Abstract
We have previously reported that the LRRC4 gene, which contains a conserved leucine-rich repeat (LRR) cassette and an immunoglobulin (Ig) IgC2 domain, is associated with glioma suppression both in vitro and in vivo. The present study provides evidence that the conspicuous absence of LRRC4 in high-grade gliomas directly contributes to the increasing tumor grade. The loss of LRRC4 in U251 cells is caused by the loss of homozygosity at chromosome 7q32-ter. It was also found that LRRC4 requires a functional LRR cassette domain to suppress U251 cell proliferation. In the LRR cassette domain, the third LRR motif of the core LRR is found to be indispensable for the function of LRRC4. The inhibitory effect of LRRC4 is accompanied by a decrease in the expression of pERK, pAkt, pNF-κBp65, signal transducer and activator of transcription protein-3 (STAT3), and mutant p53, and an increase in the expression of c-Jun NH2-terminal kinase (JNK)2 and p-c-Jun, suggesting that LRRC4 plays a major role in suppressing U251 cell proliferation by regulating the extracellular signal-regulated kinase (ERK)/Akt/NF-κBp65, STAT3, and JNK2/c-Jun pathways. In conclusion, LRRC4 may act as a novel candidate of tumor suppressor gene. Therefore, the loss of LRRC4 function may be an important event in the progression of gliomas.
INTRODUCTION
Gliomas are the most common primary malignant tumors, which account for >50% of neoplasms in the adult human central nervous system (CNS). Chromosome 7 is a frequent site of cytogenetic aberrations in human astrocytomas, and 7q contains multiple tumor suppression genes (TSGs) that are involved in the pathogenesis of head and neck tumors (Zhang et al., 2005a). In an attempt to isolate homologues of nasopharyngeal cancer-associated genes using our cDNA database, we identified a leucine-rich repeat containing 4 (LRRC4) gene, which was mapped at 7q31-32 Wang et al., 2002. LRRC4 was found to be predominantly expressed in the normal brain tissue, but it was deleted or down-regulated in primary brain tumor biopsies (up to 87.5% in gliomas) (Wang et al., 2002), and it had the potential to suppress tumorigenesis of U251 malignant glioma cells in vivo and cell proliferation in vitro by a tetracycline-inducible expression system (Zhang et al., 2005b).
The expression pattern of LRRC4 in CNS and its predicted neuronal membrane localization suggest that LRRC4 may also have signaling functions in glioma development. It has been reported that the constitutive activation of mitogen-activated protein kinases (MAPKs) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathways contribute significantly to the progression of glioblastoma multiforme, and Akt and NF-κB are constitutively activated in U251 cells (Wang et al., 2004). Activated Akt phosphorylates and activates IκB kinase (IKK), which, in turn, phosphorylates IκB and leads to its degradation and nuclear translocation of nuclear factor-κB (NF-κB). Zhang et al. (2005a) also showed that LRRC4 down-regulated the phosphorylated extracellular signal-regulated kinase (ERK) and the expression of proliferating cell nuclear antigen in U251 cells.
In the present study, we provide extensive evidence that LRRC4 acts as a candidate of TSG. To further demonstrate the functional characterization of LRRC4 domain, we constructed artificially mutants with deletion of different domains and investigated the potential molecular mechanism by which LRRC4 inhibits glioma cell proliferation.
MATERIALS AND METHODS
Tumor Samples and Cell Lines
Primary tumor samples were obtained from randomly selected cancer patients at Xiangya Hospital (Hunan, People’s Republic of China). Tumors were graded according to the revised classification of the World Health Organization (Kleihues et al., 1993). A written informed consent was obtained from each patient participating in the study.
Human glioblastoma-derived cell lines U251, U87, SF126, SF767, and BT325 and the neuroblastoma cell line M17 were obtained from the Cell Research Institute of Peking Union Medical College (Peking, China). U251, U87, M17, and BT325 were maintained in DMEM supplemented with 10% fetal calf serum (FCS) and standard antibiotics; SF126 and SF767 were cultured in minimal essential medium.
For stable transfection, cells were split at a 1:3 dilution 24 h posttransfection and were then challenged with 500 μg/ml geneticin sulfate (G418; Invitrogen, Carlsbad, CA) and left for 2–3 wk with change of the medium every 3 d, after which resistant colonies were formed by pool cloning.
For all inhibition assays, inhibitors were added to the cells, which were then kept overnight in serum-free medium pending supernatant collection. For phorbol 12-myristate 13-acetate (PMA) treatment, cells were incubated with the appropriate concentration of the inhibitor for 30 min before they were stimulated with PMA for another 10 min.
Nucleic Acid Isolation and RNA Analysis
Total RNA was isolated using the TRIzol reagent (Invitrogen). Northern blotting was performed as described previously (Zhou et al., 2004). Hybridization for Northern analysis was subsequently performed with a 32P-labeled LRRC4 probe corresponding to the full-length coding region as described previously (Zhou et al., 2004). For reverse transcription (RT)-PCR, cDNA was made using the Reverse Transcription System (Promega, Madison, WI) and oligo(dT) primers. cDNA (1 μg) was used for each PCR reaction. The following primers for LRRC4 were used: 5′-AAG CTC TTG TGG CAG GTA ACT GTG CAC-3′ (forward) and 5′-TCA TAT TTG AGT TTC CTG TAC CTT GTC CTT-3′ (reverse).
The relative quantitative TaqMan real-time PCR was performed by Shinegene Molecular Biology Technology Ltd. (Shanghai, China) with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as an endogenous control. The following primers and probes were designed with Primer Express version 1.0 software (ABI/Perkin Elmer, Foster City, CA). Casp3: Forward—GGTTCATCCAGTCGCTTTGT; Reverse—AATTCTGTTGCCACCTTTCG; Probe— fam+TGCTGAAACAGTATGCCGACAAGC+tamra. Casp8: Forward— GTTCCTGAGCCTGGACTACATT; Reverse—GATTGCTTTCCTCCAACATTCT; Probe—fam+CGCAAAGGAAGCAAGAACCCATC+tamra. hGAPDH: Forward—CCACTCCTCCACCTTTGAC; Reverse—ACCCTGTTGCTGTAGCCA; Probe—fam+TTGCCCTCAACGACCACTTTGTC+tamra.
