986. Construction and characterization of a series of multicopy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase from Tn5 as indicator. Mol. Gen. Genet. 203:468-478. [DOI] [PubMed] [Google Scholar]
- 19.Westrich, L., S. Domann, B. Faust, D. Bedford, D. A. Hopwood, and A. Bechthold. 1999. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol. Lett. 170:381-387. [DOI] [PubMed] [Google Scholar]
- 20.Wohlert, S. E., E. Wendt-Pienkowski, W. Bao, and C. R. Hutchinson. 2001. Production of aromatic minimal polyketides by the daunorubicin polyketide synthase genes reveals the incompatibility of the heterologous DpsY and JadI cyclases. J. Nat. Prod. 64:1077-1080. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita, N., T. Harada, K. Shin-ya, and H. Seto. 1998. 6-Hydroxytetrangulol, a new CPP32 protease inducer produced by Streptomyces sp. J. Antibiot. 51:79-81. [DOI] [PubMed] [Google Scholar]
- 22.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 23.Ylihonko, K., J. Hakala, J. Niemi, J. Lundell, and P. Mäntsälä. 1994. Isolation and characterization of aclacinomycin A-non-producing Streptomyces galilaeus (ATCC 31615) mutants. Microbiology 140:1359-1365. [DOI] [PubMed] [Google Scholar]
- 24.Ylihonko, K., J. Tuikkanen, S. Jussila, L. Cong, and P. Mäntsälä. 1996. A gene cluster involved in nogalamycin biosynthesis from Streptomyces nogalater: sequence analysis and complementation of early-block mutations in the anthracycline pathway. Mol. Gen. Genet. 251:113-120. [DOI] [PubMed] [Google Scholar]