The 30-μl reaction mixture contained 6 μl of 5× PCR buffer, 1.2 μl of Primer (25 pmol/μl), 0.3 μl of probe (25 pmol/μl), 1 μl of dNTPs (10 mM), 0.3 μl of Taq DNA polymerase (5 U/μl), 4 μl of Mg2+ (25 mM), 1 μl of template DNA, and 16.2 μl of diethyl pyrocarbonate-treated water. The initial denaturation was carried out at 94°C for 4 min, which was followed by amplification in 40 cycles, at 94°C for 15 s, and 60°C for 25 s, using a PTC200 real-time PCR thermocycler (MJ Research, Biozym, Germany).
One-Step PCR Mutagenesis and Constructs
Deletion mutants were obtained by reverse PCR using plasmid pGEM-T easy/LRRC4-FLAG as the templates. Pyrobest DNA polymerase (Takara, Kyoto, Japan) was used in PCR. The oligonucleotides used for PCR were selected along the opposition direction in the flank of mutants, and the primer sequences are listed in Table 1. In all cases, PCR products were ligated into pGEM-T easy, and mutants were verified by the sequencing analysis. Mutant fragments were released from pGEM-T easy with BamHI/EcoRI double digestion and subcloned into pcDNA3.1(+) to generate the appropriate expression constructs. All mutants were verified by the sequencing analysis.
Table 1.
Oligonucleotides used to create LRRC4 mutants
| LRRC4 mutant | Primer description | Primer sequence (5′–3′) |
|---|---|---|
| Δ1 LRR | Forward | CTC CAC CAC CTG GAG GTC CT |
| Δ1-2LRR | Forward | CTG GCC AGC CTC AAC ACC |
| Δ1-3LRR | Forward | CTG TCC AAG CTG CGG GAG CTC T |
| Δ1-4LRR | Forward | GTG CCC TCC CTC ATG CGC CT |
| Δ1-5LRR | Forward | CTG AGC TCC CTC AAG AAG CTC T |
| Δ1-6LRR | Forward | CTG GCT TCA CTT GTG GAA CTC |
| Δthe core LRR | Forward | CTG AGG TAC CTG GTG GAG TTG CAT |
| All of the above | Reverse | AAT ACC CTG CGG GAC CTC |
| ΔLRR cassette | Forward | GAG GTG GAC CAG GCC TCC |
| ΔLRR cassette | Reverse | CTG GGG CCC GGC TGA GGC |
| ΔIgC2 | Forward | TGC TGC CCA ATG GGA CAG TGC T |
| ΔIgC2 | Reverse | ATA CTC TCG AAG CCA CCA GGC |
| ΔTm | Forward | CGT AAG CGG CAC CAG CAG |
| ΔTm | Reverse | CTT GGT GGT CTT CAT GAC TT |
The constitutively active Akt construct (Myr Akt) was generated by appending the sequences encoding the chicken c-Src myristoylation signal and a hemagglutinin (HA) epitope to the 5′ and 3′ end of Akt1, respectively, by PCR, and incorporated in pcDNA3.1(+) vector (Xu et al., 2002). The transfection of Akt plasmid was identified with anti-HA antibody (Roche Diagnostics, Basel, Switzerland) by Western blotting.
Western Blotting Analysis
Cells were harvested at 80% confluence and lysed in the lysis buffer (1% Nonidet P-40, 50 mM Tris-HCl, pH 7.5, 50 mM NaF, 2 mM EDTA, 10% glycerol plus complete protease inhibitor mixture [Roche Diagnostics] with NaCl adjusted to 400 mM). The protein concentrations were determined using the bicinchoninic acid (Pierce Chemical, Rockford, IL) protein assay method. Cell extracts containing 50 μg of proteins were separated in 8–12% SDS-PAGE gels and electroblotted onto nitrocellulose membranes (Hyclone Laboratories, Logan, UT). The membranes were blocked using Tris-buffered saline/Tween 20 (25 mM Tris-HCl, 150 mM NaCl, pH 7.5, and 0.05% Tween 20) containing 5% nonfat milk followed by overnight incubation at 4°C with primary antibodies. After three washes, anti-horseradish peroxidase antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were added. Then, anti-α-tubulin or anti-β-actin antibody (Santa Cruz Biotechnology) was used as a loading control. Antibodies against pERKTyr-42, AktSer-473, NF-κBSer-536, STAT3Tyr-705, and STAT3Ser-727 were purchased from Cell Signaling Technology (Beverly, MA), and antibodies against c-Jun NH2-terminal kinase (JNK)1, JNK2, p-c-Jun, p53, cyclinB1, and cyclinD1 were purchased from Santa Cruz Biotechnology.
Cell Proliferation Assay
Cell proliferation was measured by the reduction of the yellow tetrazolium salt 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT) to the purple formazan. The U251 cells transfected with or without the wild-type LRRC4 or different LRRC4 mutants were cultured in 96-well plates at a density of 1 × 104 cells/well for 24, 48, and 72 h, respectively. The cells were incubated with 5 mg/ml MTT in complete DMEM for 4 h before harvest. The viable cells converted MTT to formazan, which generated a blue-purple color when dissolved in dimethyl sulfoxide. The absorbance at 570 mm was measured using an ELX-800 enzyme-linked immunosorbent assay plate reader (Bio-Tek Instruments, Winooski, VT). Relative cell numbers were calculated in sextuple for three independent experiments.
Soft Agar Assay
To evaluate the ability of individual cell lines to grow in an anchorage-independent manner, cells were plated in soft agar (Agarose 1000; Invitrogen, Carlsbad, CA) (Wang et al., 2005). In brief, a bottom layer was prepared using 1 ml of the corresponding culture medium containing 0.6% agar and 10% FCS. It was placed in 35-mm dishes and allowed to solidify. Transfected or untransfected U251 cells (2 × 104) were suspended in 50 μl of completed DMEM. The culture medium (1 ml) containing 0.33% agarose was added to the cell suspension before seeding on the dishes. Each assay was performed in triplicate. Dishes were examined twice per week, and the colonies were then counted manually after 2 wk.
Cell Cycle Analysis
Cell cultures were plated in 10-cm dishes at ∼40% confluence and allowed to grow exponentially. The adherent cells were collected by trypsinization, pooled with the nonadherent cells, and washed with phosphate-buffered saline (PBS). The cells were then fixed in 70% cold ethanol overnight at 4°C. Before the analysis, the cells were adjusted to a final density of 1 × 106 cells/ml in PBS containing RNase (1 μg/ml) and stained with 10 μg/ml propidium iodide (Roche Diagnostics, Basel, Switzerland) for 30 min at room temperature. The multiparameter analysis of 5000 cells was performed on a FACScan flow cytometer using CellQuest software (BD Biosciences, San Jose, CA).
Acridine Orange (AO)/Ethidium Bromide (EB) Staining
An AO/EB cocktail (80 μl) containing 1 ml of DMEM was added in the culture plate. Fields of stained cells were selected and focused using fluorescence microscopy (Nikon Eclipse E800; Nikon, Tokyo, Japan). Viable cells stained only with AO were bright green with intact structure; early apoptotic cells stained with AO/EB were bright green in the nucleus with red-orange chromatin. Late apoptotic cells stained with both AO and EB were red-orange with chromatin condensation (Wang and Huang, 2005).
Statistical Analysis
Differences of the variables between groups were tested by Student’s t test using SPSS 11.0 program (SPSS, Chicago, IL). A p value of <0.05 was considered statistically significant.
RESULTS
Expression Assay of LRRC4 in Gliomas and Glioblastoma Cell Lines
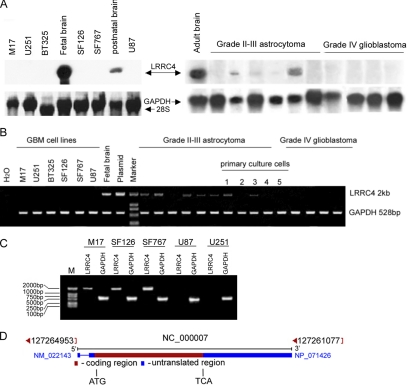
As shown in Figure 1, A and B, despite the abundant expression of LRRC4 in brain tissue, Northern blotting and RT-PCR analysis of RNA from glioblastoma cell lines (SF126, SF767, M17, BT325, U251, and U87) and glioblastoma (World Health Organization [WHO], grade IV) failed to detect any LRRC4-specific transcripts. The lower grade astrocytoma and primary culture cells (WHO, grade II–III) had low levels of or undetectable LRRC4 transcripts, as determined by RT-PCR analysis. Northern blotting analysis also revealed different expression of LRRC4 in fetal, postnatal, and adult brain tissues, suggesting a possible role of LRRC4 in the brain development.
Figure 1.
Analysis of LRRC4 expression in gliomas and glioblastoma cell lines. (A) Northern blotting analysis of LRRC4 in glioblastoma cell lines and gliomas (top). Relative levels of RNA loading are shown as methylene blue staining of 28s RNA and GAPDH (bottom). (B) RT-PCR analysis of LRRC4 expression in gliomas, primary culture glioma cells, and glioblastoma cell lines. RNA from gliomas and cell lines was amplified for 25 and 35 cycles, respectively, using GAPDH (bottom) and LRRC4 (top) primers. Water was used as the negative control, and fetal brain cDNA and pcDNA3.1(+)-LRRC4 plasmid were used as the positive control templates. Lanes 1–4, primary culture tumor cells derived from grade II–III glioma; and lane 5, primary culture tumor cells derived from grade IV glioblastoma. (C) Schematic view of LRRC4 gene. The gene is composed of two exons. Exon 2 contains the entire ORF of LRRC4. The start and the end of the coding sequence are marked by ATG and TGA, respectively. (D) ORF of LRRC4 was amplified from genomic DNA of glioblastoma cell lines (marker −2-kb DNA ladder). A 528-base pair PCR product from GAPDH was used for the normalization of genomic DNA levels.
A schematic illustration of LRRC4 gene is presented in Figure 1C. LRRC4 gene is composed of two exons and one intron. Exon 2 contains the entire open reading frame (ORF) of LRRC4. To determine whether mutations in the coding region are responsible for the nonfunctional LRRC4, we obtained the LRRC4 ORF product from genomic DNA of glioblastoma cell lines by PCR and performed the DNA sequence analysis. Sequencing of PCR products generated from SF126, SF767, and M17 cell lines showed that they all contained a synonymous point mutation at the same position (T977A). However, we failed to obtain PCR products from genomic DNA of U251 and U87 cell lines (Figure 1D).
Assessment of Proliferation-Suppressing Effect of LRRC4 and Its Mutants in U251 Cell Line
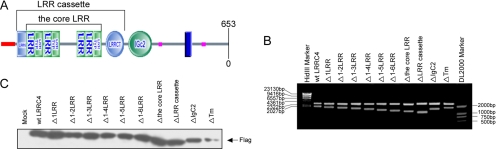
Having identified that the down-regulation of LRRC4 was common among malignant glioma cell lines and that LRRC4 was absent in U251 cell line, we chose U251 cell line to examine the effects of exogenous LRRC4 and its mutants on the proliferation in vitro. According to motif search of LRRC4 gene predicted by SMART software (http://smart.embl-heidelberg.del) (Figure 2A), 10 deletion mutants of LRRC4 were constructed by one-step PCR (Figure 2B). To assess the transfection efficiency of each FLAG-tagged construct, anti-M2 FLAG mouse monoclonal antibody (Sigma-Aldrich, St. Louis, MO) was used to detect the FLAG epitope. Ten mutant proteins were expressed at the levels equal to or lower than those of the wild-type LRRC4 (Figure 2C).
Figure 2.
(A) Schematic illustration of the various domains of the LRRC4 protein by the SMART software. LRRNT and LRRCT indicate that the cysteine-rich regions flanking the core LRRs; Tm, transmembrane region. (B) Restriction enzyme analysis of pcDNA3.1(+)LRRC4/FLAG mutants. Plasmids were digested with BamHI and EcoRI and analyzed by agarose gel electrophoresis. Wild-type (wt) LRRC4 was used as a control. (C) Constructs of pcDNA3.1(+)/LRRC4-FLAG mutants were transfected into U251 cells, and the expression of mutant proteins was detected by immunoblotting with anti-M2/FLAG antibodies.
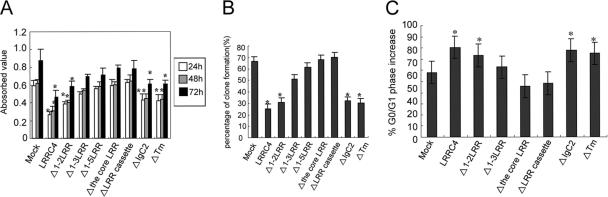
MTT assay showed that the exogenous LRRC4 significantly inhibited U251 cell proliferation in wild-type LRRC4-transfected cells (p < 0.01) (Figure 3A). Wild-type LRRC4 induced a two- to threefold decrease in the number of cells. To further test which domain is important in LRRC4-reduced U251 cell response, we transfected U251 cells with various LRRC4 mutants. It was demonstrated that 1-2LRR, IgC2, and transmembrane region (Tm) deletion constructs significantly inhibited U251 cell growth, compared with the mock-transfected cells (p < 0.01) (Figure 3A). However, Δ1-3LRR, Δ1-5LRR, Δthe core LRR, or ΔLRR cassette mutants did not inhibit U251 cell proliferation (Figure 3A).
Figure 3.
The third LRR motif of the core LRRs is indispensable for LRRC4 to inhibit U251 cell proliferation. (A) Comparison of the proliferation potential between the mock transfected U251 cells and U251 cells transfected with wild-type LRRC4 or selected LRRC4 deletion mutants by MTT assay. (B) The soft agar assay for U251 cells stably expressing wild-type LRRC4 or its mutants. (C) The cell cycle assay for U251 cells stably expressing wild-type LRRC4 or its mutants. All the experiments were repeated three times. *p < 0.01, compared with the mock-transfected control cells.
The mock-transfected U251 cells readily formed colonies in the soft agar (Figure 3B). However, wild-type LRRC4 or Δ1-2 LRR-transfected U251 cells greatly reduced the capacity to form colonies, compared with the mock-transfected cells (p < 0.01). After 10 d, cells without LRRC4 expression formed increased colonies, compared with LRRC4-expressing cells, and the growth did not slow down until after 20 d. In contrast, U251 cells transfected with other different LRR deletion mutants (Δ1-3 LRR, Δ1-5 LRR, Δthe core LRR, or ΔLRR cassette) were not inhibited in the anchorage-independent growth. Interestingly, similar colony reduction was seen in U251 cells transfected with IgC2- or Tm-deleted constructs as well as in LRRC4-expressing U251 cells.
We analyzed the cell cycle profiles of U251 cells transfected with wild-type or different mutants of LRRC4 (Figure 3C). Wild-type LRRC4, Δ1-2LRR, ΔIgC2, or ΔTm mutants caused a prominent G1 arrest (80.3, 73.2, 77.6, and 74.9%, respectively) compared with the mock-transfected cells (57.9%) (p < 0.01). U251 cells transfected with Δ1-3LRR LRRC4 construct exhibited a modest G1 (63.6%) arrest. The mutants that lacked the core LRR or ΔLRR cassette did not arrest G1 cell cycle progression (45.5 and 48.1%, respectively).
LRRC4 Inhibited the Akt/NF-κBp65 Pathway in an ERK-Dependent Manner
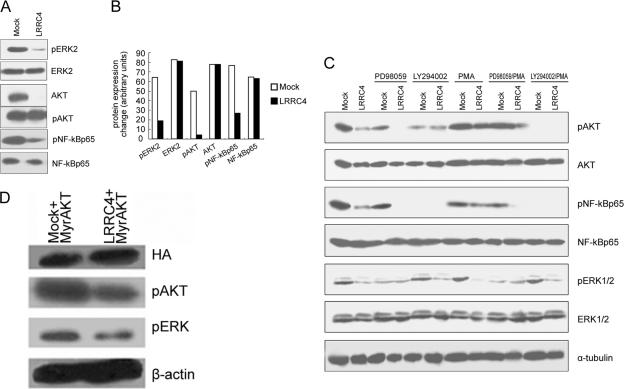
It has been shown that in some circumstances, the control of cell proliferation is through ERK-MAPK and PI3K/Akt signaling pathways (Sah et al., 2004). To investigate whether LRRC4 has effects on signaling through these pathways, we analyzed the phosphorylation status of ERKTyr-42 and AktSer-473 in the presence or absence of LRRC4. In the presence of the exogenous LRRC4, the level of pERKTyr-42 and pAktSer-473 was significantly reduced, or even undetectable. In contrast, high level of pERKTyr-42 and pAktSer-473 was detected in the mock-transfected U251 cells (Figure 4, A and B). The decrease of ERK and Akt phosphorylation signal in U251 cells expressing wild-type LRRC4 did not contribute to a decrease in their protein levels, as revealed by reprobing the immunoblot with antibodies recognizing the unphosphorylated forms of ERK and Akt (Figure 4, A and B).
Figure 4.
Analysis of the phosphorylation status of ERK2, PKB/Akt and NFκBp65 in the mock- and wild-type LRRC4-transfected U251 cells. (A) Western blotting assay showing phospho- and total protein levels of ERK2, Akt, and NFκBp65. (B) Quantitative analysis of the phospho- or total protein level change of ERK, Akt, and NF-κB by scanning densitometry. (C) Western blotting assay showing phospho- and total protein levels of ERK2, Akt, and NFκBp65 in the mock- and wild-type LRRC4-transfected U251 cells treated with PD98059, LY294002, or PMA. (D) Effect of exogenous Akt on pERK and pAkt in LRRC4/U251 cells.
It has been reported that U251 cells with high levels of activated Akt also show high levels of NF-κB activity. Phosphorylated Akt activates NF-κB either by activating IKK or by directly phosphorylating the NF-κB p65 subunit (Gupta et al., 2004). In the presence of exogenous LRRC4, pNF-κBp65 was distinctly decreased (Figure 4, A and B). In the mock-transfected U251 cells, both pAkt and pNF-κBp65 were expressed at high levels (Figure 4, A and B). When the mock or LRRC4-transfected U251 cells were treated with PI3K inhibitor,LY294002, the expression of both pAkt and pNF-κBp65 was inhibited (Figure 4C). Reintroduction of LRRC4 apparently influenced the inactivation of pNF-κBp65 through pAkt inhibition. An ERK/MAPK pathway inhibitor, PD98059, inhibited pAkt expression, whereas LY294002 did not inhibit pERK expression (Figure 4C). Moreover, PD98059 strengthened the inhibitory effect of LRRC4 on pAkt and pNF-κBp65, but not on pERK (Figure 4C). Moreover, LRRC4 still inhibited pAkt and pERK expression in LRRC4/U251 cells transfected with the Akt plasmid (Myr.Akt) (Figure 4D).
PMA-Stimulated Activation of Protein Kinase C (PKC) Activated PI3K/Akt in the Presence of LRRC4
PKC family members, which also contribute to cell proliferation, are activated by PI3K, suggesting that Akt and PKC might interact to activate signaling through the PI3K cascade (Mao et al., 2000). To determine whether the PKC activation affects Akt in transfected U251 cells, we treated these cells with PMA, a pharmacological activator of PKC. PMA induced the reactivation of pAkt, but not pERK, in LRRC4-transfected cells (Figure 4C). When the mock or LRRC4-transfected U251 cells were cotreated with PD98059 together with PMA, the expression of pAkt inhibited by LRRC4 and/or PD98059 was resumed, whereas no change occurred in pERK expression (Figure 4C). When the mock- or LRRC4-transfected U251 cells were treated with LY294002, PMA did not reverse the expression of pAkt or pERK (Figure 4C). The above-mentioned results indicate that the PMA-stimulated activation of PKC activates PI3K/Akt, but not pERK, in the presence of LRRC4, which is consistent with the fact that PKC is located downstream of ERK but upstream of PI3K/Akt.
pNF-κBp65 expression in the mock-transfected U251 cells was inhibited by LY294002, but not by PD98059 (Figure 4C). This finding indicates that pAkt directly regulates pNF-κBp65 in U251 cells, whereas pERK alone may not be able to regulate pNF-κBp65. Both PD98059 and LY294002 inhibited pNF-κBp65 expression in LRRC4-transfected U251 cells, indicating that LRRC4 controls pNF-κBp65 by inhibiting pERK and pAkt. In addition, PMA suppressed pNF-κB p65 expression (Figure 4C), suggesting that PMA may regulate NF-κB p65 through other mechanisms.
LRRC4 Required Its N-Terminal LRR Cassette Domain to Inhibit the ERK/Akt/NF-κB Pathway
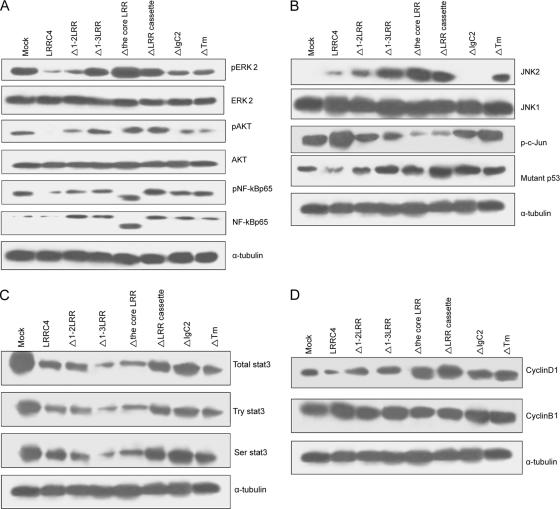
The finding that the LRR cassette domain of LRRC4 was required for LRRC4 to block U251 cell growth prompted more detailed examination. It was observed that the expression of phosphorylated ERK2 was substantially reduced in U251 cells transfected with ΔIgC2 or ΔTm mutants of LRRC4, which was not observed in cells transfected with other LRR deletion mutants (i.e., Δ1-3LRR, Δthe core LRR, or ΔLRR mutants) (Figure 5A). The Δ1-2 LRR mutation led to a modest increase in ERK2 phosphorylation (∼10%), but in 1-3LRR mutant, a similar level of pERK2 was displayed as that in the mock-transfected cells (Figure 5A). Cells transfected with Δthe core LRR or ΔLRR cassette mutants showed a significantly higher expression of pERK2 than the mock-transfected U251 cells. Moreover, a similar trend in the change of pAkt and pNF-κBp65 was observed in all of the LRR mutants, but IgC2 or Tm domain had a negligible effect (Figure 5A). Remarkably, when the core LRRs was deleted, the LRRC4 mutant mainly up-regulated NF-κB p50 expression although the underlying mechanism is unclear.
Figure 5.
Western blotting assay showing the change of the ERK/Akt/NF-κB, JNK2/c-Jun, STAT3 signaling pathways in the mock-, wild-type LRRC4, or its mutant-transfected U251 cells. (A) Protein levels of phospho-ERK2, Akt, and NFκBp65. (B) Protein levels of JNK2/p-c-Jun and mutant p53. (C) Protein level of STAT3. (D) Protein levels of cyclinD1.
LRRC4 Required LRR, IgC2, and Tm Domain to Regulate the JNK2/c-Jun/p53 or STAT3 Pathway
Many proteins containing LRR, such as NOD2 (Chen et al., 2004) and TLR6 (Takeuchi et al., 1999), regulate the JNK and NF-κB pathway. We reported that LRRC4 inhibits the NF-κB activation through its extracellular LRR cassette domain. We then investigated a potential role of LRRC4 in regulating the JNK signaling pathway in U251 cells. The expression of JNK2 was absent in mock-transfectedU251 cells and at a very low level in wild-type LRRC4-transfected cells. In U251 cells transfected with variant mutants of LRRC4, the expression of JNK2 was absent when IgC2 was deleted. In contrast, when the different LRR motifs, the LRR cassette domain, or Tm was deleted, the expression of JNK2 was increased. However, LRRC4 did not affect the JNK1 expression (Figure 5B).
U251 cell line expresses the mutant p53 (mutant at codon 273; CGT/CAT; Arg/His) (Kataoka et al., 2000). In the present study, LRRC4 activated p-c-Jun and down-regulated mutant p53. All mutant LRRC4 constructs resulted in a decrease in the expression of activated p-c-Jun and an increase in the expression of the mutant p53 (Figure 5B).
Moreover, U251 cells expressed constitutively activated STAT3. The reexpression of LRRC4 not only markedly suppressed the phosphorylation expression of Ser-727 and Tyr-705 of STAT3 but also down-regulated the total protein expression of STAT3. Deletion of LRR, IgC2, or Tm domain affected the inhibitory effect of LRRC4 on STAT3 expression, suggesting that LRRC4 requires LRR, IgC2, and Tm domain together to down-regulate Ser, Tyr phosphorylation and the total protein of STAT3 (Figure 5C).
LRRC4 Did Not Induce Apoptosis of U251 Cells
It was found that LRRC4 required LRR, IgC2, and Tm domain to inhibit cyclinD1 expression. However, LRRC4 and its mutants did not affect cyclinB1 expression (Figure 5D). The data indicated that LRRC4 arrests U251 cells in G0/G1 phase, which is consistent with the results obtained by flow cytometry. CyclinD1 expression was discrete in U251 cells transfected with IgC2 or Tm domain deletion mutants, as detected by flow cytometry.
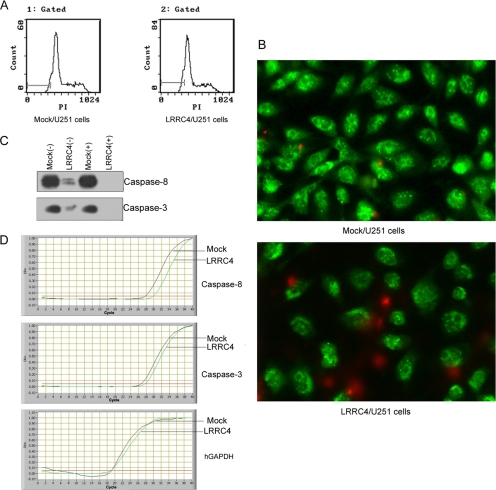
Because LRRC4 arrests U251 cells in G0/G1 phase, it is possible that LRRC4 also induces U251 cells apoptosis. We used the analysis of flow cytometry (Figure 6A) and AO/EB dual staining (Figure 6B) to detect U251 cell apoptosis. It was shown that LRRC4 induced necrosis, but not apoptosis (Figure 6B). Caspases play a key role in apoptosis. Caspase-8, which is activated by the induction of tumor necrosis factor-α, leads to the activation of caspase-3, which activates apoptotic nucleases (Miyao et al., 2006). The expression of caspase-8 and caspase-3 was reduced in U251 cells transfected with wild-type LRRC4, compared with that in the mock-transfected cells (Figure 6C). The real-time PCR showed that LRRC4 suppresses the expression of caspase-8 (p < 0.01) and caspase-3 (p < 0.05) mRNA (Table 2) (Figure 6D).
Figure 6.
Apoptosis assay of LRRC4 in the mock- and LRRC4-transfected U251cells. (A) Flow cytometry histogram displaying apoptosis apex in front of G1 phase. (B) AO/EB dual staining showing necrosis cells and apoptosis cells. (C) Western blotting image showing caspase-8 and caspase-3 at the protein level in the mock- and LRRC4-transfected U251 cells (+/− serum). β-actin is used as the loading control. (D) Real-time PCR products displaying caspase-8 and caspase-3 mRNA in the mock- and LRRC4-transfected U251 cells.
Table 2.
Real-time PCR products displaying change of caspase-8 and caspase-3 mRNA in mock- and LRRC4-transfected U251 cells
| Sample | 10,000*casp3/gapdh | 10,000*casp8/gapdh |
|---|---|---|
| Mock | 31.829758 ± 0.996821 | 15.333219 ± 0.34612 |
| LRRC4 | 9.623506 ± 0.315080* | 2.065222 ± 0.751587* |
* p < 0.01, compared with the mock-transfected control cells.
DISCUSSION
Multiple genetic alterations are involved in the development and progression of malignant brain tumors. Frequently, these events involve the functional loss of a TSG, which may occur as a result of mutation, deletion, or structural chromosome rearrangement. Our previous and present studies demonstrated that the expression of LRRC4 gene was highly specific in the brain tissue (Zhang et al., 2005a), but it was reduced or absent in the lower grade astrocytomas, and absent in glioblastomas and permanent cell lines (Figure 1). The conspicuous absence of LRRC4 in high-grade gliomas suggests that the loss of LRRC4 function may directly contribute to the increasing tumor grade and is a late event in the pathogenesis of gliomas. By searching public fluorescent in situ hybridization and comparative genomic hybridization database, we also revealed that the deletion of LRRC4 in genomic DNA occurred in ∼40% (2/5) of glioblastoma cell lines (Figure 1D) and that 7pter-14 and 7q32-ter were deleted in U251 cells (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=CancerChromosomes). Therefore, the inactivation of LRRC4 may be ascribed to the loss of homozygosity of 7q32-ter in U251 cells. Although ORF of LRRC4 was obtained from the genomic DNA of SF126, SF767, and M17 cells, the DNA content was reduced compared with that of the housekeeper gene GAPDH (Figure 1D). We speculate that there may be a homozygous deletion of 7q32 in U87 cell line, but a loss-of-heterozygosity in SF126, SF767, and M17 cell lines. Because of its distinctive expression in normal brain tissue and different grade gliomas or cell lines and its inhibitory ability to U251 cell growth in vitro and in vivo (Zhang et al., 2005b), LRRC4 is proposed to be a candidate TSG that may be involved in the pathogenesis of malignant gliomas.
LRRC4, a member of leucine-rich repeat protein superfamily, contains a conserved LRR cassette and an IgC2 domain. The LRR domain is involved in highly specific protein–protein interactions or cell adhesion via the Ras/ERK/MAPK pathway, which is one of the best-characterized signal transduction pathways. It has been reported that a cross-talk exists between the Ras/ERK/MAPK and PI3K/Akt pathway and that ERK, Akt, and NF-κB are highly activated in glioblastomas (Kaufmann and Thiel, 2001; Wang et al., 2004). The present study demonstrated that LRRC4 was responsible for the inhibition of the Akt/NF-κB signaling pathway in an ERK-dependent manner (Figure 4C). The expression of exogenous Akt did not override LRRC4-mediated growth suppression in U251/LRRC4 cells. Constitutively active Akt and ERK existed in U251 cells, but hyperactive Akt did not inhibit pERK, although pAkt promoted pRaf-1Ser-259 expression and inhibited pERK expression in certain cell lines. The reexpression of LRRC4 inhibited pAkt and pERK expression, but it did not reduce Raf-1Ser-259 phosphorylation (our unpublished data). These findings indicate that LRRC4 inhibits U251 cells proliferation through the ERK/Akt signaling pathway (Figure 4, C and D), but not through the Akt/Raf/ERK signaling pathway. Therefore, the Akt/Raf pathway is not the important signaling pathway mediated by LRRC4 in U251 cells. Moreover, our findings, for the first time, provide evidence that the reexpression of full-length LRRC4 has the potential to inhibit U251 cell growth by modulating the ERK/Akt/NF-κB signaling pathway and that this inhibitory effect of the reexpressed LRRC4 is dependent on its LRR cassette domain, but not on IgC2 or Tm domain. In the LRR cassette domain, the third LRR motif of the core LRR plays a crucial role as a “proliferation-inhibition switch” (Figure 3).
JNKs represent a subgroup of MAPKs that is activated primarily by cytokines and exposure to environmental stress (Weston and Davis, 2002). JNK1 and JNK2 perform distinct functions in regulating cellular proliferation via differential regulation of c-Jun, which is a critical regulator of cell cycle progression. JNK does not require its kinase activity to target p53, ATF2, and c-Jun ubiquitination and degradation (Yin et al., 2004). Mdm2 and JNK are the cellular proteins whose direct association with p53 results in p53 ubiquitination and its subsequent degradation. JNK-p53 complexes are preferentially found in G0/G1, whereas Mdm2–p53 complexes are primarily found in S and G2/M phases of the cell cycle (Yin et al., 2004). In the present study, the reexpression of LRRC4 results in the up-regulation of both JNK2 and p-c-Jun expression and the down-regulation of mutant p53 expression. U251 cells are arrested in G0/G1. The IgC2 domain is indispensable for LRRC4 to up-regulate JNK2 expression. Both the LRR cassette and Tm domain restricts IgC2 to up-regulate JNK2 expression. The IgC2 domain has been implied to interact with the Ig domains of other Ig superfamily members (Chandra et al., 2003). Thus, it is possible for LRRC4 to participate in inhibition of tumor cell immune escape via interactions between its extracellular IgC2 domain with other Ig domains. In addition, all LRR motifs or domains of LRRC4 are required for p-c-Jun and p53 expression. These findings suggest that other molecules may regulate the expression of c-Jun and p53 in LRRC4-transfected U251 cells.
STAT3, a latent transcription factor activated by aberrant cytokine or growth factor signals, participates in pathogenesis of several human cancers by inducing cell proliferation and inhibiting apoptosis, and it is frequently activated in astrocytomas (Konnikova et al., 2003). The expression of a dominant-negative mutant STAT3 protein or the treatment with AG490 (a Janus tyrosine kinase 2 inhibitor) markedly reduces the proliferation of U251 cells by inhibiting the constitutive activation of STAT3 (Rahaman et al., 2002). In the present study, the reexpression of LRRC4 markedly reduces STAT3 Tyr-705 and Ser-727 phosphorylation and the expression at the protein level in U251 cells. When the LRR cassette or IgC2 domain is deleted, the inhibitory effect of LRRC4 on STAT3 expression is partially abolished, indicating that LRRC4 requires N, C terminal LRR and the IgC2 domain to inhibit STAT3 Tyr-705 and Ser-727 phosphorylation and protein expression. In addition, LRRC4 may also regulate the STAT3 expression through the ERK and/or Akt signaling pathway.
The structure characteristics also suggest that LRRC4 may be a receptor kinase or an adhesion receptor. LRRC4 may interact with extracellular components or cytokines to regulate ligand presentation to receptors, facilitate cytokine–receptor interaction, change receptor internalization dynamics, prevent receptor desensitization, or directly contribute to the signal transduction with its intracellular domain. The reexpression of LRRC4 down-regulates the gene expression, of many growth factors and receptors such as platelet-derived growth factor (PDGF) (PDGF-A and -B), PDGF receptor (PDGFR)-α and -β (our unpublished data), epidermal growth factor (EGF), and EGF receptor (EGFR) (Zhang et al., 2005b). In addition, the conditioned medium isolated from LRRC4-expressing U251 glioma cells does not inhibit the growth of the mock-transfected U251 cells. Thus, we postulate that LRRC4 may down-regulate PDGF and EGF autocrine signaling by PDGFR and EGFR mediating the proliferation and the survival signaling pathway (ERK/Akt/NF-κB, STAT3, and JNK2/p-c-Jun/p53), and thus block U251 cells in G0/G1, and induce U251 cell growth arrest and differentiation. LRRC4 does not directly induce the secretion of any growth inhibitory cytokines. A future study on the interactions among LRRC4, cytokines, cytokine receptors, and transgenic or knockout phenotypes will help to gain a deeper insight into the biology of this molecule.
In conclusion, LRRC4, as a new candidate TSG for gliomas, may be involved in the pathogenesis of gliomas and the transmission of complex growth suppressive signals.
ACKNOWLEDGMENTS
This study was supported by National Science Foundation of China Grants 30500192, 30330560, 30500444, and 30500295 and Hunan Province Natural Sciences Foundations of China Grant 05JJ40059.
Abbreviations used:
- Akt
protein kinase B
- ERK
extracellular signal-regulated kinase
- IKK
IκB kinase
- JNK
c-Jun NH2-terminal kinase
- LRR
leucine-rich repeat
- LRRC4
leucine-rich repeat containing 4
- MTT
tetrazolium salt 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide
- ORF
open reading frame
- PCNA
proliferating cell nuclear antigen
- PI3K
phosphatidylinositol 3-kinase
- STAT3
signal transducer and activator of transcription protein-3
- TSG
tumor suppression gene.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1082) on May 24, 2006.
REFERENCES
- Chandra S., Ahmed A., Vaessin H. The Drosophila IgC2 domain protein Friend-of-Echinoid, a paralogue of Echinoid, limits the number of sensory organ precursors in the wing disc and interacts with the Notch signaling pathway. Dev. Biol. 2003;256:302–316. doi: 10.1016/s0012-1606(03)00038-1. [DOI] [PubMed] [Google Scholar]
- Chen C. M., Gong Y., Zhang M., Chen J. J. Reciprocal cross-talk between Nod2 and TAK1 signaling pathways. J. Biol. Chem. 2004;279:25876–25882. doi: 10.1074/jbc.M400682200. [DOI] [PubMed] [Google Scholar]
- Gupta D., Syed N. A., Roesler W. J., Khandelwal R. L. Effect of overexpression and nuclear translocation of constitutively active PKB-alpha on cellular survival and proliferation in HepG2 cells. J. Cell. Biochem. 2004;93:513–525. doi: 10.1002/jcb.20200. [DOI] [PubMed] [Google Scholar]
- Kataoka Y., Murley J. S., Patel R., Grdina D. J. Cytoprotection by WR-1065, the active form of amifostine, is independent of p53 status in human malignant glioma cell lines. Int. J. Radiat. Biol. 2000;76:633–639. doi: 10.1080/095530000138295. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Thiel G. Epidermal growth factor and PDGF induce expression of Egr-1, a zinc finger transcription factor, in human malignant glioma cells. J. Neurol. Sci. 2001;189:83–91. doi: 10.1016/s0022-510x(01)00562-7. [DOI] [PubMed] [Google Scholar]
- Kleihues P., Burger P. C., Scheithauer B. W. The new WHO classification of brain tumours. Brain Pathol. 1993;3:255–268. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Konnikova L., Kotecki M., Kruger M. M., Cochran B. H. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M., Fang X., Lu Y., Lapushin R., Bast R. C., Jr, Mills G. B. Inhibition of growth-factor-induced phosphorylation and activation of protein kinase B/Akt by atypical PKC in breast cancer cells. Biochem. J. 2000;352:475–482. [PMC free article] [PubMed] [Google Scholar]
- Miyao M., Shinoda H., Takahashi S. Caspase-3, caspase-8, and nuclear factor-kappaB expression in human cholesteatoma. Otol. Neurotol. 2006;27:8–13. doi: 10.1097/01.mao.0000180482.34545.b8. [DOI] [PubMed] [Google Scholar]
- Rahaman S. O., Harbor P. C., Chernova O., Barnett G. H., Vogelbaum M. A., Haque S. J. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- Sah J. F., Balasubramanian S., Eckert R. L., Rorke E. A. Epigallocatechin-3-gallate inhibits epidermal growth factor receptor signaling pathway. Evidence for direct inhibition of ERK1/2 and Akt kinases. J. Biol. Chem. 2004;279:12755–12762. doi: 10.1074/jbc.M312333200. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Kawai T., Sanjo H., Copeland N. G., Gilbert D. J., Jenkins N. A., Takeda K., Akira S. TLR 6, A novel member of an expanding toll-like receptor family. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang H., Zhang W., Huang H. J., Liao W. S., Fuller G. N. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab. Investig. 2004;84:941–951. doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- Wang J. R., Qian J., Dong L., Li X. L., Tan C., Li J., Zhang B. C., Zhou J., Li G. Y. Identification of LRRC4, a novel member of Leucine-rich repeat (LRR) superfamily, and its expression analysis in brain tumor. Prog. Biochem. Biophys. 2002;29:233–239. [Google Scholar]
- Wang L. NGX6 gene inhibits cell proliferation and plays a negative role in EGFR pathway in nasopharyngeal carcinoma cells. J. Cell. Biochem. 2005;95:64–73. doi: 10.1002/jcb.20393. [DOI] [PubMed] [Google Scholar]
- Wang Y. K., Huang Z. Q. Protective effects of icariin on human umbilical vein endothelial cell injury induced by H2O2 in vitro. Pharmacol. Res. 2005;52:174–182. doi: 10.1016/j.phrs.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Weston C. R., Davis R. J. The JNK signal transduction pathway. Curr. Opin. Genet. 2002;12:14–21. doi: 10.1016/s0959-437x(01)00258-1. [DOI] [PubMed] [Google Scholar]
- Xu J., Liu D., Songyang Z. The Role of Asp-462 in Regulating Akt Activity. J. Cell. Biochem. 2002;277:35561–35566. doi: 10.1074/jbc.M203805200. [DOI] [PubMed] [Google Scholar]
- Yin Z. M., Sima J., Wu Y. F., Zhu J., Jiang Y. The effect of C-terminal fragment of JNK2 on the stability of p53 and cell proliferation. Cell Res. 2004;14:434–438. doi: 10.1038/sj.cr.7290244. [DOI] [PubMed] [Google Scholar]
- Zhang Q. Expression and functional characterization of LRRC4, a novel brain-specific member of the LRR superfamily. FEBS Lett. 2005a;579:3674–3682. doi: 10.1016/j.febslet.2005.05.058. [DOI] [PubMed] [Google Scholar]
- Zhang Q. H., Wang L. L., Cao L., Peng C., Li X. L., Tang K., Li W. F., Liao P., Wang J. R., Li G. Y. Study of a novel brain relatively specific gene LRRC4 involved in glioma tumorigenesis suppression using the Tet-on system. Acta Biochim. Biophys. Sin. 2005b;37:532–540. doi: 10.1111/j.1745-7270.2005.00079.x. [DOI] [PubMed] [Google Scholar]
- Zhou J. BRD7, a novel bromodomain gene, inhibits G1-S progression by transcriptionally regulating some important molecules involved in ras/MEK/ERK and Rb/E2F pathways. Cell Physiol. 2004;200:89–98. doi: 10.1002/jcp.20013. [DOI] [PubMed] [Google Scholar]